Profiling sensory neuron microenvironment after peripheral and central axon injury reveals key pathways for neural repair
Figures
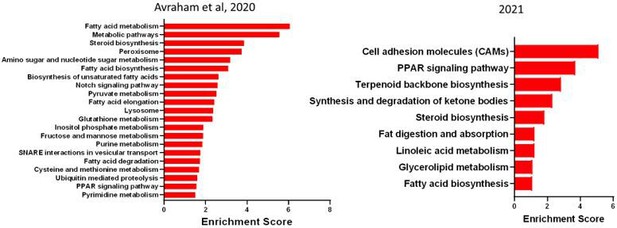
DRG cells respond differently following peripheral and central axon injuries.
(A) Representative TEM images of a DRG section showing neuronal cell bodies (pseudo-colored in purple) its enveloping SGC (pseudo-colored in turquoise) and other non-neuronal cells (pseudo-colored in orange). n = 4 biologically independent animals. Scale bar:20 µm (B) Actl6bCre mice crossed with Sun1GFP show expression of GFP in all neuronal cell somas, co-labeled with the unique neuronal marker ISL1 (magenta). n = 4 biologically independent animals, Scale bar: 50 µm (C) Flow cytometry analysis of dissociated DRG cells from Actl6b Cre:Sun1GFP mice. Scatter plot of fluorescence intensities of live Hoechst + cells (x axis) and GFP+ (y axis). 12.5% of Hoechst + cells are also GFP+ positive. n = 3 biologically independent animals. (D) Diagram of mouse peripheral and central injury models. (E) Schematic of the experimental design for scRNAseq. (F) t-SNE plot of 25,154 cells from L4,L5 dissociated naïve and injured mouse DRG. 9 distinct cell clusters were assigned based on known marker genes. (G) Fraction of each cell type within naive (6343 cells), SNC (4735 cells), DRC (7199 cells) and SCI (7063 cells) conditions. n = 2 (NAI,DRC,SCI) and n = 1 (SNC) biologically independent experiments. (H) t-SNE plots of DRG cells separated by the different injury conditions, colored by cell type. (I) Heatmap of the number of differentially regulated genes in each cell type and injury condition (FDR ≤ 0.05, fold-change ≥ 2).
-
Figure 1—source data 1
Source files for scRNAseq analysis; top DEG for cell clustering and cluster counts.
- https://cdn.elifesciences.org/articles/68457/elife-68457-fig1-data1-v1.xlsx
-
Figure 1—source data 2
Significant ligand-receptor interactions (p-value < 0.05).
- https://cdn.elifesciences.org/articles/68457/elife-68457-fig1-data2-v1.xlsx
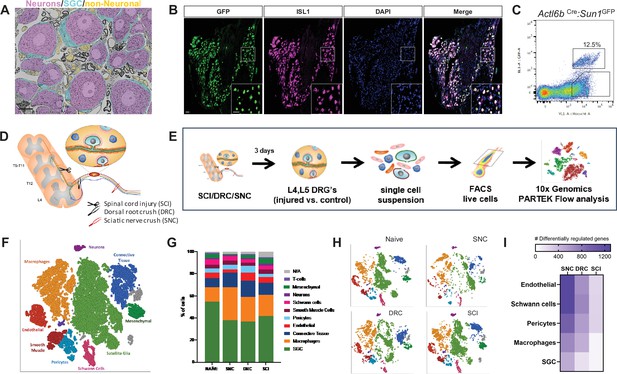
scRNAseq analysis of naïve and injured mouse DRG.
(A) t-SNE plots of mouse DRG cells separated by injury condition and colored by biological replicate. (B) t-SNE plot of naïve and injured mouse DRG cells where 19 distinct cell clusters were identified by unbiased, Graph based clustering. (C) t-SNE plot of naïve and injured mouse DRG cells colored by injury condition. (D) t-SNE overlay for expression of marker genes for different cell populations including Pecam/Cd31 for endothelial cells, Col1a1 for connective tissue, Ncmap for Schwann cells, Isl1 for neurons, Kcnj8 for pericytes, Pln for smooth muscle cells, Fabp7 for SGC and Aif1/Iba1 for Macrophages. The relative levels of expression are presented as a red color gradient on the left. (E) Cell-cell interaction network analysis (CellPhoneDB) and visualization (Cytoscape). Nodes represent cell clusters, and the node size correlates with the relative cell counts in the cluster. Significant cell-cell interactions represent the edges in the network. The width and transparency of the edges correlate with the number of interactions and the arrow indicates the directionality of ligand/receptor interaction.
