A signal capture and proofreading mechanism for the KDEL-receptor explains selectivity and dynamic range in ER retrieval
Figures
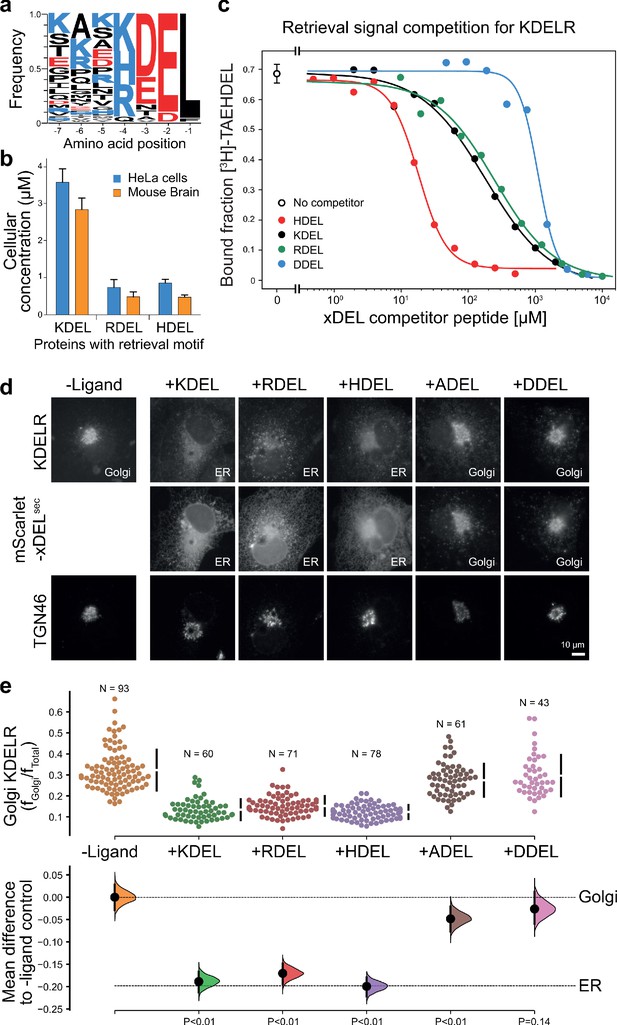
ER retrieval signal abundance and affinity are not correlated.
(a) Sequence logos for ER resident proteins with C-terminal KDEL retrieval signals and variants thereof calculated using frequency or protein abundance (Itzhak et al., 2017; Itzhak et al., 2016). (b) Combined cellular concentrations of ER resident proteins with canonical KDEL, RDEL, and HDEL retrieval sequences in HeLa cells and mouse brain. (c) Competition binding assays for [3H]-TAEHDEL and unlabelled TAEKDEL, TAERDEL, and TAEHDEL to the KDEL receptor. IC50 values for the competing peptides were used to calculate the apparent KDwith the Cheng-Prusoff equation (Cheng and Prusoff, 1973). (d) Endogenous KDEL receptor redistribution was measured in COS-7 cells in the absence (-ligand) or presence of K/R/H/A/DDEL (mScarlet-xDELsec). TGN46 was used as a Golgi marker. Scale bar is 10 µm. (e) The mean difference for K/R/H/A/DDEL comparisons against the shared no ligand control are shown as Cummings estimation plots. The individual data points for the fraction of KDEL receptor fluorescence in the Golgi are plotted on the upper axes with sample sizes and p values.
-
Figure 1—source data 1
Source data for the ligand-induced KDELR receptor retrieval assays in Figure 1e.
- https://cdn.elifesciences.org/articles/68380/elife-68380-fig1-data1-v3.xlsx
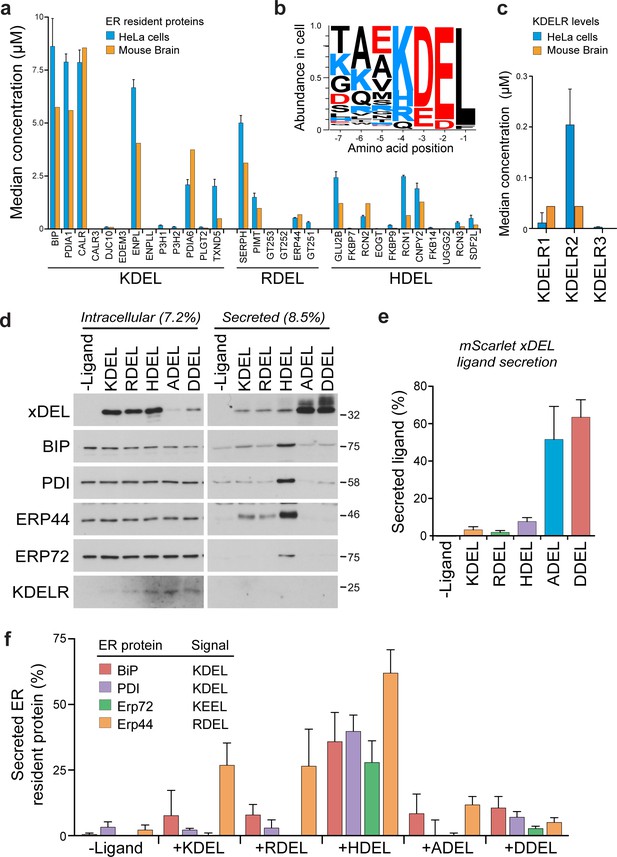
Abundance of ER resident proteins and chaperones in human cells and mouse brain.
(a) The mean concentration of ER resident chaperones with the indicated ER retrieval sequence variant is plotted in the bar graph (Itzhak et al., 2017; Itzhak et al., 2016). (b) Combined cellular concentrations of ER resident proteins with canonical KDEL, RDEL, and HDEL retrieval sequences in HeLa cells and mouse brain. (c) The mean concentration of KDELR1, KDELR2, and KDELR3 in HeLa cells and mouse brain is plotted in the bar graph (Itzhak et al., 2017; Itzhak et al., 2016). (d) Cells and media collected from HeLa S3 cells expressing the xDEL variants (mScarlet-xDELsec) indicated in the figure were Western blotted for resident ER chaperones and KDELR. (e) A bar graph of xDEL secretion showing mean ± SEM (n = 3). (f). Endogenous ER chaperone secretion was measured by western blotting after challenge with different retrieval signals, and plotted as a bar graph showing mean ± SEM (n = 3).
-
Figure 1—figure supplement 1—source data 1
Source data for analysis of ER protein levels in Figure 1—figure supplement 1a and c.
- https://cdn.elifesciences.org/articles/68380/elife-68380-fig1-figsupp1-data1-v3.xlsx
-
Figure 1—figure supplement 1—source data 2
Source data for the western blots in Figure 1—figure supplement 1d showing the regions taken for the figure.
Individual blot files are provided as a ZIP archive.
- https://cdn.elifesciences.org/articles/68380/elife-68380-fig1-figsupp1-data2-v3.pdf
-
Figure 1—figure supplement 1—source data 3
Source data for analysis of ER protein levels in Figure 1—figure supplement 1e and f.
