Protective function and durability of mouse lymph node-resident memory CD8+ T cells
Figures
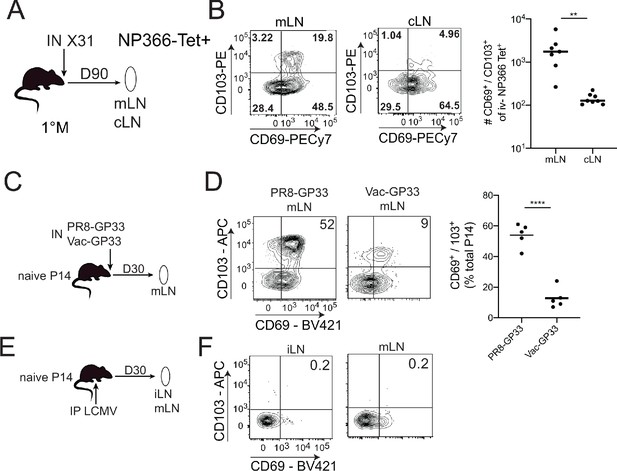
CD103+ memory CD8+ T cells are generated in draining lymph nodes (LNs) during localized but not systemic infections.
(A) C57BL/6 mice were infected IN with X31 (H3N2); mice were sacrificed 90 days post-infection; non-draining cervical lymph nodes (cLN) or lung-draining mediastinal lymph nodes (mLNs) were harvested and analyzed by flow cytometry. (B) Representative plots of % of CD69 and CD103 expression (left) in NP366 tetramer+ IV- memory CD8+ T cells from the cLN or mLN and cumulative data (right). n = 3–5 mice/group. Representative of three independent experiments. Bars denote mean values, dots represent independent mice. ****p<0.0001, Students t-test. (C) Mice were seeded with 104 naive P14 cells and infected IN with either PR8-GP33 (H1N1) or Vac-GP33. 30 days post-infection, draining mLNs were isolated and CD69+ CD103+ P14 Trm populations were evaluated (D). Representative plots (left), cumulative data (right). Representative of two independent experiments, n = 5 mice/group. Error bars represent mean ± SD. ****p<0.0001, Students t-test. (E) Mice were seeded with 104 naive P14 cells and infected IP with LCMV Armstrong. 30 days post-infection LNs (mLN and iLN) were isolated and evaluated for the frequency (F) of CD69+/CD103+ P14 s.
-
Figure 1—source data 1
Source data for Figure 1B.
- https://cdn.elifesciences.org/articles/68662/elife-68662-fig1-data1-v1.xlsx
-
Figure 1—source data 2
Source data for Figure 1D.
- https://cdn.elifesciences.org/articles/68662/elife-68662-fig1-data2-v1.xlsx
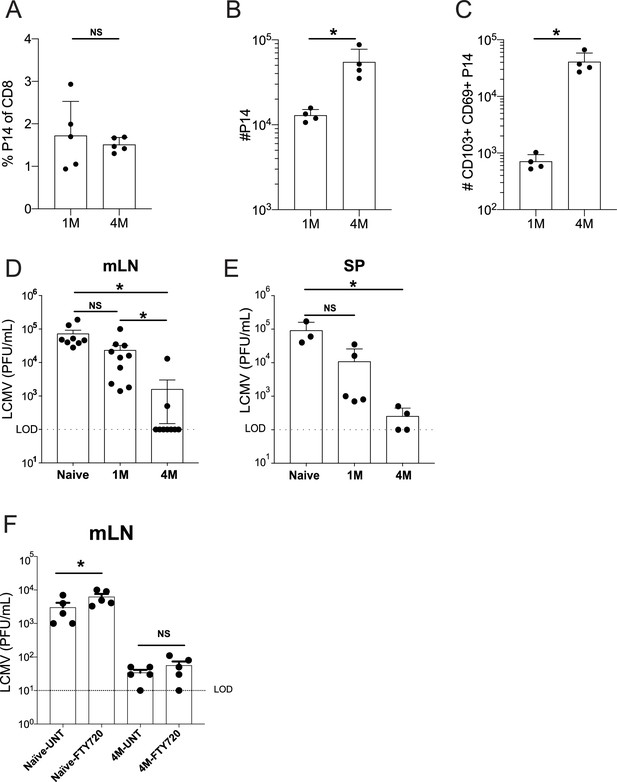
Lung-draining LN Trm cells mediate local protective immunity.