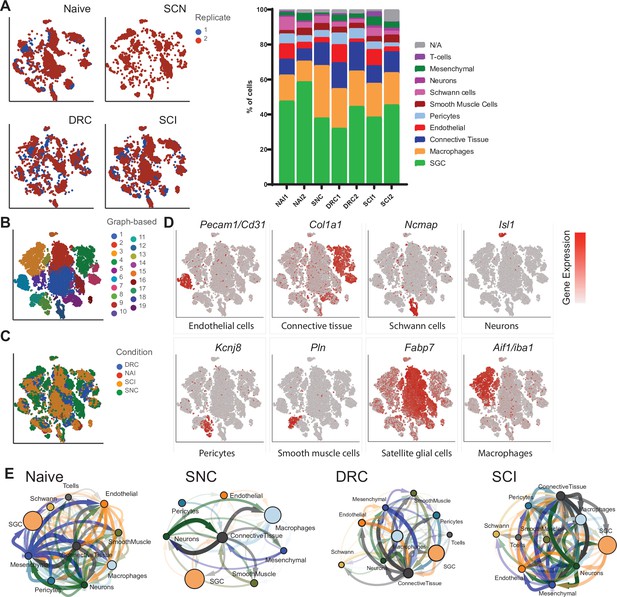
Molecular changes in non-neuronal cells in response to central and peripheral injuries.
(A) Representative images of mouse DRG sections injected with Lycopersicon esculentum (Tomato) Lectin (magenta), labeling blood vessels and immunostained with FABP7 (green) labeling SGC. Scale bar: 50 µm. (B) Representative TEM images of DRG sections focusing on blood vessels with the surrounding endothelial (pseudo-colored in purple) and pericytes (pseudo-colored in turquoise) n = 4 biologically independent animals. Scale bar:2 µm (C) t-SNE plot of DRG endothelial cells colored by injury condition. (D) t-SNE plot of DRG pericytes colored by injury condition. (E) Heatmap of Adherens junction (AJ) and Tight Junction (TJ) genes expression in endothelial cells by z-score for all injury conditions. (F) Heatmap of Adherens junction (AJ) and Tight Junction (TJ) genes expression in pericytes by z-score for all injury conditions. (G) Pathway analysis (KEGG 2019) of differentially upregulated (red) and downregulated (blue) genes in the endothelial cell cluster. n = 2 biologically independent experiments. (FDR ≤ 0.05, fold-change ≥2). (H) Pathway analysis (KEGG 2019) of differentially upregulated (red) and downregulated (blue) genes in the pericyte cluster. n = 2 biologically independent experiments. (FDR ≤ 0.05, fold-change ≥ 2). (I) t-SNE plot of DRG Schwann cells colored by injury condition. (J) Heatmap of fold change expression for selected repair Schwann cell genes after SNC, DRC and SCI compared to naïve. (K) Pathway analysis (KEGG 2019) of differentially upregulated (red) and downregulated (blue) genes in the Schwann cell cluster. n = 2 biologically independent experiments (FDR ≤ 0.05, fold-change ≥ 2).
-
Figure 2—source data 1
Source files for scRNAseq analysis; DEG in endothelial cells, pericytes and Schwann cells in response to peripheral and central injuries (FDR ≤ 0.05, fold-change ≥ 2), repair Schwann cell genes expression across injuries.
- https://cdn.elifesciences.org/articles/68457/elife-68457-fig2-data1-v1.xlsx
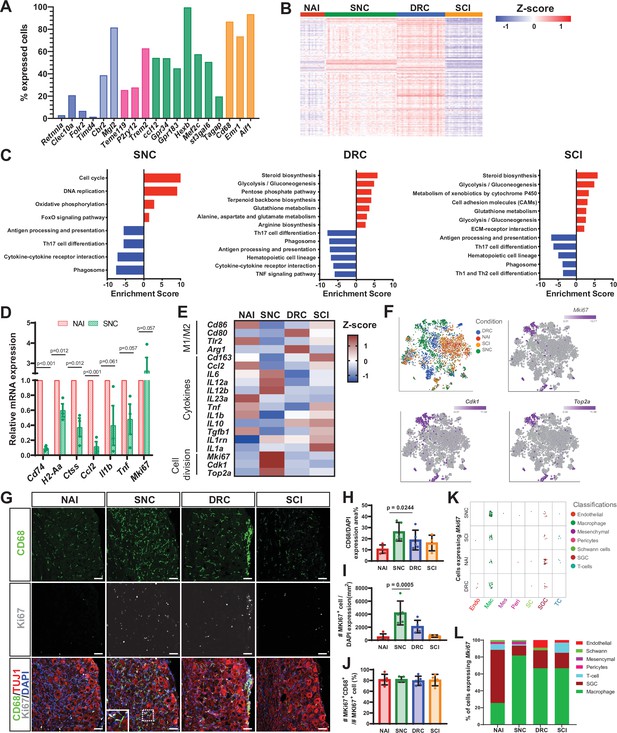
Macrophages undergo distinct transcriptional changes in response to central and peripheral injuries.