- https://cdn.elifesciences.org/articles/68380/elife-68380-fig1-figsupp1-data3-v3.xlsx
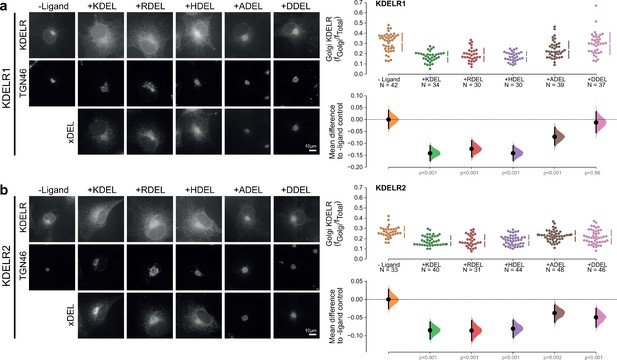
Retrieval specificity of KDELR1 and KDELR2.
(a) Distribution of human KDELR1 and (b) KDELR2 was measured in COS-7 cells in the absence (-ligand) or presence of K/R/H/A/DDEL retrieval signals (K/R/H/A/DDELsec). TGN46 was used as a Golgi marker. Scale bar is 10 µm. The fraction of KDEL receptor localised to the Golgi was measured before (no ligand) and after challenge with different retrieval signals as indicated. Golgi signal for KDEL receptor and ligand intensity are shown on the scatter plots with sample sizes. Effect sizes are shown as the mean difference for K/R/H/A/DDEL comparisons against the shared -ligand control with sample sizes and p-values.
-
Figure 1—figure supplement 2—source data 1
Source data for the ligand-induced KDELR receptor retrieval assays in Figure 1—figure supplement 2.
- https://cdn.elifesciences.org/articles/68380/elife-68380-fig1-figsupp2-data1-v3.xlsx
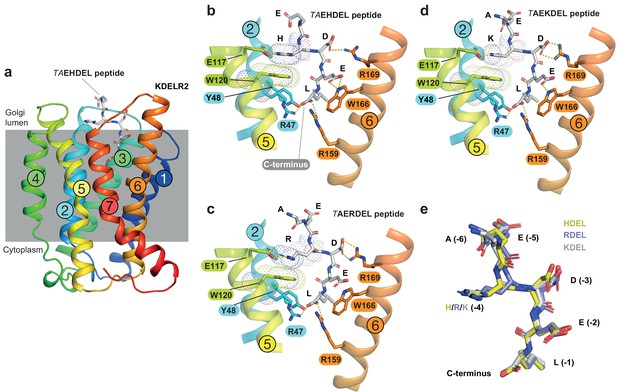
Structures of the KDEL receptor bound to HDEL and RDEL retrieval signals.
(a) Crystal structure of chicken KDELR2 viewed from the side with the transmembrane helices numbered and coloured from N-terminus (blue) to C-terminus (red). The predicted membrane-embedded region of the receptor is indicated by a grey shaded box, with labels at the luminal and cytoplasmic faces. The TAEHDEL peptide is shown in stick format, coloured grey. (b) Close up views of bound TAEHDEL (this study), (c) TAERDEL (this study), and (d) TAEKDEL (PDB:6I6H) peptides bound to the receptor are shown with contributing side chains labelled. Hydrogen bonds are indicated as dashed lines. The molecular orbitals of W120 and the −4 histidine on the peptide are shown as a dotted surface. (e) Superposition of the HDEL, RDEL, and KDEL peptides reveals near identical binding position within the receptor. Retrieval signal side chains are numbered counting down from the C-terminus.
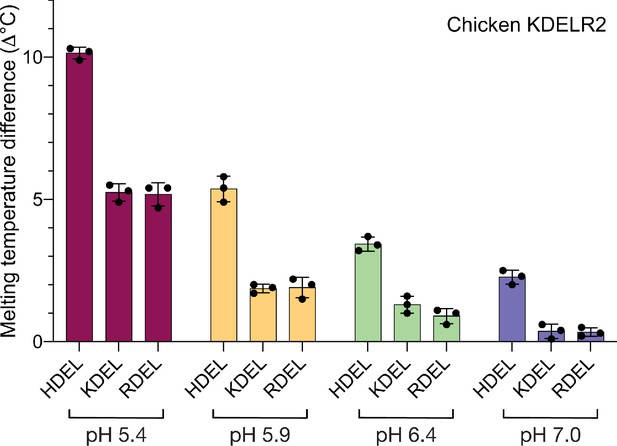
Analysis of pH-dependent interaction of HDEL, KDEL, and RDEL signals with KDELR2.
Thermal stability of chicken KDEL2 was measured at pH 5.4, 5.9, 6.4, and 7.0 in the presence of TAEHDEL, TAEKDEL, and TAERDEL peptides. The difference in melting temperature to a no ligand control is plotted in the bar graph as mean ± SEM (n = 3).
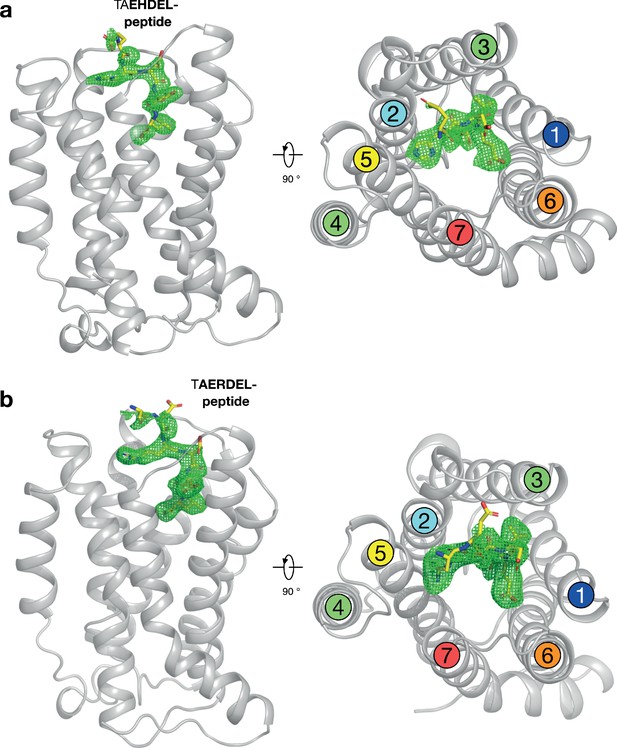
Polder difference density electron density maps for HDEL and RDEL peptides.
(a) The structure of the KDELR bound to the TAEHDEL peptide is shown as in Figure 2a. The mFo-DFc difference electron density used for model building is displayed (green mesh), contoured at 3σ. (b) Equivalent maps calculated for the RDEL peptide.
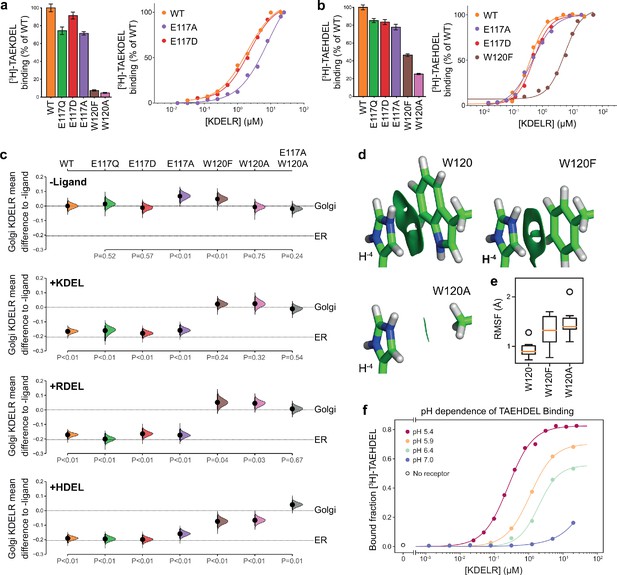
Roles of KDEL receptor E117 and W120 in retrieval signal binding and function in cells.