Mice were seeded with 104 naive or 105 3M P14 cells and IN infected with PR8-GP33 virus. At 50 days post-infection, frequency of 1M or 4M P14s were measured in the peripheral blood (A). At 100 days post-infection, mLNs were harvested and the numbers of total (B) and CD103+ CD69+ (C) P14s were determined. Representative of three independent experiments, n = 4–5 mice/group. Error bars represent mean ± SD, *p<0.05, **p<0.01, Students t-test in (A–C). LCMV challenge (D,E). Naïve, 1M, or 4M mice were infected with LCMV-Armstrong (2.0 × 105 PFU/mouse i.p.); 72 hr post LCMV challenge, mLN (D) and SP (E) were harvested and individually evaluated for LCMV titers by plaque assay. FTY720 treatment impact on LCMV challenge (F). Naïve, 1M, or 4M mice were infected with LCMV-Armstrong (2.0 × 105 PFU/mouse i.p.) and treated with vehicle or FTY720 daily for 72 hr. 72 hr post LCMV challenge, mLN (F) were harvested and individually evaluated for LCMV titers by plaque assay. Dotted line denotes limit of detection. One representative of 2–3 independent experiments is shown, n = 3–5 mice/group. Error bars represent mean ± SEM (G) or mean ± SD (H). NS = not significant, *p<0.05, one-way ANOVA.
-
Figure 2—source data 1
Source data for Figure 2A.
- https://cdn.elifesciences.org/articles/68662/elife-68662-fig2-data1-v1.xlsx
-
Figure 2—source data 2
Source data for Figure 2B.
- https://cdn.elifesciences.org/articles/68662/elife-68662-fig2-data2-v1.xlsx
-
Figure 2—source data 3
Source data for Figure 2C.
- https://cdn.elifesciences.org/articles/68662/elife-68662-fig2-data3-v1.xlsx
-
