Binding blockade between TLN1 and integrin β1 represses triple-negative breast cancer
Figures
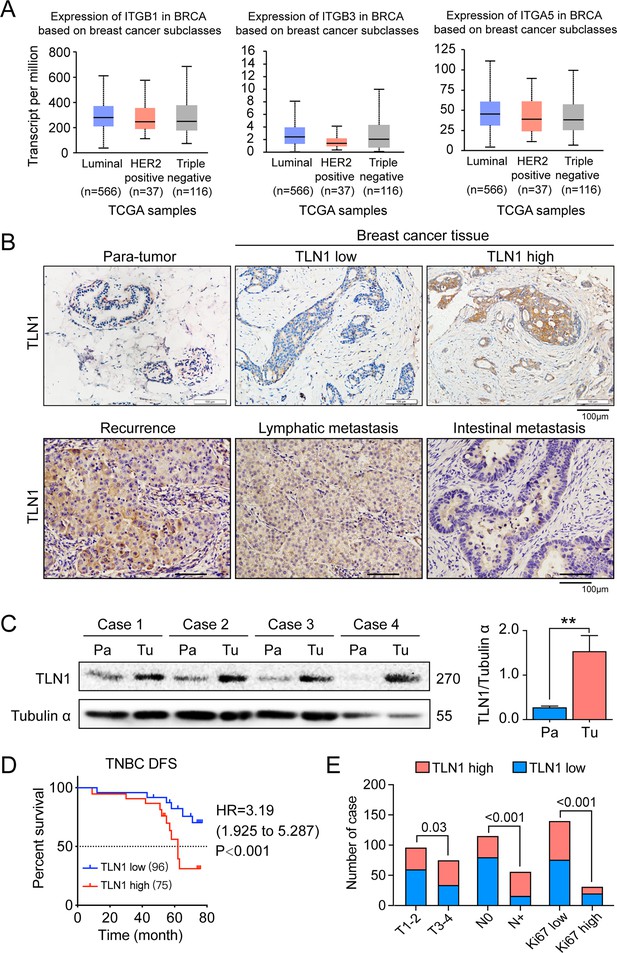
TLN1 upregulation is associated with poor disease-free survival (DFS) in triple-negative breast cancer (TNBC).
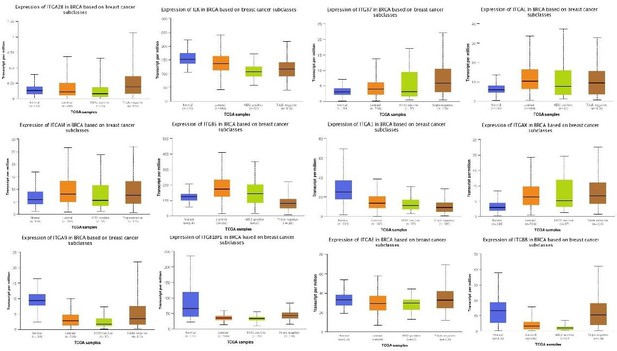
(A) The expression of ITGB1, ITGB3, and ITGA5 in breast cancer subclasses and normal breast tissue using The Cancer Genome Atlas (TCGA) samples (n = 114 of normal samples, n = 556, 37, and 116 of luminal, human epidermal growth factor receptor 2 [HER2] positive and TNBC samples, respectively; p-value is the result of comparison with normal samples, respectively). (B) Representative immunohistochemistry images of TLN1 expression in TNBC tissue, chest wall recurrence, lymphatic metastasis, and intestinal metastasis (scale bar, 100 µm). (C) Western blot analysis of TLN1 expression in fresh TNBC and para-cancerous tissues (n = 4, p = 0.009). (D) The DFS of 171 TNBC patients in the cohort was estimated by the Kaplan-Meier method, and the difference between groups with high and low TLN1 expression was compared within each set of patients listed and analysed by log-rank analysis (HR = 3.19, p < 0.001). (E) Chi-square analysis of high or low TLN1 expression with T-stage, N-stage, and Ki67 index. Data are either presented as representative images or expressed as the mean ± standard error of the mean (SEM) of each group. p < 0.01 was indicated by **.
-
Figure 1—source data 1
TLN1 upregulation is associated with poor DFS in TNBC.