(A) Fraction of uninjured cells expressing selected genes in the macrophage cluster. snMac2 (blue), snMac1 (light blue), specific microglia genes (pink), CNS microglia/macrophages (green) and common macrophage markers (orange). (B) Heatmap of gene expression profile in macrophages by z-score for all injury conditions. (C) Pathway analysis (KEGG 2019) of differentially upregulated (red) and downregulated (blue) genes in the Macrophage cell cluster. n = 2 biologically independent experiments. (FDR ≤ 0.05, fold-change ≥ 2). (D) DRG qPCR analysis of DEG in macrophages after SNC compared to Naive. (E) Heatmap of M1, M2 macrophage markers, selected cytokines and proliferation marker gene expression by z- score for all injury conditions. (F) t-SNE plots of mouse DRG macrophages colored by injury condition and t-SNE overlay for expression of proliferation marker genes in pooled macrophage cluster from all injury conditions. (G) Representative images of immunofluorescence staining of DRG sections labeled with CD68 (green), MKI67 (white) and TUJ1 (red) from naïve mice, SNC, DRC and SCI injuries n = 5 biologically independent animals. Scale bar: 50 µm. (H) Quantification of area with CD68 expressing cells. (I) Quantification of MKI67 expressing cells normalized to DAPI. (J) Quantification of the percentage of cells expressing both MKI67 and CD68 out of all MKI67 positive cells. n = 5 (NAI,SNC,DRC) and n = 4 (SCI) biologically independent animals. (H-J) One-way analysis of variance (ANOVA) followed by Bonferroni’s multiple comparisons test. Data are presented as mean values ± SD. (K) Plot of cells expressing Mki67, colored by cell type, for all injury conditions. Every dot represents one cell. (6< log gene counts). (L) Quantification of the percentage of cells expressing Mki67, colored by cell type, in all injury conditions.
-
Figure 3—source data 1
Source files for scRNAseq analysis; Macrophage markers in the macrophage cluster, DEG in macrophages (FDR ≤ 0.05, fold-change ≥ 2) and MkKi67 expression.
Image analysis of DRG sections, qPCR raw data.
- https://cdn.elifesciences.org/articles/68457/elife-68457-fig3-data1-v1.xlsx
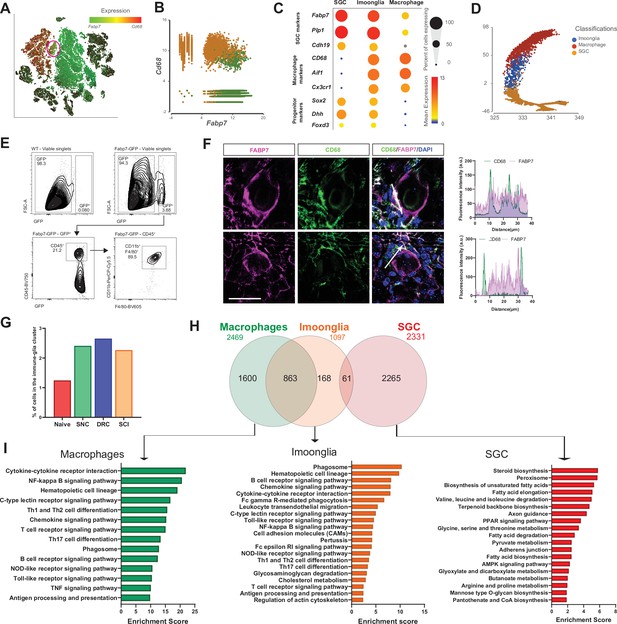
A subset of macrophages expressing glial markers is increased by injury.