Figure 2—source data 4
Source data for Figure 2D.
- https://cdn.elifesciences.org/articles/68662/elife-68662-fig2-data4-v1.xlsx
-
Figure 2—source data 5
Source data for Figure 2E.
- https://cdn.elifesciences.org/articles/68662/elife-68662-fig2-data5-v1.xlsx
-
Figure 2—source data 6
Source data for Figure 2F.
- https://cdn.elifesciences.org/articles/68662/elife-68662-fig2-data6-v1.xlsx
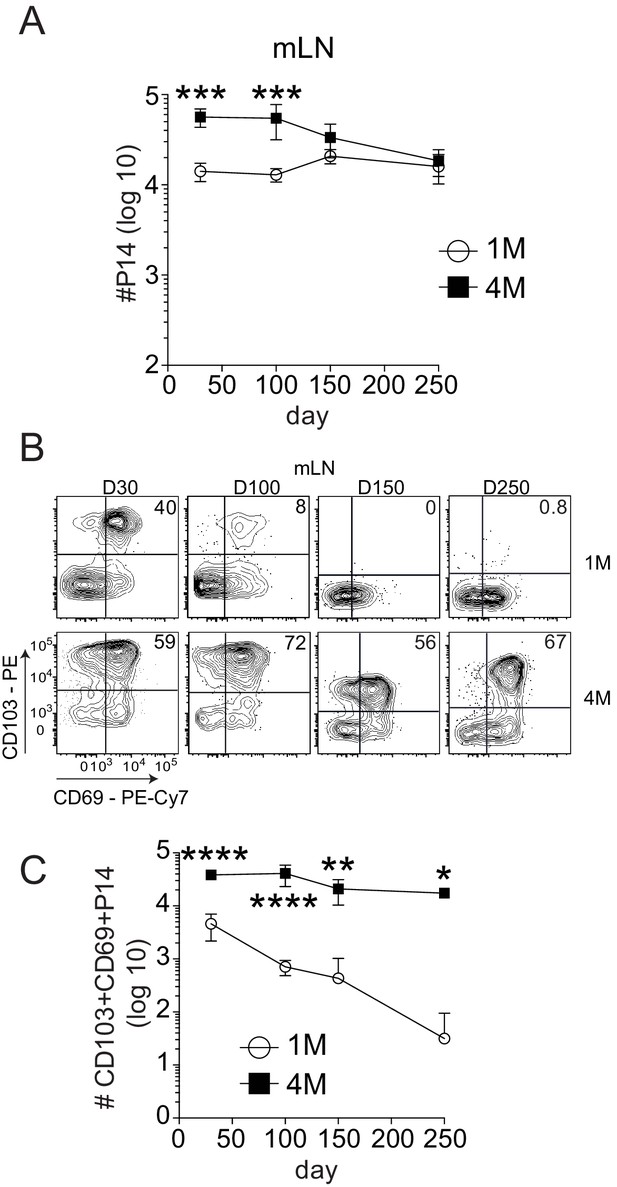
Repeated antigen stimulation extends the survival of LN Trms.
Mice were seeded with 104 naive or 105 3M P14 cells and IN infected with PR8-GP33 virus. At indicated time points, mLN (A) was harvested and total numbers of 1M (white) and 4M (black) P14 cells were evaluated. Representative of three independent experiments, n = 4 mice/group/time point. Error bars represent mean ± SD. ***p<0.001, ****p<0.0001, two-way ANOVA with Sidak’s multiple comparison test. Representative plots (B) and cumulative results (C) of 1M and 4M CD69+ CD103+ P14 Trm cells in mLN evaluated at indicated time points. Representative of three independent experiments, n = 4 mice/group/time point. Error bars represent mean ± SD. *p<0.05, **p<0.01, ****p<0.0001, two-way ANOVA with Sidak’s multiple comparison test.
-
Figure 3—source data 1
Source data for Figure 3A.
- https://cdn.elifesciences.org/articles/68662/elife-68662-fig3-data1-v1.xlsx
-
Figure 3—source data 2
Source data for Figure 3C.
- https://cdn.elifesciences.org/articles/68662/elife-68662-fig3-data2-v1.xlsx
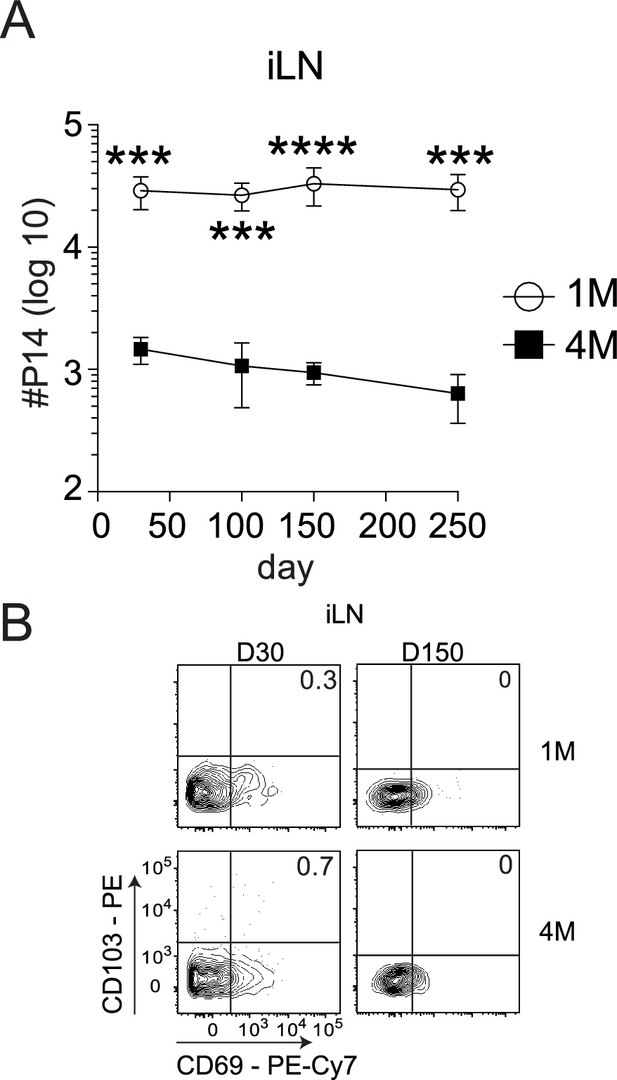
CD103+ LN Trm cells are not present within non-draining iLN.