- https://cdn.elifesciences.org/articles/68481/elife-68481-fig1-data1-v3.zip
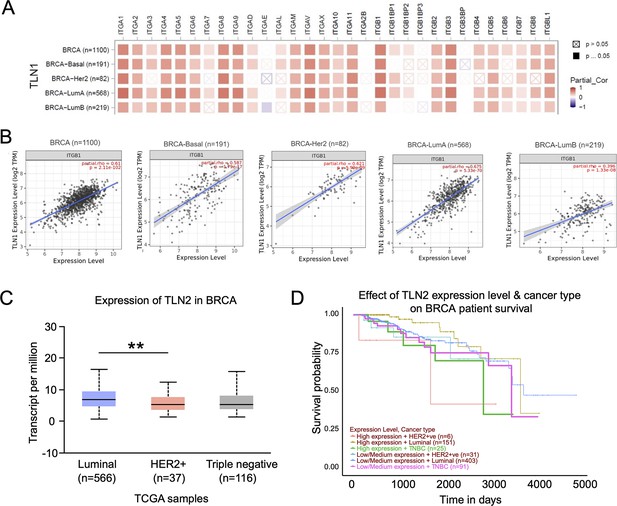
Analysis of TLN2 expression in a dataset from the The Cancer Genome Atlas (TCGA) database.
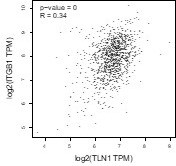
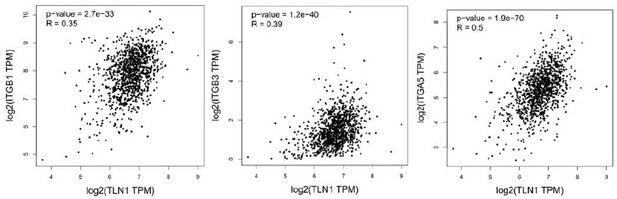
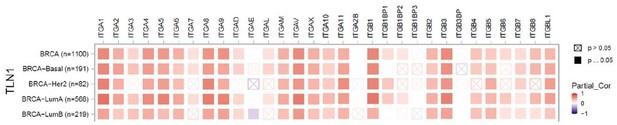
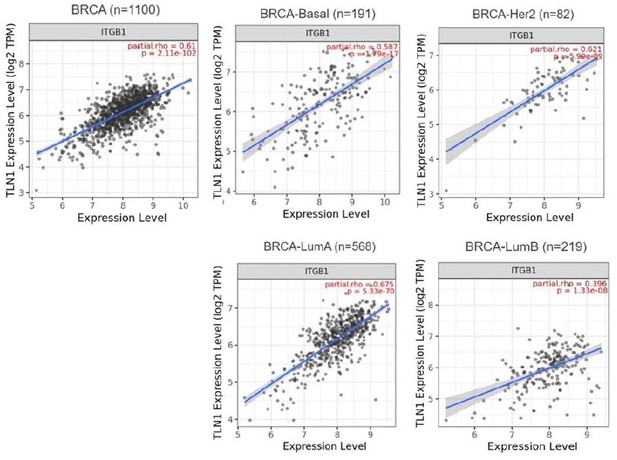
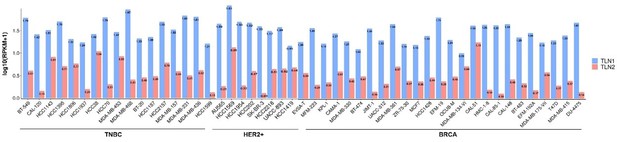
(A–B) Correlation between TLN1 and the integrin family in different types of the TCGA-BRCA dataset obtained from TIMER database. (C) TLN2 mRNA levels were significantly lower in breast cancer tissue than in normal breast tissue (p < 0.01 for all), and except for comparison between luminal and human epidermal growth factor receptor 2 (HER2) positive subtypes (p = 0.006), there was no difference in the levels among luminal and triple-negative breast cancer (TNBC), HER2 positive and TNBC subtypes. (D) Disease-free survival (DFS) analysis of patients stratified according to their TLN2 expression. Comparisons were made among the three types of breast cancer.
-
Figure 1—figure supplement 1—source data 1
Analysis of TLN2 expression in a dataset from the TCGA database.
- https://cdn.elifesciences.org/articles/68481/elife-68481-fig1-figsupp1-data1-v3.zip

Silencing TLN1 promotes apoptosis and inhibits tumour growth of MDA-MB-231 cells.
(A) TLN1 expression in the indicated breast cancer cell lines, using western blotting. (B) The efficiency of TLN1 silencing with shRNA in MDA-MB-231 cells was evaluated by western blotting. (C) Silencing TLN1 inhibited the proliferation of MDA-MB-231 cells (scale bar, 20 µm, p < 0.05 at 48 hr and p < 0.01 at 72 hr). (D) Silencing TLN1 enhanced spontaneous apoptosis in MDA-MB-231 cells, which was detected by Flowcyto. (E) Silencing TLN1 increased apoptosis-related vacuoles (red arrows), which was detected by transmission electron microscopy (TEM). (F) Silencing TLN1 reduced the growth of xenografted triple-negative breast cancer (TNBC) tumours in NOD/SCID mice (n = 8 per group). Data are presented as representative images, flow cytometric plots, or the mean ± standard error of the mean (SEM) of each group from three separate experiments. *p < 0.05, **p < 0.01 vs. the NC group.