(a) Normalised binding of [3H]-TAEKDEL and (b) [3H]-TAEHDEL signals to purified WT and the indicated E117 and W120 mutant variants of chicken KDELR2. Bar graphs show mean binding ± SEM (n = 3). Line graphs show titration binding assays. (c) The fraction of WT, E117, and W120 mutant KDEL receptor localised to the Golgi in COS-7 cells was measured before (no ligand) and after challenge with different retrieval signals (K/R/HDEL) as indicated. Effect sizes are shown as the mean difference for K/R/HDEL comparisons against the shared -ligand control with sample sizes and p-values. Also see Figure 3—figure supplements 1—source data 1 files. (d) The π-π interactions between W120 and the histidine were visualised using reduced density gradient analysis. The wild-type W120 exhibit stronger π-π interactions compared with W120F, while W120A shows no π-π interactions. (e) When W120 is changed to phenylalanine, the protonated histidine has a higher root mean squared fluctuation (RMSF) in the binding pocket, which is further increased for the W120A substitution. (f) Binding of [3H]-TAEHDEL to the KDEL receptor was measured at pH 5.4–7.0 and is plotted as a function of receptor concentration.
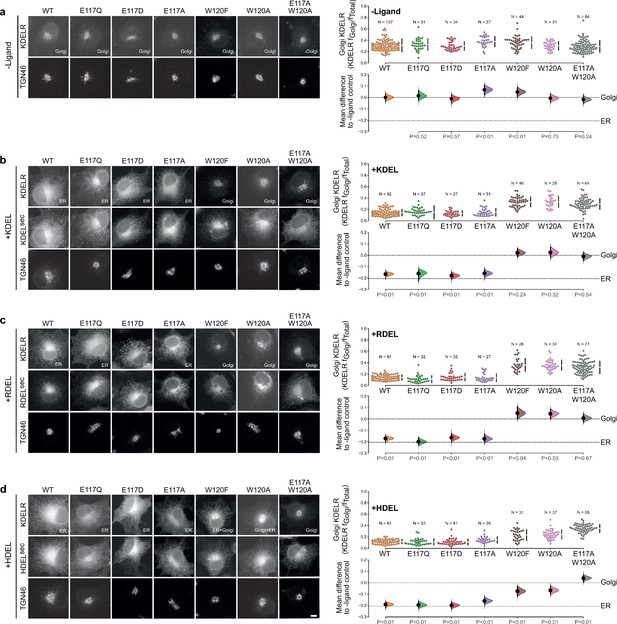
Effect of KDEL receptor E117 and W120 mutants on retrieval signal function in cells.
(a) Distribution of WT, E117, and W120 mutant KDEL receptors was measured in COS-7 cells in the absence (-ligand) or presence of (b) KDEL, (c) RDEL, or (d) HDEL retrieval signals (K/R/HDELsec). TGN46 was used as a Golgi marker. Scale bar is 10 µm. The fraction of WT, E117 and W120 mutant KDEL receptor localised to the Golgi was measured before (no ligand) and after challenge with different retrieval signals (K/R/HDEL) as indicated. Golgi signal for KDEL receptor and ligand intensity are shown on the scatter plots with sample sizes. Effect sizes are shown as the mean difference for K/R/HDEL comparisons against the shared -ligand control with sample sizes and p-values. The Cumming estimation plots for this data are used in main Figure 3c.
-
Figure 3—figure supplement 1—source data 1
Source data for the ligand-induced KDELR receptor retrieval assays in Figure 3 and Figure 3—figure supplement 1.
- https://cdn.elifesciences.org/articles/68380/elife-68380-fig3-figsupp1-data1-v3.xlsx
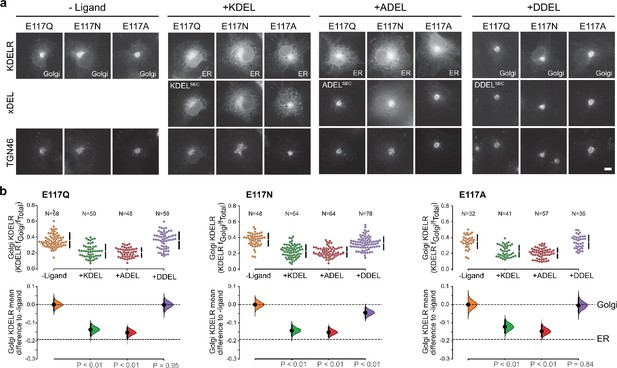
KDEL receptor E117 mutants show reduced selectivity for retrieval signals.
(a) E117Q, E117N, or E117A mutant KDEL receptors were tested for K/A/DDEL-induced redistribution from Golgi to ER in COS-7 cells. KDEL receptor distribution was followed in the absence (-ligand) or presence of K/A/DDELsec. TGN46 was used as a Golgi marker. Scale bar is 10 µm. (b) The fraction of E117Q, E117N or E117A mutant KDEL receptor localised to the Golgi was measured before (no ligand) and after challenge with different retrieval signals (K/A/DDEL). Effect sizes are shown as the mean difference for K/A/DDEL comparisons against the shared -ligand control with sample sizes and p values. .
-
Figure 4—source data 1
Source data for the ligand-induced KDELR receptor retrieval assays in Figure 4.
- https://cdn.elifesciences.org/articles/68380/elife-68380-fig4-data1-v3.xlsx
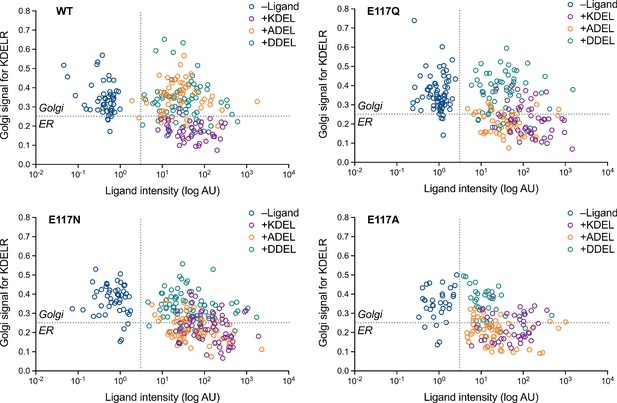
Effect of ligand levels on the response of KDEL receptor E117 mutants to KDEL, ADEL, and DDEL signals.
Distribution of WT, E117A, E117Q, and E117N mutant KDEL receptors was measured in COS-7 cells in the absence (-ligand) or presence of KDEL, ADEL, and DDEL retrieval signals. The fraction of WT, E117 mutant KDEL receptor localised to the Golgi was measured before (no ligand) and after challenge with different retrieval signals. Ligand intensity was also measured and plotted against the Golgi fraction of KDEL receptor.
-
Figure 4—figure supplement 1—source data 1
Source data for the ligand-induced KDELR receptor retrieval assays in Figure 4—figure supplement 1.