Mice were seeded with 104naive P14 or 105 3M P14 cells and IN infected with PR8-GP33 virus. At indicated time points, iLN (A) were harvested and total numbers of 1M (white) and 4M (black) P14 cells were evaluated. Representative of three independent experiments, n = 4 mice/group/time point. Error bars represent mean ± SD. ***p<0.001, ****p<0.0001, two-way ANOVA with Sidak’s multiple comparison test. Representative plots of 1M and 4M CD69+ CD103+ P14 Trm cells in iLN (B) evaluated 30 and 150 days post-infection. Representative of three independent experiments, n = 4 mice/group/time point.
-
Figure 3—figure supplement 1—source data 1
Source data for Figure 3—figure supplement 1A.
- https://cdn.elifesciences.org/articles/68662/elife-68662-fig3-figsupp1-data1-v1.xlsx
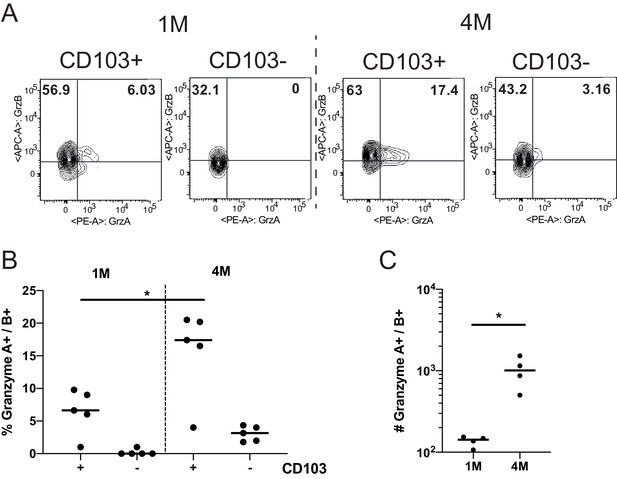
Repeated antigen stimulation increases granzyme production of LN Trms.
Mice were seeded with 104 naive or 105 3M P14 cells and IN infected with PR8-GP33 virus. At >60 days post-infection, mLNs were harvested, stimulated with cognate (GP33) peptide in the presence of BFA for 5 hr, and ICS was performed to evaluate the frequency of GrzA+ and GrzB+ fractions of IV− CD103− or CD103+ 1M or 4M by flow cytometry. Representative flow plots (A), cumulative frequencies (B), and total numbers per mLN (C) are shown. Representative of two independent experiments, n = 4–5 mice/group. Error bars represent mean ± SD. *p<0.05, one-way ANOVA in (B), *p<0.05, Students t-test in (C).
-
Figure 4—source data 1
Source data for Figure 4B.
- https://cdn.elifesciences.org/articles/68662/elife-68662-fig4-data1-v1.xlsx
-
Figure 4—source data 2
Source data for Figure 4C.