-
Figure 2—source data 1
Silencing TLN1 promotes apoptosis and inhibits tumour growth of MDA-MB-231 cells.
- https://cdn.elifesciences.org/articles/68481/elife-68481-fig2-data1-v3.zip
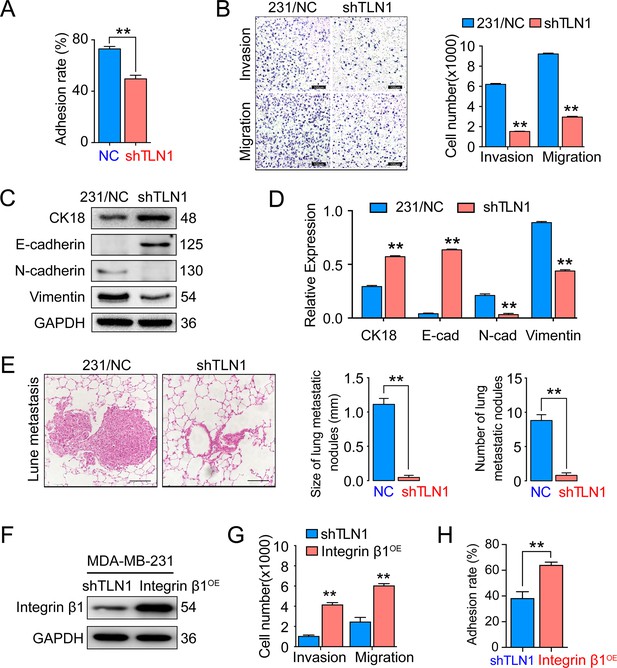
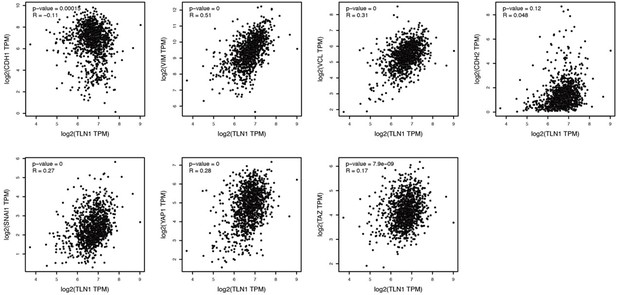
Silencing TLN1 reduces triple-negative breast cancer (TNBC) cell motility by blocking epithelial-mesenchymal transformation (EMT).
(A) TLN1 silencing decreased the adhesion of MDA-MB-231 cells (n = 3, p < 0.01). (B) Transwell assay was used to evaluate the migration and invasion of MDA-MB-231/NC and shTLN1 cells (n = 3, p < 0.01; scale bar, 100 µm). (C and D) The relative expression level of CK18, E-cadherin, N-cadherin, and vimentin relative to those of GAPDH in MDA-MB-231/NC and shTLN1 cells, using western blotting (n = 3, p < 0.01, respectively). (E) Haematoxylin and eosin (H&E) staining of mitigated lung metastasis derived from MDA-MB-231/NC and shTLN1 tumours in NOD/SCID mice (n = 5–8 per group; scale bar, 50 µm). (F) Western blot of integrin β1 overexpression in MDA-MB-231/shTLN1 cells. (G) Transwell analysis of invasion and migration following integrin β1 overexpression. (H) Adhesion assay after integrin β1 overexpression. Data are representative images or expressed as the mean ± standard error of the mean (SEM) of each group from three separate experiments for in vitro and 5–8 per group for in vivo studies. *p < 0.05, **p < 0.01 vs. the control group.
-
Figure 3—source data 1
Silencing TLN1 reduces TNBC cell motility by blocking EMT.
- https://cdn.elifesciences.org/articles/68481/elife-68481-fig3-data1-v3.zip
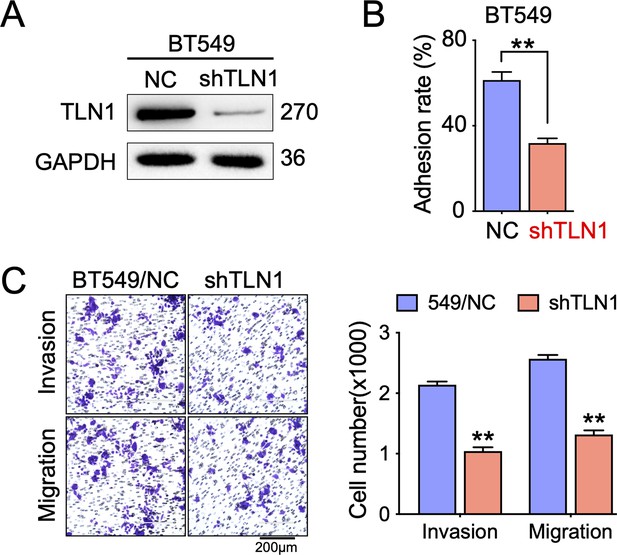
Silencing TLN1 in BT549 cell line reduces adhesion, invasion, and migration.