- https://cdn.elifesciences.org/articles/68380/elife-68380-fig4-figsupp1-data1-v3.xlsx
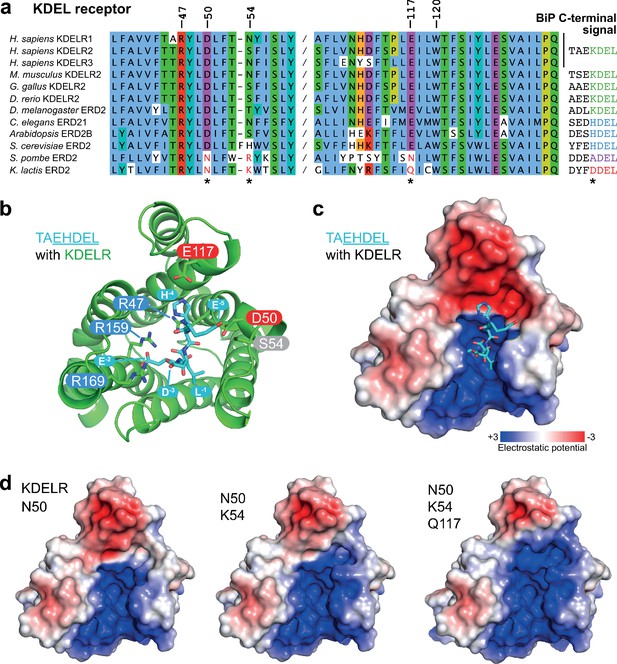
Charge distribution across the luminal entrance to the KDEL receptor binding pocket.
(a) KDEL receptor sequence alignment showing two regions centred around amino acid D50 and W120 of the human proteins. Cognate retrieval signal variants are shown to the right of the alignment. (b) The structure of the KDEL receptor with bound TAEHDEL highlighting key residues involved in ligand binding and variant residues D50, N54, and E117. (c) The charged surface for the WT KDEL receptor and (d) N50, N50/K54 and N50/K54/Q117 mutants is shown.
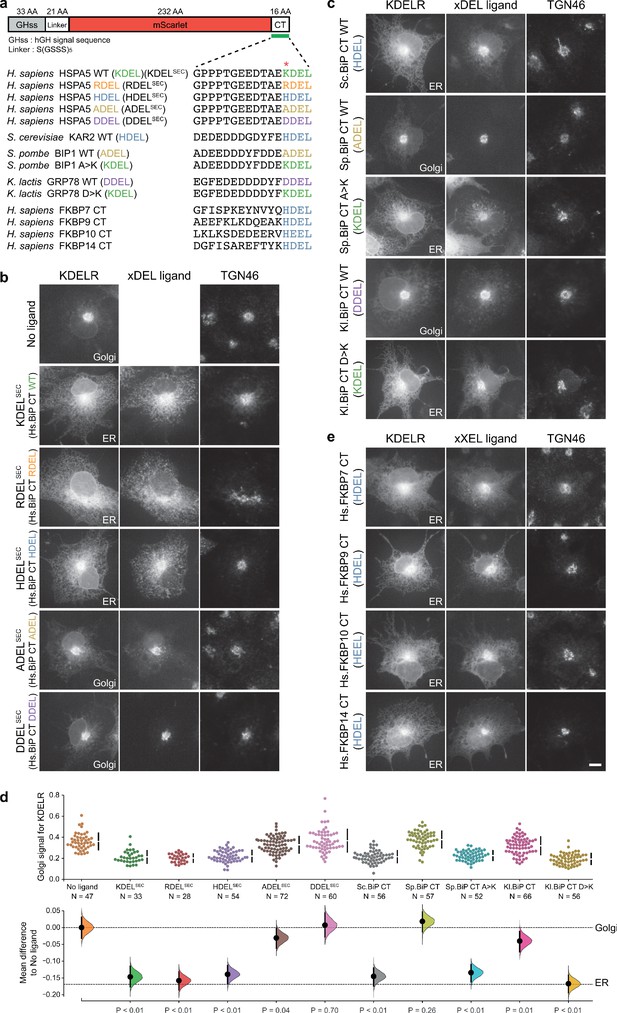
Comparison of human and yeast ER retrieval signals.
(a) Schematic of the ER retrieval construct showing the human growth hormone signal sequence (hGHss), linker, mScarlet fluorescent protein, and 16 amino acid extension carrying a C-terminal (CT) retrieval signal from known human and yeast ER proteins. A sequence alignment shows the conservation of the retrieval signal. (b) WT KDEL receptor distribution was followed in the absence (-ligand) or presence of human BIP derived signals (K/R/H/A/DDELsec). (c) WT KDEL receptor distribution was followed in COS-7 cells in the absence (-ligand) or presence of yeast BIP derived signals K/R/HA/DDELsec. (d) The fraction of KDEL receptor localised to the Golgi was measured before (no ligand) and after challenge with the different retrieval signals tested in b. and c. Effect sizes are shown as the mean difference for retrieval signal comparisons against the shared -ligand control with sample sizes and p-values. (e) WT KDEL receptor distribution was followed in COS-7 cells in the absence (-ligand) or presence of ER retrieval signals from human FKBP family proteins. In all image panels, TGN46 was used as a Golgi marker and the scale bar is 10 µm.
-
Figure 5—figure supplement 1—source data 1
Source data for the ligand-induced KDELR receptor retrieval assays in Figure 5—figure supplement 1.
- https://cdn.elifesciences.org/articles/68380/elife-68380-fig5-figsupp1-data1-v3.xlsx
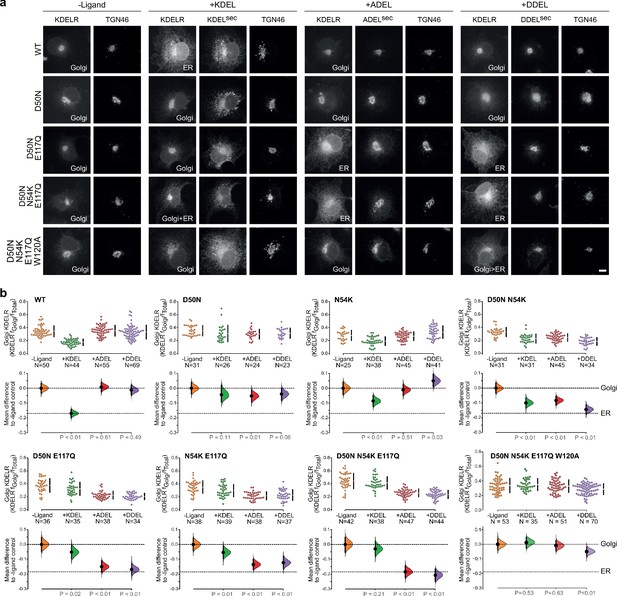
Re-engineering the selectivity of the human KDEL receptor for ADEL and DDEL signals.
(a) WT and a series of ‘K. lactis’-like mutant KDEL receptors were tested for K/A/DDEL-induced redistribution from Golgi to ER in COS-7 cells. KDEL receptor distribution was followed in the absence (-ligand) or presence of K/A/DDELsec. TGN46 was used as a Golgi marker. Scale bar is 10 µm. (b) The fraction of WT and mutant KDEL receptor localised to the Golgi was measured before (no ligand) after challenge with different retrieval signals (K/A/DDEL). Effect sizes are shown as the mean difference for K/A/DDEL comparisons against the shared -ligand control with sample sizes and p values.
-
Figure 6—source data 1
Source data for the ligand-induced KDELR receptor retrieval assays in Figure 6.