(A) t-SNE plot of cells from all injury conditions overlay for expression of Cd68 (red) and Fabp7 (green). (B) Plotting cells in the macrophage (orange) and SGC (green) clusters for expression of Cd68 (y-axis) and Fabp7 (x-axis). (C) Dot plot of macrophage, glial and progenitor marker genes expression in the macrophage, SGC and Imoonglia clusters. The percentage of cell expressing the gene is calculated as the number of cells in each cluster express the gene ( > 0 counts) divided by the total number of cells in the respective cluster. Expression in each cluster is calculated as mean expression of the gene relative to the highest mean expression of that gene across all clusters. (D) Trajectory analysis of macrophage, SGC and Imoonglia cell clusters. (E) Flow cytometry analysis of DRG cells from Fabp7creER:Sun1GFP mice, stained with the macrophage marker genes CD45, CD11b and F4/80 n = 2 (F4/80) n = 1 (CD45,CD11b,F4/80) biologically independent animals. (F) Representative confocal images of immunofluorescence staining of DRG sections labeled with CD68 (green) and FABP7 (magenta). Fluorescence intensity for CD68 and FABP7 was measured along the arrow line. Scale bar: 50 µm (G) Fraction of cells in the Imoonglia cluster by injury condition. n = 2 biologically independent experiments. (H) Venn diagram of differentially expressed genes in the Imoonglia cluster (1,097 genes) was compared to top differentially expressed genes in the macrophage cluster (2,469 genes) and the SGC (2,331 genes) (FDR ≤ 0.05, fold-change ≥ 2). (I) Pathway analysis (KEGG 2019) of differentially expressed genes in Macrophages, Imoonglia and SGC.
-
Figure 4—source data 1
Source files for scRNAseq analysis, flow cytometry additional experiments.
- https://cdn.elifesciences.org/articles/68457/elife-68457-fig4-data1-v1.xlsx
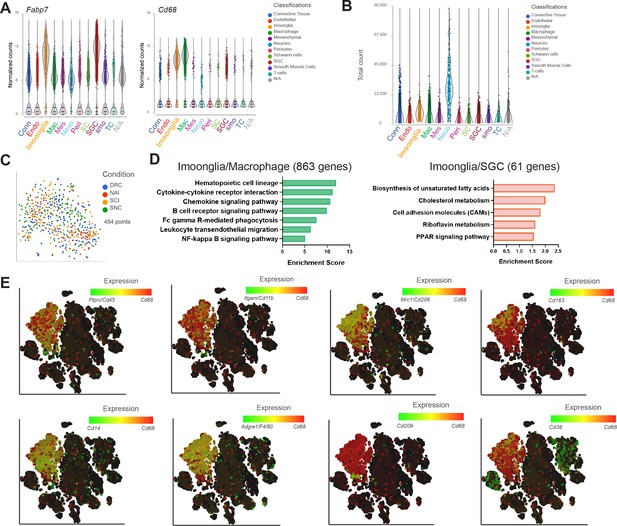
Imoonglia- a subset of macrophage expressing glia markers.
(A) Violin plots illustrate the expression signatures of Cd68 and Fabp7 in distinct cell populations of naïve mouse DRG cells. (B) Violin plot of total counts in all cell clusters. (C) t-SNE plot of pooled Imoonglia cells from all injury conditions. (D) Pathway analysis (KEGG 2019) of shared genes in Imoonglia and Macrophage cluster (863 genes) and Imoonglia and SGC (61 genes) (FDR ≤ 0.05, fold-change ≥ 2). (E) t-SNE plot of cells from all injury conditions overlay for expression of Cd68 (red) and various macrophage/myeloid markers genes (green).