(A) The efficiency of TLN1 silencing with shRNA in BT549 cells was evaluated by western blotting. (B) TLN1 silencing decreased the adhesion of BT549 cells. (C) Transwell assay was used to evaluate the migration and invasion of BT549/NC and shTLN1 cells (scale bar, 200 µm). **p < 0.01 vs. the NC group.
-
Figure 3—figure supplement 1—source data 1
Silencing TLN1 in BT549 cell line reduces adhesion, invasion and migration.
- https://cdn.elifesciences.org/articles/68481/elife-68481-fig3-figsupp1-data1-v3.zip
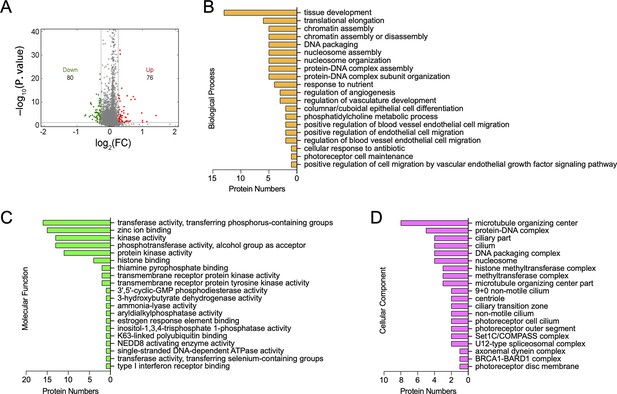
Tandem mass tag analysis of differentially expressed proteins (DEPs) between MDA-MB-231/NC and shTLN1 cells.
(A) Volcano plot of DEPs between MDA-MB-231/NC and shTLN1 cells; 76 were significantly upregulated (red), whereas 80 were significantly downregulated (green). Abscissa is the log2 value of the multiple of difference, and ordinate is the −log10 value of p-value. (B–D) Enriched terms of DEP profiles comparing MDA-MB-231/NC to shTLN1 cells detected by tandem mass tag. Gene Ontology (GO) functional enrichment analysis of DEPs involved in the biological process, cellular component, and molecular mechanism.
-
Figure 3—figure supplement 2—source data 1
Tandem mass tag analysis of differentially expressed proteins (DEPs) between MDA-MB-231/NC and shTLN1 cells.
- https://cdn.elifesciences.org/articles/68481/elife-68481-fig3-figsupp2-data1-v3.zip
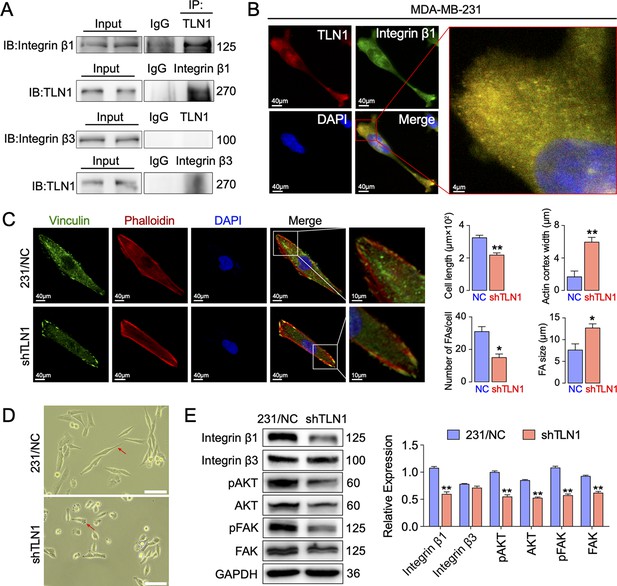
Silencing TLN1 reduces focal adhesion (FA) dynamic formation and integrin β1-mediated signalling in MDA-MB-231 cells via loss of interactions with integrin β1.