- https://cdn.elifesciences.org/articles/68380/elife-68380-fig6-data1-v3.xlsx
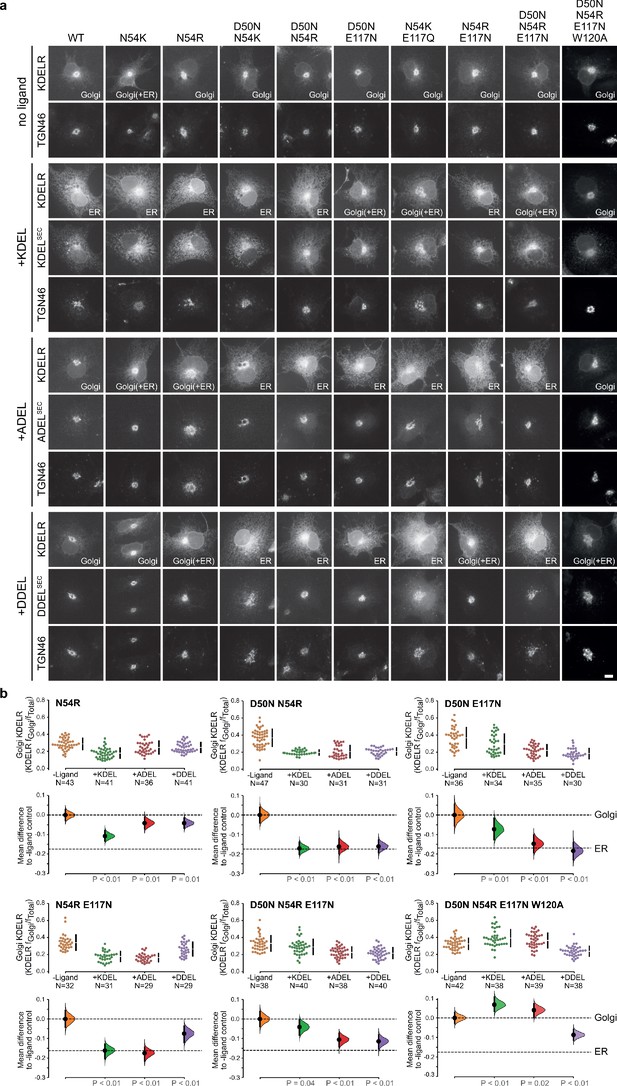
Extended analysis of human KDEL receptor selectivity.
(a) WT KDEL receptors and a series of S. pombe like mutants were tested for K/A/DDEL-induced redistribution from Golgi to ER in COS-7 cells. KDEL receptor distribution was followed in the absence (-ligand) or presence of K/A/DDELsec. TGN46 was used as a Golgi marker. Scale bar is 10 µm. (b) The fraction of WT and mutant KDEL receptor localised to the Golgi was measured before (no ligand) after challenge with different retrieval signals (K/A/DDEL). Effect sizes are shown as the mean difference for K/A/DDEL comparisons against the shared -ligand control with sample sizes and p values.
-
Figure 6—figure supplement 1—source data 1
Source data for the ligand-induced KDELR receptor retrieval assays in Figure 6—figure supplement 1.
- https://cdn.elifesciences.org/articles/68380/elife-68380-fig6-figsupp1-data1-v3.xlsx
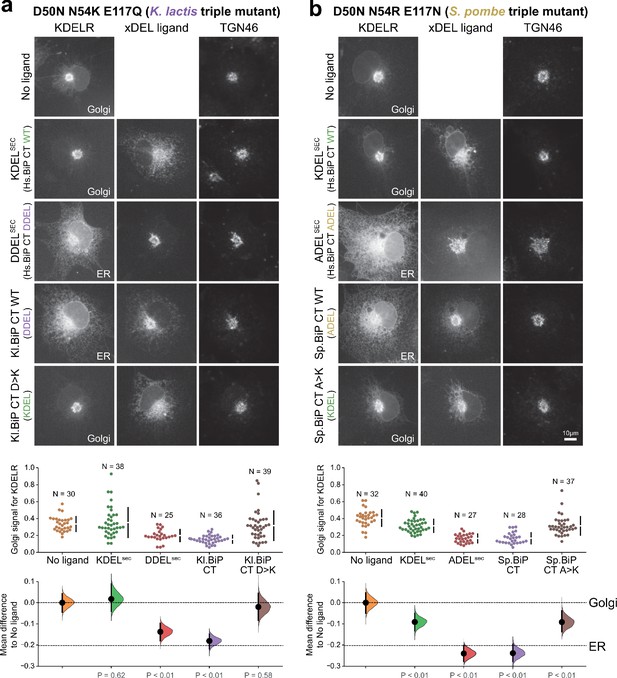
Retrieval specificity of ‘K. lactis’ and ‘S. pombe’ triple mutant KDEL receptors.
(a) Triple mutant ‘K. lactis’ and (b) ‘S. pombe’-like KDEL receptors were tested for K/A/DDEL-induced redistribution from Golgi to ER in COS-7 cells. KDEL receptor distribution was followed in the absence (-ligand) or presence of the indicated ligands. TGN46 was used as a Golgi marker. Scale bar is 10 µm. The fraction of WT and mutant KDEL receptor localised to the Golgi was measured before (no ligand) after challenge with different retrieval signals.
-
Figure 6—figure supplement 2—source data 1
Source data for the ligand-induced KDELR receptor retrieval assays in Figure 6—figure supplement 2.
- https://cdn.elifesciences.org/articles/68380/elife-68380-fig6-figsupp2-data1-v3.xlsx
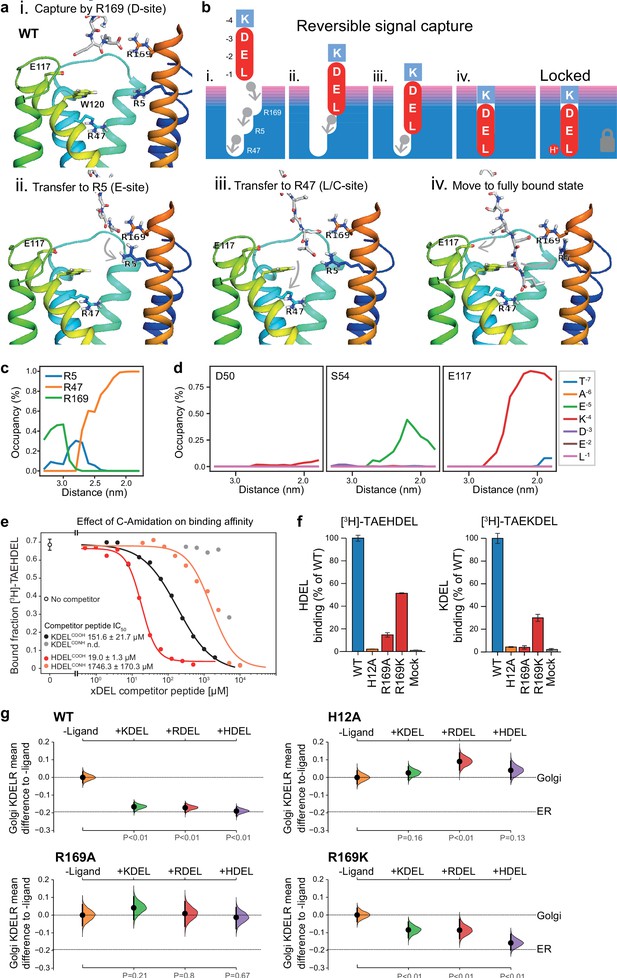
Mechanism for initial retrieval signal capture by the KDEL receptor.