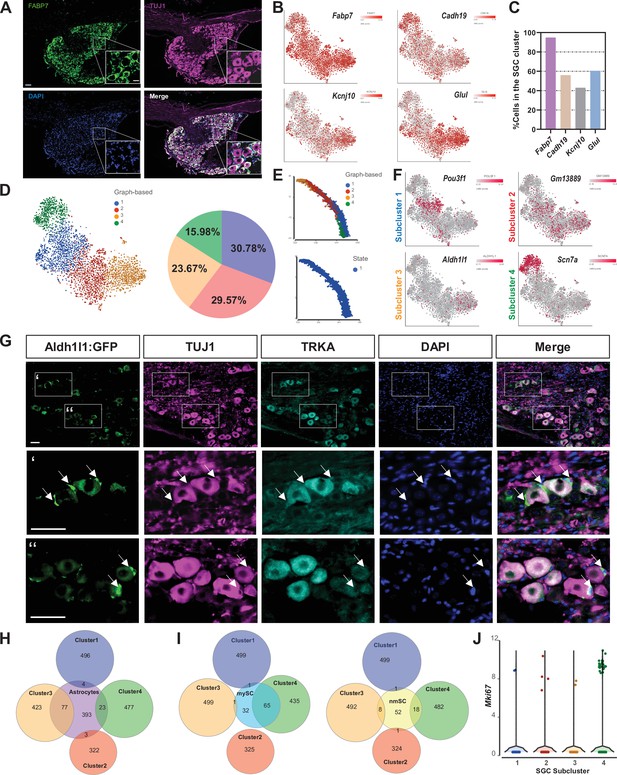
SGC represent a diverse cell population.
(A) Representative images of immunofluorescence staining of DRG sections labeled with FABP7 (green) and TUJ1 (magenta). n = 4 biologically independent animals. Scale bar: 100 µm, zoomed image: 50 µm (B) t-SNE overlay for expression of SGC marker genes in pooled SGC cluster from naïve mice. (C) Fraction of cells in the SGC cluster expressing the SGC marker genes Fabp7, Cadh19, Kcnj10, and Glul. (6< log gene counts). (D) t-SNE plot of SGC cluster colored by subclusters (unbiased, Graph based clustering) with quantification of the fraction of cells in the different SGC subclusters out of total number of naïve SGC. (E) Trajectory analysis of SGC subclusters. (F) t-SNE overlay for expression of top differentially expressed genes in SGC subclusters. (G) Representative images of immunofluorescence staining of DRG sections from Aldh1l1::Rpl10a-Egfp mice (green) labeled with TUJ1 (magenta) and TRKA (cyan). n = 4 biologically independent animals. Scale bar 50 µm (H) Venn diagram comparing signature genes in SGC subclusters and astrocytes. (I) Venn diagrams comparing signature genes in SGC subclusters with myelinating (mySC) and non-myelinating (nmSC) Schwann cells markers. (J) Violin plot for expression of Mki67 across SGC subclusters.
-
Figure 5—source data 1
Source files for scRNAseq analysis; SGC marker genes expression, DEG in SGC subclusters (FDR ≤ 0.05, fold-change ≥2), Astrocytes and Schwann cells.
- https://cdn.elifesciences.org/articles/68457/elife-68457-fig5-data1-v1.xlsx
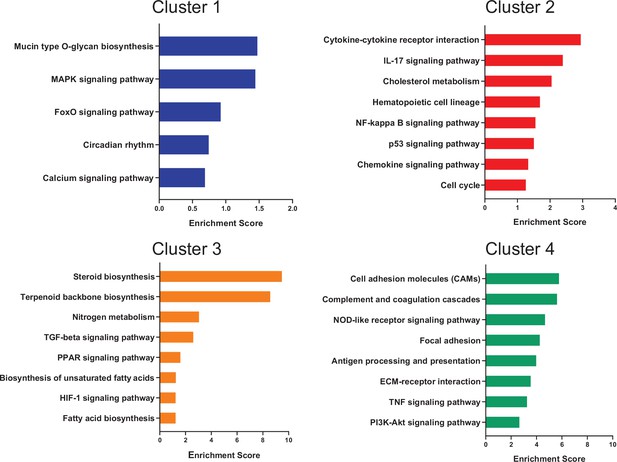
Unique enriched pathways of SGC subclusters.
Enriched signaling pathways (KEGG 2019) for top differentially expressed genes in each SGC subcluster.