(A) Immunoprecipitation analysis of the interaction of TLN1 with integrin β1 or integrin β3 in MDA-MB-231 cells. (B) Immunofluorescence images of TLN1 and integrin β1 in MDA-MB-231 cells (scale bar, 40 µm; 5 µm for enlarged image). (C) Immunofluorescence confocal microscopy analysis of FAs and the actin cortex in MDA-MB-231/NC and shTLN1 cells (scale bar, 40 µm; 10 µm for enlarged image). (D) Light microscopy images of MDA-MB-231/NC and shTLN1 cells (scale bar, 100 µm). (E) Western blot analysis of the relative levels of integrin β1, integrin β3, total AKT, total FAK, phosphorylated AKT (Ser473), and phosphorylated FAK (Tyr397) in MDA-MB-231/NC and shTLN1 cells. GAPDH used as the control to evaluate relative expression. Data are presented as representative images or the mean ± standard error of the mean (SEM) of each group from three separate experiments. **p < 0.01 vs. the NC group.
-
Figure 4—source data 1
Silencing TLN1 reduces FA dynamic formation and integrin β1-mediated signalling in MDA-MB-231 cells via loss of interactions with integrin β1.
- https://cdn.elifesciences.org/articles/68481/elife-68481-fig4-data1-v3.zip
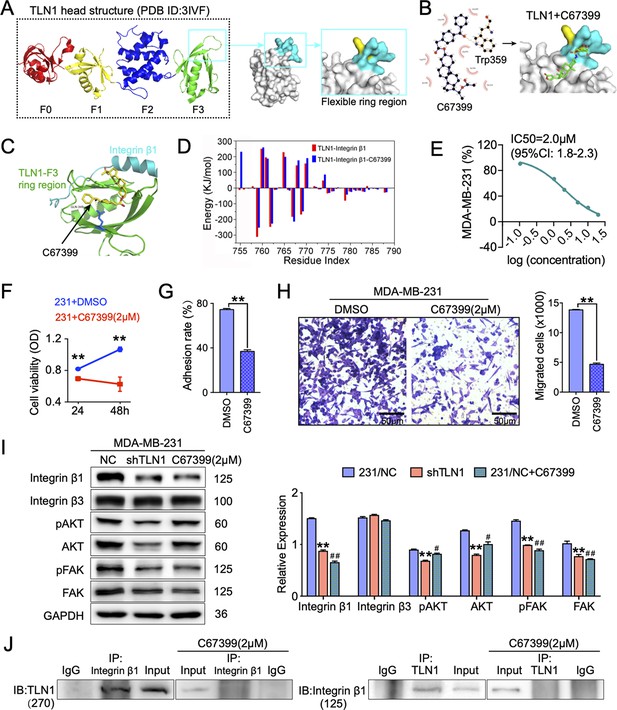
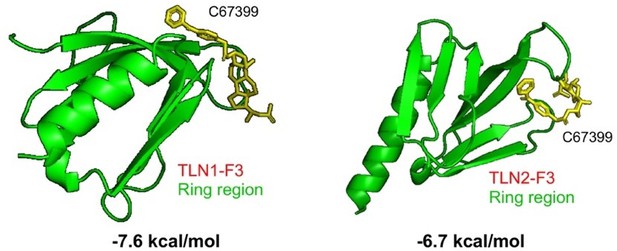
C67399 blocks TLN1-integrin β1 binding and attenuates the malignant behaviours of MDA-MB-231 cells.
(A) Complete structure of the human TLN1 ‘head’ with the F0, F1, F2, and F3 domains, with the predicted interaction between the TLN1 F3 domain (PDB ID: 3IVF) and integrin β1 at far right. The targeting of the flexible ring region of the F3 domain of the TLN1 head structure was labelled in cartoon mode and the head structure was labelled in surface mode using PyMOL software. The yellow- and paon-labelled regions indicate the hydrophobic ring structure of the F3 domain of TLN1, with the yellow-labelled region representing the K324 residue. (B) Docking mode diagram of the chemical molecule C67399 and TLN1; the hydrophobic pocket formed by the flexible ring of C67399 and the F3 domain was screened by FIPSDock. (C) The main favourable contributions to the binding of C67399 came from hydrophobic contacts with Thr354, Ala389, Gln390, Ala393, Ile396, Asp397, and Ile398, using a closer analysis of the complex structure and energy term. (D) Molecular dynamic simulations of the electrostatic interactions between C67399 and Trp359. (E) The dose-response curve of C67399 in MDA-MB-231 cells using CCK-8 kit (IC50 = 2.0 µM). (F–H) C67399 treatment significantly reduced the viability(F), adhesion(G), and migration(H) of MDA-MB-231 cells. (I) Western blot analysis of the relative levels of integrin β1, integrin β3, total AKT, total FAK, phosphorylated AKT (Ser473), and phosphorylated FAK (Tyr397) in MDA-MB-231/NC and shTLN1 cells, as well as in MDA-MB-231/NC treated with C67399 (2 µM for 48 hr). GAPDH used as the control to evaluate relative expression. (J) Immunoprecipitation analysis of TLN1 and integrin β1 in the presence or absence of C67399 treatment (2 µM for 48 hr). Data are shown as representative images, charts, or the mean ± standard error of the mean (SEM) of each group from three separate experiments. #p < 0.05, ##p < 0.01, and **p < 0.01 vs. the NC group.