- https://cdn.elifesciences.org/articles/68662/elife-68662-fig4-data2-v1.xlsx
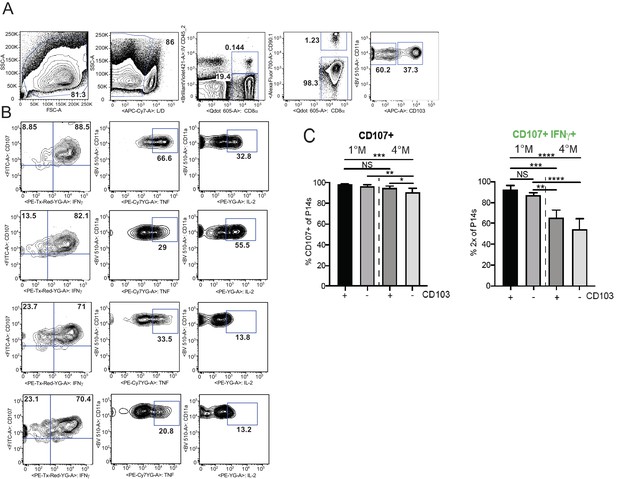
Repeated influenza stimulation reduces cytokine production, but does not affect degranulation capacity of LN Trm cells.
Mice were seeded with 104naive P14 or 105 3M P14 cells and IN infected with PR8-GP33 virus. At 60 days post-infection, mLN and spleen were harvested and cells analyzed for peptide-stimulated cytokine production and degranulation. (A) Gating strategy for analysis of IV− P14s. Single-cell suspensions were stimulated with cognate peptide (GP33) in the presence of BFA for 5 hr and ICS was performed to evaluate the frequency of CD107+, IFNγ+, TNF+, and IL-2+ fractions of IV− CD103− or CD103+ 1M or 4M by flow cytometry. Representative flow plots of IFNg, TNF, and IL-2 (B), cumulative bar graphs denoting frequencies of CD107+ (C, left) and CD107+ IFNg+ (C, right) are displayed. Representative of two independent experiments, n = 4–5 mice/group. NS = not significant, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001; two-way ANOVA with Sidak’s multiple comparison test.
-
Figure 4—figure supplement 1—source data 1
Source data for Figure 4—figure supplement 1C.
- https://cdn.elifesciences.org/articles/68662/elife-68662-fig4-figsupp1-data1-v1.xlsx
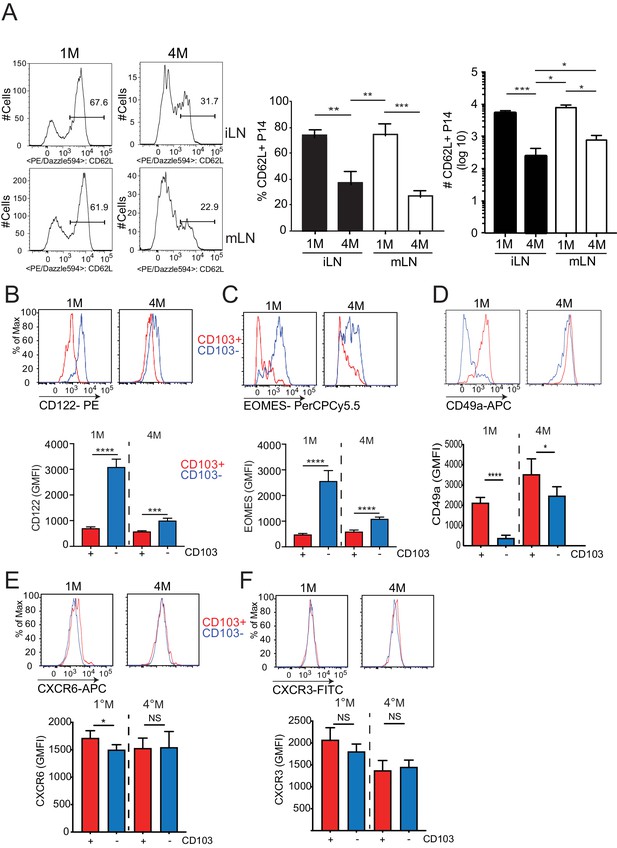
Repeated antigen stimulation alters the phenotype of LN Trm cells.