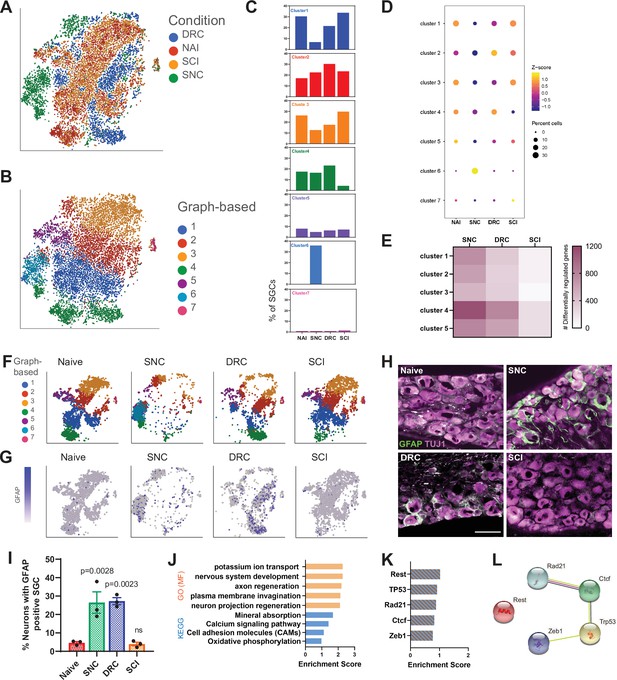
A distinct SGC cluster appears in response to peripheral nerve injury.
(A) t-SNE plot of pooled SGC from naïve and injured mice, colored by injury condition. (B) t-SNE plot of pooled SGC from naïve and injured mice, colored by unbiased clustering. (C) Quantification of the fraction of cells for each subcluster in the different injury conditions. (D) Dot plot of SGC subclusters representation in the different injury conditions by z-score. The percentage of cells in a subcluster is divided by the total number of cells in the respective condition. (E) Heatmap of the number of differentially regulated genes in each SGC subcluster and injury condition (FDR ≤ 0.05, fold-change ≥ 2). (F) t-SNE plots of pooled SGC colored by unbiased clustering, separated by injury condition. (G) t-SNE plots overlay for Gfap expression (blue), separated by injury condition. (H) Representative images of immunofluorescence staining of DRG sections labeled for GFAP (green) and TUJ1 (magenta) from naïve, SNC, DRC, and SCI conditions. n = 3 biologically independent animals. Scale bar: 100 µm (I) Quantification of the percentage of neurons with GFAP (green) positive SGC around them out of all TUJ1-positive neurons (magenta). n = 3 biologically independent animals. One-way analysis of variance (ANOVA) followed by Bonferroni’s multiple comparisons test. Data are presented as mean values ± SEM (J) Enriched signaling pathways (GO Molecular Function and KEGG 2019) for top differentially expressed genes in subcluster 6. (K) Enriched TF (ENCODE and ChEA) in top differentially expressed genes in subcluster 6. (L) Protein-protein interaction of top TF expressed in subcluster 6 (STRING).
-
Figure 6—source data 1
Source files for scRNAseq analysis; SGC subtypes injury marker genes, DEG in SGC subclusters in all injuries (FDR ≤ 0.05, fold-change ≥2), cluster distribution.
GFAP image analysis in DRG sections.
- https://cdn.elifesciences.org/articles/68457/elife-68457-fig6-data1-v1.xlsx
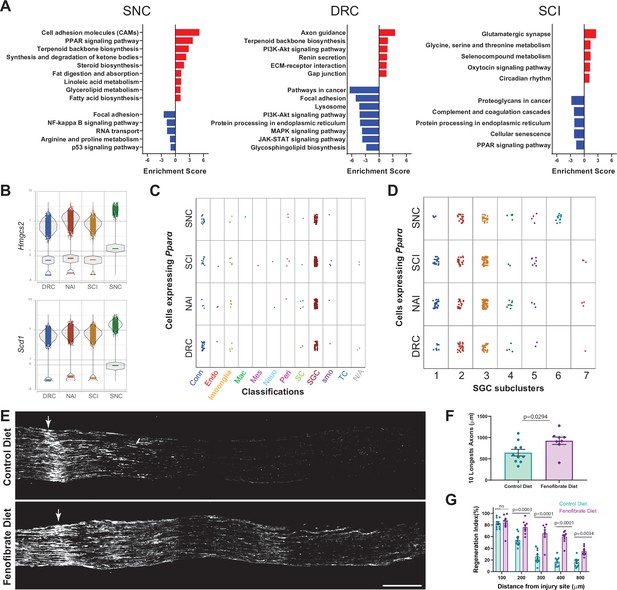
Activation of PPARα with fenofibrate increases axon regeneration after dorsal root crush.