-
Figure 5—source data 1
C67399 blocks TLN1–integrin β1 binding and attenuates the malignant behaviours of MDA-MB-231 cells.
- https://cdn.elifesciences.org/articles/68481/elife-68481-fig5-data1-v3.zip
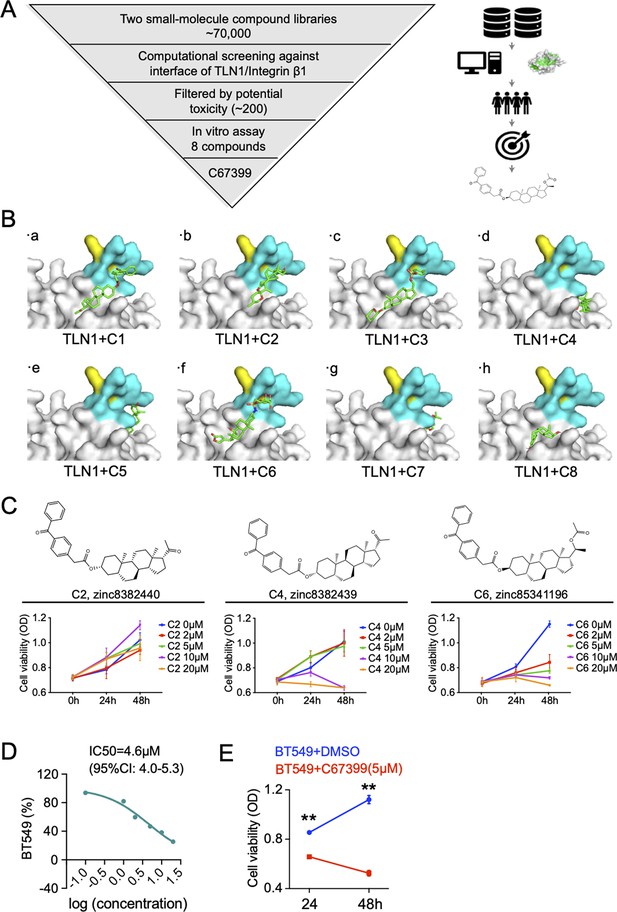
The process of computational screening small-molecule compounds targeting TLN1.
(A) Schematic diagram of computational screening small-molecule compounds targeting TLN1. (B) The top eight small molecular compounds in affinity score when targeting TLN1. (C) Drug sensitivity test of three drugs (C2, C4, and C6 [C67399]) with the best solubility of eight small molecular compounds to MDA-MB-231 cells at 0, 2, 5, 10, 20 µM using CCK-8 kit. (D) The dose-response curve of C67399 in BT549 cells using CCK-8 kit (IC50 = 4.6 µM). (E) C67399 treatment significantly reduced the viability of BT549 cells, which was detected by CCK-8 kit.
-
Figure 5—figure supplement 1—source data 1
The process of computational screening small molecule compounds targeting TLN1.
- https://cdn.elifesciences.org/articles/68481/elife-68481-fig5-figsupp1-data1-v3.zip
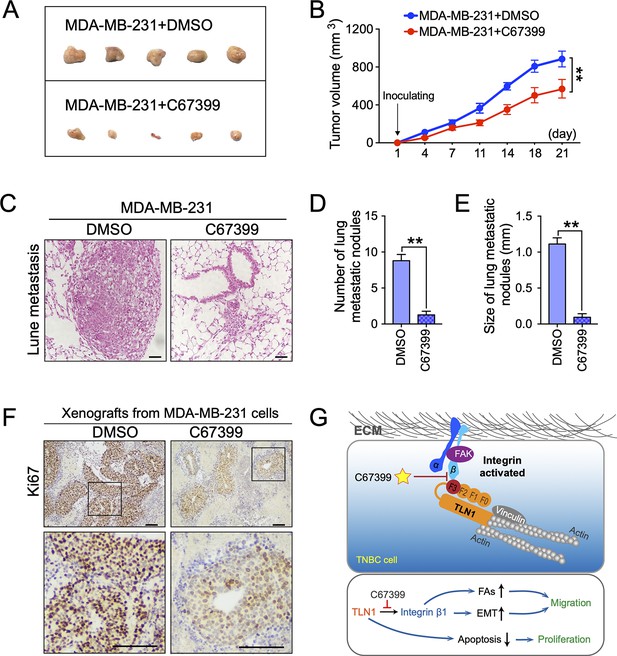
C67399 inhibits the growth and metastasis of implanted triple-negative breast cancer (TNBC) cells.