Mice were seeded with 104naive P14 or 105 3M P14 cells and IN infected with PR8-GP33 virus. At >90 days post-infection, iLN (top) and mLN (bottom) were harvested and the frequency of 1M and 4M P14 cells evaluated. (A) Representative data examining CD62L expression; cumulative frequency data (middle); cumulative total number data (right). Representative of three independent experiments, n = 4 mice/group. Error bars represent mean ± SD. ***p<0.001, ****p<0.0001 one-way ANOVA. At >75 days post-infection, mLN were harvested from 1M and 4M mice and the relative expressions of CD122 (B), Eomes (C), CD49a (D), CXCR6 (E), and CXCR3 (F) by CD103+ (red) or CD103– (blue) P14s were evaluated. Representative histograms (top) and cumulative data (bottom) are displayed. Representative of three independent experiments, n = 4–5 mice/group. Error bars represent mean ± SD. NS = not significant, *p<0.05, ****p<0.0001, one-way ANOVA.
-
Figure 4—figure supplement 2—source data 1
Source data for Figure 4—figure supplement 2A.
- https://cdn.elifesciences.org/articles/68662/elife-68662-fig4-figsupp2-data1-v1.xlsx
-
Figure 4—figure supplement 2—source data 2
Source data for Figure 4—figure supplement 2B.
- https://cdn.elifesciences.org/articles/68662/elife-68662-fig4-figsupp2-data2-v1.xlsx
-
Figure 4—figure supplement 2—source data 3
Source data for Figure 4—figure supplement 2C.
- https://cdn.elifesciences.org/articles/68662/elife-68662-fig4-figsupp2-data3-v1.xlsx
-
Figure 4—figure supplement 2—source data 4
Source data for Figure 4—figure supplement 2D.
- https://cdn.elifesciences.org/articles/68662/elife-68662-fig4-figsupp2-data4-v1.xlsx
-
Figure 4—figure supplement 2—source data 5
Source data for Figure 4—figure supplement 2E.
- https://cdn.elifesciences.org/articles/68662/elife-68662-fig4-figsupp2-data5-v1.xlsx
-
Figure 4—figure supplement 2—source data 6
Source data for Figure 4—figure supplement 2F.
- https://cdn.elifesciences.org/articles/68662/elife-68662-fig4-figsupp2-data6-v1.xlsx
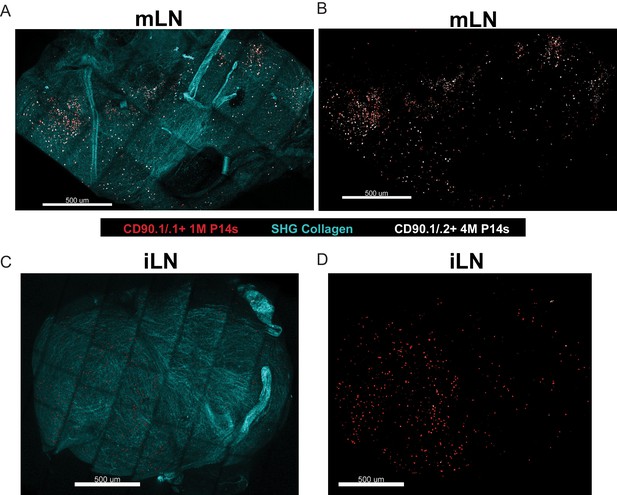
Repeated antigen stimulation alters localization of LN Trm cells.
(A–D) Mice containing mixed congenically distinct populations of 1M (CD90.1/.1, seeded with 104 naive P14) and 4M (CD90.1/.2 seeded with 105 3M P14) P14s were injected with bolus IV administration of CD90.1-PE (red) and CD90.2-APC (white). Approximately 5 hr post-injection, organs were isolated and two-photon microscopy was performed on whole mLN (A–B) or iLN (C–D) explants ex vivo. The LN surrounding collagen capsule (pseudocolored blue) was captured with secondary harmonic generation (SHG). Representative of two independent experiments, n = 3–4 mice/group.