(A) Enriched signaling pathways (KEGG 2019) of differentially upregulated (red) and downregulated (blue) genes in the SGC cluster. n = 2 biologically independent experiments. (FDR ≤ 0.05, fold-change ≥ 2). (B) Violin plots for expression of PPARα target genes Hmgcs2 and Scd1 across all injuries. (C) Plot of Pparα expressing cells across all injury conditions. (D) Plot of Pparα expressing cells in SGC subclusters across all injury conditions. (E) Representative longitudinal sections of dorsal roots 3 days after injury from mice fed with fenofibrate or control diet, stained for SCG10. Arrows indicate the crush site, Scale Bar: 100 µm. (F) Length of the longest 10 axons was measured in 10 sections for each nerve. Unpaired t-test. n = 10 (control diet) and n = 8 (Fenofibrate diet) biologically independent animals. Data are presented as mean values ± SD (G) Regeneration index was measured as SGC10 intensity normalized to the crush site. Two-way ANOVA followed by Bonferroni’s multiple comparisons test. n = 10 (control diet) and n = 8 (Fenofibrate diet) biologically independent animals. Data are presented as mean values ± SEM.
-
Figure 7—source data 1
Source files for scRNAseq analysis; DEG in SGC (FDR ≤ 0.05, fold-change ≥ 2), PPARαexpression in all cell types.
Image analysis for axon regeneration.
- https://cdn.elifesciences.org/articles/68457/elife-68457-fig7-data1-v1.xlsx
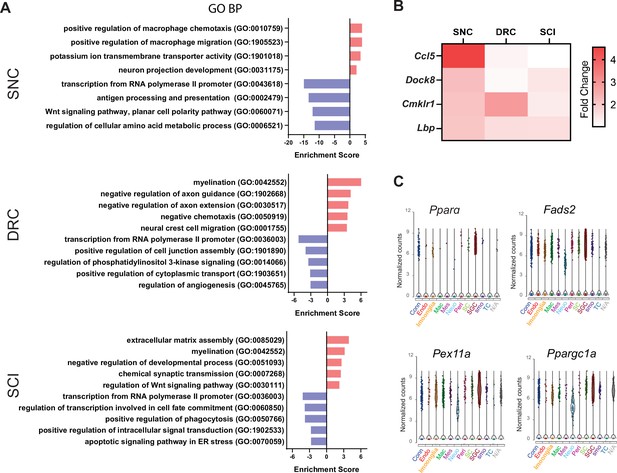
Distinct response of SGC to peripheral vs. central injuries.
(A) Enriched biological processes (Richner et al., 2018) for differentially up and downregulated genes in SGC after SNC, DRC and SCI. (B) Heatmap of genes with positive regulation of macrophage chemotaxis and migration, color coded by fold change expression. (C) Violin plots of Pparαand PPARαtarget genes across cell types.
Videos
3D video of Imoonglia Glia.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (mice, C57Bl/6) | Sun1-sfGFP-myc | Mo et al., 2015 | R26-CAG-LSL-Sun1-sfGFP-myc | from Dr. Harrison Gabel |
| Strain, strain background (M. musculus, C57Bl/6) | Actl6bCre | Zhan et al., 2015 | from Dr. Harrison Gabel | |
| Strain, strain background (M. musculus, C57Bl/6) | Fabp7creER | Maruoka et al., 2011 | from Dr. Toshihiko Hosoya | |
| Strain, strain background (M. musculus, C57Bl/6) | Aldh1l1::Rpl10a-Egfp | Doyle et al., 2008 | B6;FVB-Tg(Aldh1l1-EGFP/Rpl10a) JD130Htz/J | from Dr. Joseph Dougherty. |
| Antibody | Rabbit polyclonal anti Glial Fibrillary Acidic Protein | Agilent | Cat# Z033429-2 | IF (1:500) |
| Antibody | Rabbit polyclonal anti Fatty acid binding protein 7 | Thermo Fisher Scientific | Cat# PA5-24949, RRID:AB_2542449 | IF (1:1000) |
| Antibody | Rabbit polyclonal anti STMN2/ SCG10 | Novus | Cat# NBP1-49461, RRID:AB_10011569 | IF (1:1000) |
| Antibody | Mouse monoclonal anti Tubulin beta-3 chain | BioLegend | Cat# 802001, RRID:AB_291637 | IF (1:1000) |
| Antibody | Rabbit polyclonal anti MKI67 | Abcam | cat# ab15580 | IF (1:500) |
| Antibody | Mouse monoclonal anti CD68 | Bio-Rad | Cat# MCA1957 clone;FA-11 | IF (1:1000) |
| Antibody | Rat monoclonal anti EMR1/F4/80-PE | eBioscience | Cat# 5010786 | FC (1:1000) |