(a) Images depicting the key stages (i.-iv.) of TAEKDEL binding to the wild-type (WT) KDEL receptor simulated using molecular dynamics. Initial engagement of the C-terminus to R169 (i) is followed by transfer to R5 (ii), shortly followed by interaction of E −2 with R169 (iii). Finally, R47 engages the C-terminus allowing D −3 to interact with R169 (iv). See also Video 1. (b) A carton model depicting the key stages of retrieval signal binding and final pH-dependent locked state. (c) Occupancy of the hydrogen bonds between the C-terminus of the KDEL retrieval signal and R5, R47, and R169 is plotted as a function of signal position within the binding pocket. (d) The occupancy of potential hydrogen bonds between the different positions of the KDEL retrieval signal and D50, S54, and E117 is plotted as a function of signal position within the binding pocket. (e) Competition binding assays for [3H]-TAEHDEL and unlabelled TAEKDEL and TAEHDEL with a free (COOH) or amidated (CONH) C-terminus to chicken KDELR2 showing IC50 values for the competing peptides. (f) Normalised binding of [3H]-TAEHDEL and [3H]-TAEKDEL signals to the purified WT H12A, R169A, or R169K mutant chicken KDELR2. A mock binding control with no receptor indicates the background signal. (g) Distribution of WT, H12A, R169A, and R169K KDEL receptors was measured in COS-7 cells in the absence (-ligand) or presence of K/R/HDELsec. The mean differences for K/R/HDEL comparisons against the shared no ligand control are shown with sample sizes and p values. See also Figure 7—figure supplement 1 with accompanying source data.
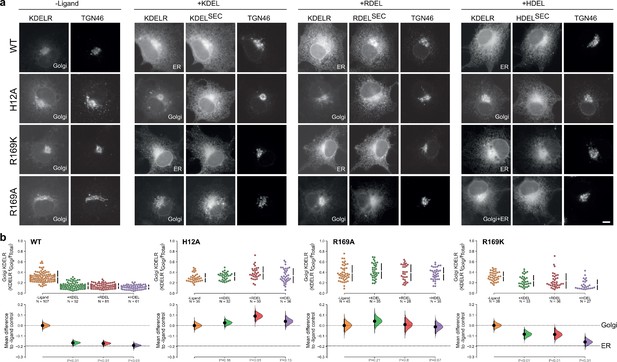
R169 plays a crucial role in signal recognition.
(a) Distribution of WT, H12A, R169A, and R169K KDEL receptors was measured in COS-7 cells in the absence (-ligand) or presence of K/R/HDEL (mScarlet-xDELsec). TGN46 was used as a Golgi marker. Scale bar is 10 µm. (b) The raw data for the fraction of KDEL receptor fluorescence in the Golgi is plotted on the upper axes with sample sizes, with effect sizes shown in the lower graphs.
-
Figure 7—figure supplement 1—source data 1
Source data for the ligand-induced KDELR receptor retrieval assays in Figure 7 and Figure 7—figure supplement 1.
- https://cdn.elifesciences.org/articles/68380/elife-68380-fig7-figsupp1-data1-v3.xlsx
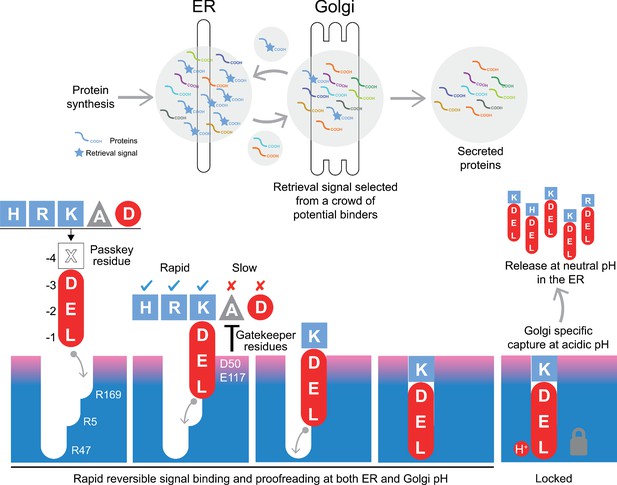
A combined proofreading and relay handover model for signal capture by the KDEL receptor.
Newly synthesised secretory and ER luminal proteins are translocated into the ER and on to the Golgi. Those proteins with C-terminal retrieval signals are captured by the KDELR receptor and returned to the ER. Other proteins with different C-terminal sequences move on to be secreted. The retrieval signal can be broken down into two sections: the variable −4 passkey position and the −1 to −3 positions with free carboxyl-terminus. Signals are initially captured through their free carboxyl-terminus by the receptor R169. This is then handed over to R5 and finally R47 in a relay mechanism. Sequences are proofread for the residue at the −4 position by gatekeeper residues D50 and E117. Unwanted signal variants are rejected. Only signals that completely enter the binding pocket and engage R47 can undergo pH dependent capture and return to the ER.
Videos
Stepwise engagement of the KDEL signal with the KDEL receptor.
Molecular dynamics simulation of TAEKDEL binding to the KDEL receptor simulated using molecular dynamics.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Escherichia coli) | XL1-Blue Competent Cells | Agilent Technologies | 200249 | Used to prepare plasmid DNA |
| Strain, strain background (Saccharomyces cerevisiae) | Bj5460 | ATCC | 208285 | Used for KDELR protein expression |
| Cell line (African green monkey) | COS-7 kidney fibroblast-like cell line | ATCC | CRL-1651 | ER retrieval assays |
| Cell line (Homo-sapiens) | HeLa S3 cervical adenocarcinoma | ATCC | CCL-2.2 | Protein secretion assays |
| Antibody | TGN46 sheep polyclonal | Bio-rad (AbD Serotec) | AHP500G | IF (1:1000) |
| Antibody | GRP78 BiP rabbit polyclonal | Abcam | ab21685 | WB (1:1000) |
| Antibody | PDI rabbit polyclonal | ProteinTech | #11245–1 | WB (1:1000) |
| Antibody | ERp72 rabbit monoclonal | Cell Signalling Technology | #5033S | WB (1:1000) |
| Antibody | ERp44 rabbit monoclonal | Cell Signalling Technology | # 3798S | WB (1:1000) |
| Antibody | KDEL receptor mouse monoclonal | Enzo Life Sciences | ADI-VAA-PT048 | IF (1:1000) WB (1:1000) |
| Antibody | RFP mouse monoclonal | Chromotek | 6G6 | WB (1:1000) Detects mScarlet on Western blot |
| Antibody | Donkey anti-Mouse IgG Alexa Fluor 488 | Invitrogen | A-21202 | IF (1:2000) Secondary |
| Antibody | Donkey anti-Sheep IgG Alexa Fluor 647 | Invitrogen | A-21448 | IF (1:2000) Secondary |
| Antibody | Peroxidase-AffiniPure Donkey Anti-Rabbit IgG | Jackson Immuno Research | 711-035-152-JIR | WB (1:2000) Secondary |
| Antibody | Peroxidase-AffiniPure Donkey Anti-Mouse IgG | Jackson Immuno Research | 711-035-152-JIR | WB (1:2000) Secondary |
| Antibody | Peroxidase-AffiniPure Donkey Anti-Sheep IgG | Jackson Immuno Research | 713-035-147-JIR | WB (1:2000) Secondary |
| Recombinant DNA reagent | pcDNA3.1 hGHss-mScarlet-H. sapiens BiP639-654 | Bräuer et al., 2019 | KDELSEC | PMID:30846601 |
| Recombinant DNA reagent | pcDNA3.1 hGHss-mScarlet-H. sapiens BiP639-654 K651R | This paper | RDELSEC | Material and methods. Available from Barr lab |
| Recombinant DNA reagent | pcDNA3.1 hGHss-mScarlet-H. sapiens BiP639-654 K651H | This paper | HDELSEC | Material and methods. Available from Barr lab |
| Recombinant DNA reagent | pcDNA3.1 hGHss-mScarlet-H. sapiens BiP639-654K651A (ADELSEC) | This paper | ADELSEC | Material and methods. Available from Barr lab |
| Recombinant DNA reagent | pcDNA3.1 hGHss-mScarlet-H. sapiens BiP639-654K651D (DDELSEC) | This paper | DDELSEC | Material and methods. Available from Barr lab |