-
Figure 5—source data 1
Source data for Figure 5A–D.
- https://cdn.elifesciences.org/articles/68662/elife-68662-fig5-data1-v1.pptx
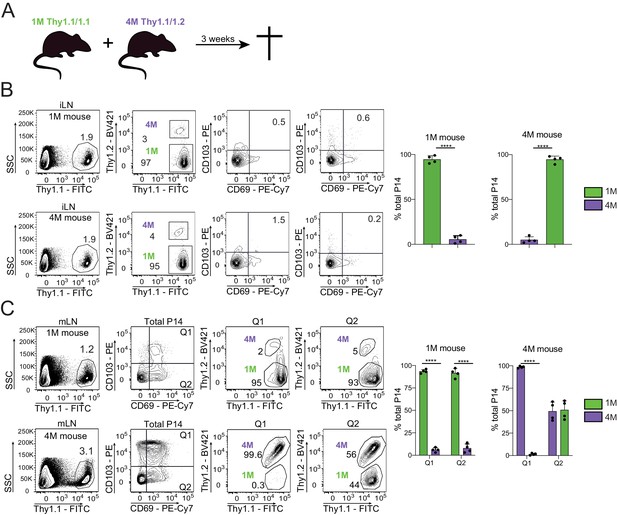
Residential nature of 1M and 4M LN Trm cells primed by influenza infection.
(A) 90 days after IN PR8-GP33 infection, mice bearing Thy1.1/1.1 1M P14 (green; 1M mice seeded with 104 naive P14) cells were joined by parabiotic surgery with mice bearing Thy1.1/1.2 4M P14 (purple, 4M mice seeded with 105 3M P14). Three weeks later parabionts were analyzed. (B) Abundance of 1M (green) and 4M (purple) P14 cells in iLNs of 1M (top row) and 4M (bottom row) parabiotic mice. Representative plots (left), cumulative data (right). Representative of two independent experiments, n = 4 parabionts/experiment. Error bars represent mean ± SD. ****p<0.0001, t-test. (C) Abundance and distribution of 1M (green) and 4M (purple) Trm P14 cells expressed as a % of the total Trm population (CD69+/CD103+) in mLN of 1M (top row) and 4M (bottom row) parabiotic mice. Representative plots (left), cumulative data (right). Representative of two independent experiments, n = 4 parabionts/experiment. Error bars represent mean ± SD. Two-way ANOVA with Sidak’s multiple comparison test. Q1(1M) vs Q1(4M) ****p<0.0001; Q2(1M) vs Q2(4M) ****p<0.0001; Q1(1M) vs Q1(4M) ****p<0.0001.
-
Figure 6—source data 1
Source data for Figure 6B.
- https://cdn.elifesciences.org/articles/68662/elife-68662-fig6-data1-v1.xlsx
-
Figure 6—source data 2
Source data for Figure 6C.
- https://cdn.elifesciences.org/articles/68662/elife-68662-fig6-data2-v1.xlsx
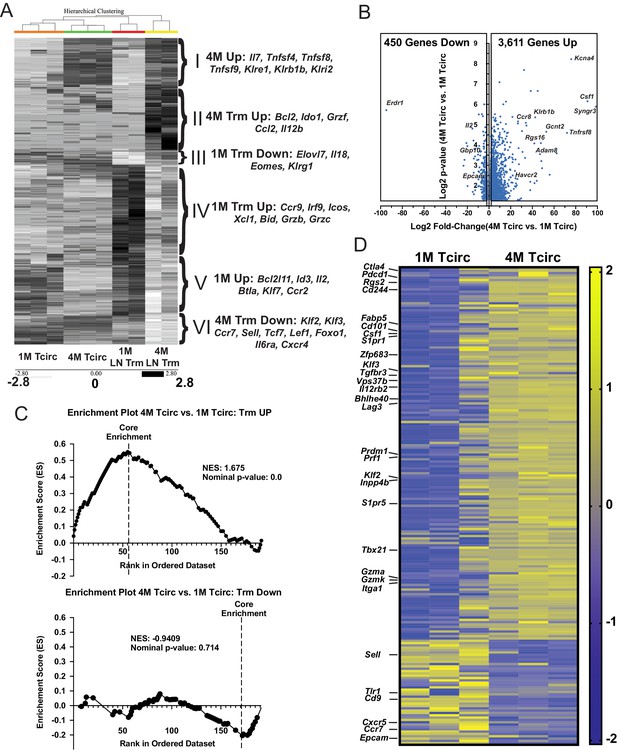
Splenic 4M cells express a core Trm signature.