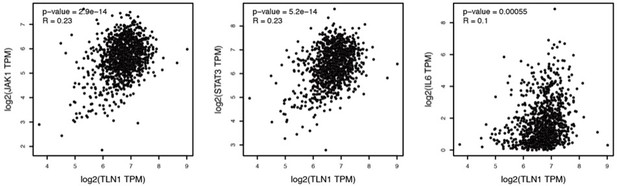
MDA-MB-231 cells were injected into fat pad or tail vein of NOD/SCID mice to establish tumour model. Mice were intravenously injected with 1.75 mg/kg C67399, twice a week for 3 weeks. (A and B) The tumour volume of xenografts derived from MDA-MB-231 cells treated with or without C67399 (n = 5–8 per group, p < 0.01). (C–E) The number and size of lung metastatic tumour nodules in mice of MDA-MB-231 cells with or without C67399 treatment (n = 5–8 per group, p < 0.01). (F) Immunohistochemical staining for Ki67 of xenografted tumours derived from MDA-MB-231 cells in the presence or absence of C67399 treatment. (G) Diagram illustrating the function of TLN1 in TNBC. TLN1 could bind and activate integrin β1 in TNBC cells. On the one hand, it can regulate the dynamic formation and maturation of focal adhesions (FAs), induce epithelial-mesenchymal transformation (EMT), and promote tumour metastasis; on the other hand, it can promote tumour proliferation by inhibiting apoptosis. A small-molecule C67399 was developed to inhibit the binding of TLN1 to integrin β1, as well as a series of integrin β1-related pathways, and ultimately inhibit the malignancy of TNBC.
-
Figure 6—source data 1
C67399 inhibits the growth and metastasis of implanted TNBC cells.
- https://cdn.elifesciences.org/articles/68481/elife-68481-fig6-data1-v3.zip
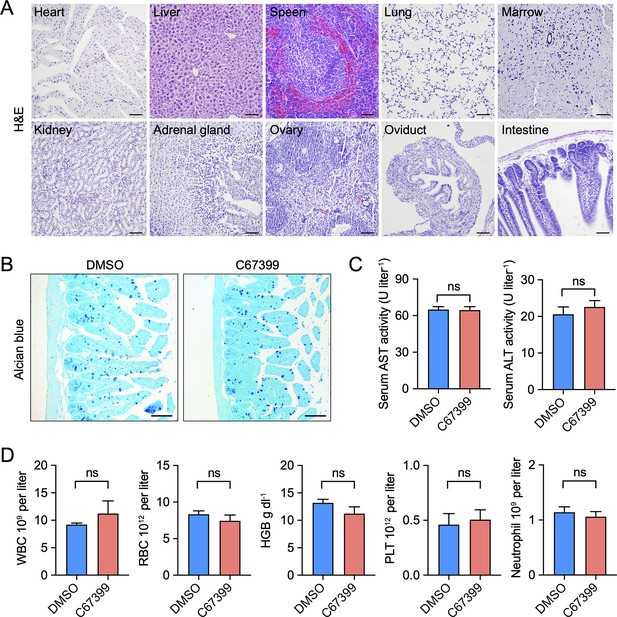
Toxicity of C67399 in mice.
(A) Haematoxylin and eosin (H&E) staining was used to observe structural changes to evaluate drug toxicity. Tissue sections of the heart, liver, spleen, lungs, kidneys, and other organs from mice treated with 1.75 mg/kg C67399 twice per week for 3 weeks, and no toxic reactions were observed (n = 5). (B–D) Alcian blue staining of intestine, alanine aminotransferase (ALT), and aspartate aminotransferase (AST) activities in mice serum, and blood cell counts difference between DMSO group and C67399 group (n = 5).
-
Figure 6—figure supplement 1—source data 1
Toxicity of C67399 in mice.