| Antibody | Mouse Recombinant anti CD16/CD32 | Biolegend | Cat# 158,302 Clone QA17A34 | FC (1:50) |
| Antibody | Rat monoclonal anti EMR1/ F4/80-BV605 | Biolegend | Cat# 123,133 Clone BM8 | FC (1:200) |
| Antibody | Rat monoclonal anti CD11b/ITGAM-PerCP-Cy5.5 | Biolegend | Cat# 101,228 Clone M1/70 | FC (1:200) |
| Antibody | Rat monoclonal anti CD45-BV750 | BD Biosciences | Cat# 746,947 Clone 30-F11 | FC (1:200) |
| Sequence-based reagent | Rpl13a Forward | PrimerBank | PCR primer ID 334688867c2 | AGCCTACCAGAAAGTTTGCTTAC |
| Sequence-based reagent | Rpl13aReverse | PrimerBank | PCR primer ID 334688867c2 | GCTTCTTCTTCCGATAGTGCATC |
| Sequence-based reagent | Cd74Forward | PrimerBank | PCR primer ID 13097486a1 | AGTGCGACGAGAACGGTAAC |
| Sequence-based reagent | Cd74Reverse | PrimerBank | PCR primer ID 13097486a1 | CGTTGGGGAACACACACCA |
| Sequence-based reagent | H2-AaForward | PrimerBank | PCR primer ID 31981716a1 | TCAGTCGCAGACGGTGTTTAT |
| Sequence-based reagent | H2-AaReverse | PrimerBank | PCR primer ID 31981716a1 | GGGGGCTGGAATCTCAGGT |
| Sequence-based reagent | CtssForward | PrimerBank | PCR primer ID 10946582a1 | CCATTGGGATCTCTGGAAGAAAA |
| Sequence-based reagent | CtssReverse | PrimerBank | PCR primer ID 10946582a1 | TCATGCCCACTTGGTAGGTAT |
| Sequence-based reagent | Ccl2 Forward | PrimerBank | PCR primer ID 6755430a1 | TTAAAAACCTGGATCGGAACCAA |
| Sequence-based reagent | Ccl2 Reverse | PrimerBank | PCR primer ID 6755430a1 | GCATTAGCTTCAGATTTACGGGT |
| Sequence-based reagent | Il1b Forward | PrimerBank | PCR primer ID 6680415a1 | GCAACTGTTCCTGAACTCAACT |
| Sequence-based reagent | Il1b Reverse | PrimerBank | PCR primer ID 6680415a1 | ATCTTTTGGGGTCCGTCAACT |
| Sequence-based reagent | Tnf Forward | PrimerBank | PCR primer ID 7305585a1 | CCCTCACACTCAGATCATCTTCT |
| Sequence-based reagent | Tnf Reverse | PrimerBank | PCR primer ID 7305585a1 | GCTACGACGTGGGCTACAG |
| Sequence-based reagent | Mki67 Forward | PrimerBank | PCR primer ID 1177528a1 | ATCATTGACCGCTCCTTTAGGT |
| Sequence-based reagent | Mki67 Reverse | PrimerBank | PCR primer ID 1177528a1 | GCTCGCCTTGATGGTTCCT |
| Commercial assay or kit | High Capacity cDNA Reverse Transcription kit | Applied Biosystems | Cat# 4368814 | |
| Commercial assay or kit | Gel Bead and Library Kit | 10 x Genomics | GemCode Single-Cell 3′ | |
| Chemical compound, drug | Lycopersicon esculentum (tomato) lectin | Vector lab | Cat# DL-1178–1 | 100 ul |
| Chemical compound, drug | Trizol | Thermo Fisher | Cat #15596026 | |
| Chemical compound, drug | Tamoxifen | Envigo Teklad | TD.130858 | Chow pellet 500 mg per kg |
| Chemical compound, drug | Fenofibrate | Envigo Teklad | Sigma Cat# F6020 | Chow pellet 0.2% |
| Chemical compound, drug | PowerUp SYBR Green master mix | Thermo Fisher | Cat #a25742 | |
| Software, algorithm | Partek Flow | Partek | Build version 9.0.20.0417 | |
| Software, algorithm | Nikon-NIS Elements | Nikon | Version 4.60 | |
| Software, algorithm | Prism | GraphPad | Prism8 | |
| Software, algorithm | Fiji | ImageJ | ||
| Software, algorithm | FlowJo | Tree Star |