| Recombinant DNA reagent | pcDNA3.1 hGHss-mScarlet-S. cerevisiae BiP667-682 | This paper | Yeast BiP | Material and methods. Available from Barr lab |
| Recombinant DNA reagent | pcDNA3.1 hGHss-mScarlet-S. pombe BiP648-663 | This paper | S. pombe BiP | Material and methods. Available from Barr lab |
| Recombinant DNA reagent | pcDNA3.1 hGHss-mScarlet-S. pombe BiP648-663 A660K | This paper | S. pombe BiP A > K | Material and methods. Available from Barr lab |
| Recombinant DNA reagent | pcDNA3.1 hGHss-mScarlet-K. lactis BiP664-679 | This paper | K. lactis BIP | Material and methods. Available from Barr lab |
| Recombinant DNA reagent | pcDNA3.1 hGHss-mScarlet-K. lactis BiP664-679 D676K | This paper | K.lactis BiP D > K | Material and methods. Available from Barr lab |
| Recombinant DNA reagent | pcDNA3.1 hGHss-mScarlet-H. sapiens FKBP7207-222 | This paper | FKBP7 | Material and methods. Available from Barr lab |
| Recombinant DNA reagent | pcDNA3.1 hGHss-mScarlet-H. sapiens FKBP9555-570 | This paper | FKBP9 | Material and methods. Available from Barr lab |
| Recombinant DNA reagent | pcDNA3.1 hGHss-mScarlet-H. sapiens FKBP10567-582 | This paper | FKBP10 | Material and methods. Available from Barr lab |
| Recombinant DNA reagent | pcDNA3.1 hGHss-mScarlet-H. sapiens FKBP14196-211 | This paper | FKBP14 | Material and methods. Available from Barr lab |
| Recombinant DNA reagent | pEF5/FRT human KDELR1-GFP | Bräuer et al., 2019 | KDELR1 | PMID:30846601 |
| Recombinant DNA reagent | pEF5/FRT human KDELR2-GFP | This paper | KDELR2 | Material and methods. Available from Barr lab |
| Recombinant DNA reagent | pEF5/FRT human KDELR1 H12A-GFP | Bräuer et al., 2019 | H12A | Expression in mammalian cells for functional assays; PMID:30846601 |
| Recombinant DNA reagent | pEF5/FRT human KDELR1 D50N-GFP | This paper | D50N | Material and methods. Available from Barr lab |
| Recombinant DNA reagent | pEF5/FRT human KDELR1 N54K-GFP | This paper | N54K | Material and methods. Available from Barr lab |
| Recombinant DNA reagent | pEF5/FRT human KDELR1 N54R-GFP | This paper | N54R | Material and methods. Available from Barr lab |
| Recombinant DNA reagent | pEF5/FRT human KDELR1 E117Q-GFP | This paper | E117Q | Material and methods. Available from Barr lab |
| Recombinant DNA reagent | pEF5/FRT human KDELR1 E117D-GFP | This paper | E117D | Material and methods. Available from Barr lab |
| Recombinant DNA reagent | pEF5/FRT human KDELR1 E117A-GFP | This paper | E117A | Material and methods. Available from Barr lab |
| Recombinant DNA reagent | pEF5/FRT human KDELR1 E117N-GFP | This paper | E117N | Material and methods. Available from Barr lab |
| Recombinant DNA reagent | pEF5/FRT human KDELR1 W120F-GFP | This paper | W120F | Material and methods. Available from Barr lab |
| Recombinant DNA reagent | pEF5/FRT human KDELR1 W120A-GFP | This paper | W120A | Material and methods. Available from Barr lab |
| Recombinant DNA reagent | pEF5/FRT human KDELR1 R169K-GFP | This paper | R169K | Material and methods. Available from Barr lab |
| Recombinant DNA reagent | pEF5/FRT human KDELR1 R169A-GFP | This paper | R169A | Material and methods. Available from Barr lab |
| Recombinant DNA reagent | pEF5/FRT human KDELR1 E117A/W120A-GFP | This paper | E117A/W120A | Material and methods. Available from Barr lab |
| Recombinant DNA reagent | pEF5/FRT human KDELR1 D50N/N54K-GFP | This paper | D50N/N54K | Material and methods. Available from Barr lab |
| Recombinant DNA reagent | pEF5/FRT human KDELR1 D50N/N54R-GFP | This paper | D50N/N54R | Material and methods. Available from Barr lab |
| Recombinant DNA reagent | pEF5/FRT human KDELR1 D50N/E117Q-GFP | This paper | D50N/E117Q | Material and methods. Available from Barr lab |
| Recombinant DNA reagent | pEF5/FRT human KDELR1 D50N/E117N-GFP | This paper | D50N/E117N | Material and methods. Available from Barr lab |
| Recombinant DNA reagent | pEF5/FRT human KDELR1 N54K/E117Q-GFP | This paper | N54K/E117Q | Material and methods. Available from Barr lab |
| Recombinant DNA reagent | pEF5/FRT human KDELR1 D50N/N54K/E117Q-GFP | This paper | D50N/N54K/E117Q | Material and methods. Available from Barr lab |
| Recombinant DNA reagent | pEF5/FRT human KDELR1 N54R/E117N-GFP | This paper | N54R/E117N | Material and methods. Available from Barr lab |
| Recombinant DNA reagent | pEF5/FRT human KDELR1 D50N/N54R/E117N-GFP | This paper | D50N/N54R/E117N | Material and methods. Available from Barr lab |
| Recombinant DNA reagent | pEF5/FRT human KDELR1 D50N/N54K/E117Q/W120A-GFP | This paper | D50N/N54K/E117Q/W120A | Material and methods. Available from Barr lab |
| Recombinant DNA reagent | pEF5/FRT human KDELR1 D50N/N54R/E117N/W120A-GFP | This paper | D50N/N54R/E117N/W120A | Material and methods. Available from Barr lab |
| Recombinant DNA reagent | pDDGFP-Leu2d-GgKDELR2 | Addgene | 123618 | Protein expression in yeast for biochemical assays and structures |
| Recombinant DNA reagent | pDDGFP-Leu2d-GgKDELR2_H12A | This paper | KDELR2_H12A | Material and methods. Available from Newstead lab |
| Recombinant DNA reagent | pDDGFP-Leu2d-GgKDELR2_E117A | This paper | KDELR2_E117A | Material and methods. Available from Newstead lab |
| Recombinant DNA reagent | pDDGFP-Leu2d-GgKDELR2_E117D | This paper | KDELR2_E117D | Material and methods. Available from Newstead lab |
| Recombinant DNA reagent | pDDGFP-Leu2d-GgKDELR2_E117Q | This paper | KDELR2_E117Q | Material and methods. Available from Newstead lab |
| Recombinant DNA reagent | pDDGFP-Leu2d-GgKDELR2_E127A | This paper | KDELR2_E127A | Material and methods. Available from Newstead lab |
| Recombinant DNA reagent | pDDGFP-Leu2d-GgKDELR2_E127Q | This paper | KDELR2_E127Q | Material and methods. Available from Newstead lab |
| Recombinant DNA reagent | pDDGFP-Leu2d-GgKDELR2_W120A | This paper | KDELR2_W120A | Material and methods. Available from Newstead lab |
| Recombinant DNA reagent | pDDGFP-Leu2d-GgKDELR2_W120F | This paper | KDELR2_W120F | Material and methods. Available from Newstead lab |