- https://cdn.elifesciences.org/articles/68481/elife-68481-fig6-figsupp1-data1-v3.zip
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo sapiens) | MDA-MB-231 | ATCC | ATCC Cat# HTB-26, RRID:CVCL_0062 | |
| Cell line (Homo sapiens) | MCF-7 | ATCC | ATCC Cat# CRL-3435, RRID:CVCL_A4AQ | |
| Cell line (Homo sapiens) | BT-549 | ATCC | ATCC Cat# HTB-122, RRID:CVCL_1092 | |
| Cell line (Homo sapiens) | SK-BR-3 | ATCC | ATCC Cat# HTB-30, RRID:CVCL_0033 | |
| Transfected construct (human) | TLN1 shRNA #1 | Sangon Biotech | Transfected construct (human) | |
| Antibody | Anti-Vinculin (Rabbit polyclonal) | Proteintech, Rosemont, IL | Cat# 26520–1-AP, RRID:AB_2868558 | WB (1:1000), IF(1:200) |
| Antibody | Anti-E-cadherin (Rabbit monoclonal) | Cell Signaling Technology, Danvers, MA | Cat# 3195, RRID:AB_2291471 | WB (1:1000) |
| Antibody | Anti-N-cadherin (Rabbit polyclonal) | Cell Signaling Technology, Danvers, MA | Cat# 4061, RRID:AB_10694647 | WB (1:1000) |
| Antibody | Anti-Tubulin α (Rabbit monoclonal) | Cell Signaling Technology, Danvers, MA | Cat# 2125, RRID:AB_2619646 | WB (1:5000) |
| Antibody | Anti-CK18 (mouse monoclonal) | Cell Signaling Technology, Danvers, MA | Cat# 4546, RRID:AB_2134843 | WB (1:1000) |
| Antibody | Anti-FAK (Rabbit polyclonal) | Cell Signaling Technology, Danvers, MA | Cat# 3285, RRID:AB_2269034 | WB (1:1000) |
| Antibody | Anti-GAPDH (Rabbit polyclonal) | Proteintech, Rosemont, IL | Cat# 10494–1-AP, RRID:AB_2263076 | WB (1:5000) |
| Antibody | Anti-AKT1 (Rabbit polyclonal) | Proteintech, Rosemont, IL | Cat# 10176–2-AP, RRID:AB_2224574 | WB (1:1000) |
| Antibody | Anti-integrin β3 (Rabbit polyclonal) | Proteintech, Rosemont, IL | Cat# 18309–1-AP, RRID:AB_2128759 | WB (1:1000) |
| Antibody | Anti-p-AKT1 (sc-81433) (mouse monoclonal) | Santa Cruz Biotechnology, Dallas, TX | Cat# sc-81433, RRID:AB_1125472 | WB (1:1000) |
| Antibody | Anti-p-FAK (Tyr397) (Rabbit polyclonal) | Immunoway, Plano, TX | Cat# YP0739, RRID:AB_2904589 | WB (1:1000) |
| Antibody | Anti-integrin β1 (Rabbit polyclonal) | Sino Biological, China | Cat# 100562-T46, RRID:AB_2895614 | WB (1:1000), IF (1:200) |
| Antibody | Anti-TLN1 (Rabbit monoclonal) | Cell Signaling Technology, Danvers, MA | Cat# 4021, RRID:AB_2204018 | WB (1:1000), IF (1:200), IHC(1:500) |
| Antibody | Goat Anti-Rabbit IgG H&L Alexa Fluor 488 (Goat polyclonal) | Abcam, Waltham, MA | Cat# ab150077, RRID:AB_2630356 | IF (1:400) |
| Antibody | Goat Anti-Mouse IgG H&L Alexa Fluor 647 (Goat polyclonal) | Abcam, Waltham, MA | Cat# ab150115, RRID:AB_2687948 | IF (1:400) |
| Commercial assay or kit | CCK-8 kit | Dojindo, Japan | CK04 | |
| Commercial assay or kit | Cell adhesion detection kit | Best Bio, Nanjing, China | BB-48120 | |
| Commercial assay or kit | Annexin-V and propidium iodide (PI) kit | BD, San Diego, CA | 559763 | |
| Commercial assay or kit | EdU kit | Ribobio, China | C10327 | |
| Chemical compound, drug | C67399 | Chemdiv Compond, San Diego, CA | 4903–2135 | C38H48O5, 584.79 Da |
| Software, algorithm | SPSS | SPSS | RRID:SCR_002865 | |
| Software, algorithm | GraphPad Prism | GraphPad Prism | RRID:SCR_002798 | |
| Software, algorithm | ImageJ | ImageJ | RRID:SCR_003070 | |
| Software, algorithm | PyMOL | PyMOL | RRID:SCR_000305 | |
| Software, algorithm | FIPSDock | FIPSDock | PMID:22961860 | |
| Other | DAPI stain | UE, China | D4054 | (5 µg/ml) |
| Other | Rhodamine-labelled phalloidin stain (TRITC Phalloidin) | Solarbio, China | Cat# CA1610, RRID:AB_2904593 | (100 nM) |
| Other | Protein A/G agarose beads | Santa Cruz Biotechnology, Dallas, TX | Cat# sc-2003, RRID:AB_10201400 | (50 µl/sample) |