| Recombinant DNA reagent | pDDGFP-Leu2d-GgKDELR2_R169A | This paper | KDELR2_R169A | Material and methods. Available from Newstead lab |
| Recombinant DNA reagent | pDDGFP-Leu2d-GgKDELR2_R169K | This paper | KDELR2_R169K | Material and methods. Available from Newstead lab |
| Peptide, recombinant protein | TEV Protease | Merck | T4455-10KU | |
| Peptide, recombinant protein | 3H-TAEHDEL | Cambridge Research Biochemicals | custom synthesis | 185 MBq 106 Ci/mmol |
| Peptide, recombinant protein | 3H-TAEKDEL | Cambridge Research Biochemicals | custom synthesis | 185 MBq 128 Ci/mmol |
| Peptide, recombinant protein | TAEHDEL | Cambridge peptides | custom synthesis | |
| Peptide, recombinant protein | TAEKDEL | Cambridge peptides | custom synthesis | |
| Peptide, recombinant protein | TAERDEL | Cambridge peptides | custom synthesis | |
| Peptide, recombinant protein | TAEDDEL | Cambridge peptides | custom synthesis | |
| Peptide, recombinant protein | TAEKDEL-CONH | Cambridge peptides | custom synthesis | C-amidated peptide variant. |
| Peptide, recombinant protein | TAEHDEL-CONH | Cambridge peptides | custom synthesis | C-amidated peptide variant. |
| Chemical compound, drug | Sodium phosphate monobasic (NaH2PO4) | Sigma | S8282 | |
| Chemical compound, drug | Sodium phosphate dibasic (Na2HPO4) | Sigma | 71640 | |
| Chemical compound, drug | Sodium periodate (NaIO4) | Sigma | 311448 | |
| Chemical compound, drug | 16% (w/v) Formaldehyde | Thermo Fisher Scientific | 28908 | |
| Chemical compound, drug | Saponin | Sigma | S7900 | |
| Chemical compound, drug | L-Lysine monohydrochloride | Sigma | 62929 | |
| Chemical compound, drug | Mowiol 4–88 | Millipore | 475904 | |
| Chemical compound, drug | Trichloroacetic acid | Sigma | T6399 | |
| Chemical compound, drug | Dodecyl maltoside (DDM) | Glycon | D97002-C | |
| Chemical compound, drug | Cholesteryl hemisuccinate (CHS) | Sigma | C6512 | |
| Chemical compound, drug | Monoolein | Sigma | M7765 | |
| Software, algorithm | Metamorph 7.5 | Molecular Dynamics Inc | http://www.moleculardevices.com | Microscope image data acquisition |
| Software, algorithm | Fiji 2.0.0-rc-49/1.52i | NIH Image | http://fiji.sc/ | Microscope image data analysis |
| Software, algorithm | GraphPad Prism 7 | GraphPad Software | http://www.graphpad.com | Graph plotting |
| Software, algorithm | R | R project for statistical computing | https://www.r-project.org | Statistical analysis and graph plotting |
| Software, algorithm | Adobe Illustrator CC | Adobe Systems Inc | http://www.adobe.com | Figure preparation |
| Software, algorithm | Adobe Photoshop CC | Adobe Systems Inc | http://www.adobe.com | Figure preparation |
| Software, algorithm | COOT | Emsley et al., 2010 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot | Macromolecular structure model building; PMID:20383002 |
| Software, algorithm | PyMOL | Schrodinger | https://pymol.org/2 | Molecular visualisation |
| Software, algorithm | Buster | Global Phasing | https://www.globalphasing.com | Structure refinement |
| Software, algorithm | GROMACS | Abraham et al., 2015 | https://www.gromacs.org | Molecular dynamics |
| Software, algorithm | GMX_lipid17.ff: GROMACS | Wu and Biggin, 2020 | http://doi.org/10.5281/zenodo.3610470 | Port of the Amber LIPID17 force field |
| Software, algorithm | MDAnalysis 1.0 | SciPy2016 | https://conference.scipy.org/proceedings/scipy2016/oliver_beckstein.html | Analysis of molecular dynamics simulations |
| Software, algorithm | Modeller 9.21 | Webb and Sali, 2016 | https://salilab.org/modeller/ | PMID:27322406 |
| Other | Ultima Gold Scintillation Fluid | Perkin Elmer | 6013326 | |
| Other | HisPurTM | Thermo Fisher Scientific | 25214 | |
| Other | HisTrap HP | Cytiva | 17-5248-01 | |
| Other | Superdex 200 10/300 GL | Cytiva | 28-9909-44 | |
| Other | Ultra-15 Centrifugal Filter Unit, 50K NMWC | Amicon | UFC905024 | |
| Other | Yeast Drop Out media -Ura | Formedium | DCS0169 | |
| Other | Yeast Drop Out media -Leu | Merck | Y1376-20G | |
| Other | Tunair Flasks | Sigma | Z710822-4EA | |
| Other | Dulbecco's modified Eagle's medium | Thermo Fisher Scientific | 31966–047 | |
| Other | Foetal Bovine Serum | Sigma | F9665 | |
| Other | TrypLE Express Enzyme | Thermo Fisher Scientific | 12605036 | |
| Other | Opti-MEM | Thermo Fisher Scientific | 11058021 | |
| Other | EZ-PCR Mycoplasma Test Kit | Geneflow | K1-0210 | |
| Other | TransIT-LT1 | Mirus Bio LLC | MIR 2306 | |
| Other | ECL western blotting reagent | Cytiva | RPN2106 |
Calculated pKa values for retrieval signal -4 side chains.
| WT | W120F | W120A | |
|---|---|---|---|
| HDEL | 8.9+/-0.5 | 7.6+/-0.3 | 6.5+/-0.1 |
| KDEL | 11.1+/-0.4 | 9.7+/-0.2 | 9.0+/-0.1 |
Additional files
-
Supplementary file 1
Crystallographic data collection statistics.
Values in parentheses are for the highest resolution shell.
- https://cdn.elifesciences.org/articles/68380/elife-68380-supp1-v3.docx
-
Supplementary file 2
Free energy differences for HDEL protonation states and KDEL.
The free energy difference is expressed in kcal/mol and the sum is the ensemble free energy difference between KDEL and HDEL. HID signifies that the histidine is protonated at δ nitrogen and HIE means that the histidine is protonated at ε nitrogen, while HIP means that both positions are protonated. The occupancy of protonation state is computed at pH seven and expressed as a percentage (%).
- https://cdn.elifesciences.org/articles/68380/elife-68380-supp2-v3.docx
-
Supplementary file 3
Cation-π and π-π contributions between receptor W120 and the −4 histidine in the retrieval signal.
The cation-π interaction is the energy resulting from induction calculated at the level of sSAPT0/jun-cc-pVDZ. The π-π interaction is the sum of exchange and correlation energy. The sum is the sum of cation-π and π-π energy.
- https://cdn.elifesciences.org/articles/68380/elife-68380-supp3-v3.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/68380/elife-68380-transrepform-v3.docx