Mice were seeded with 104 naive P14 or 105 3M P14 cells and IN infected with PR8-GP33 virus. At 22–30 days post-infection, IV exclusion was performed and negatively enriched pooled groups of spleens (3–5 spleens/sample, n = 3) or mLNs (15–25 mLNs/sample, n = 2) were stained for CD8α, CD90.1, CD69, and CD103. Bulk RNAseq was performed on RNA from sort-purified spleen samples (20k IV−, CD69−/CD103− cells/sample) or mLN Trm samples (2–5k IV− CD69+/CD103+ cells/sample). (A) Heatmaps of 1300 most differentially expressed genes (log2FC > 1.5, p<0.05) between 1M and 4M LN Trms are plotted from the four respective groups of samples. The six core signature sets of genes offset to the right were derived from unbiased hierarchical clustering. (B) Volcano plot of 4061 differentially expressed genes between 1M and 4M splenic memory P14 cells (log2FC > 1.5, p<0.05). (C) GSEA of core Trm genes defined in Table 2 from splenic 1M and 4M populations separated into respective upregulated (top) and downregulated (bottom) gene sets in regard to annotated expression in Trms. (D) Heatmap of a core set of selected Trm genes (as in C) within 1M and 4M splenic populations.
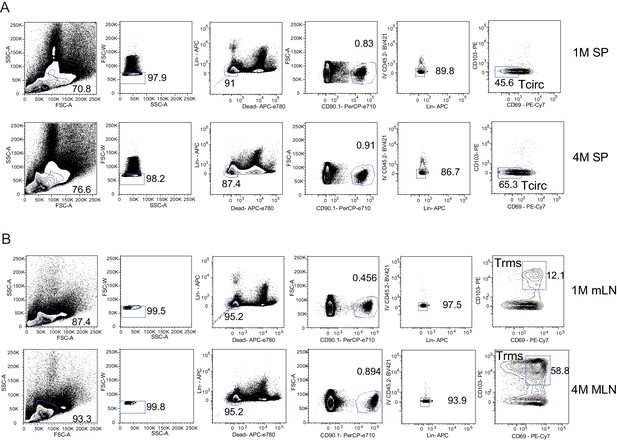
Gating strategy of FACS for RNAseq.
Mice were seeded with 104 naive P14 or 105 3M P14 cells and IN infected with PR8-GP33 virus. At 23–30 days post-infection, mLN and spleen were pooled, harvested, negatively enriched, and sorted for bulk RNAseq. (A) Gating strategy for analysis of 1M or 4M splenic IV− Tcirc P14s. (B) Gating strategy for analysis of 1M or 4M IV− mLN Trm P14s.
Tables
Samples for RNAseq, cell number, and RNA quality.
| RNAseq sample | Information | ||||
|---|---|---|---|---|---|
| Sample info | (pg/µL) RNA concentration | Sample volume (µl) | Total ng, RNA per tube | (RIN) RNA Integrity Number as determined by Agilent | RNA ratio |
| 1M Sp DN #1 | 618 | 17 | 10.506 | N/A | 2 |
| 1M SP DN #2 | 678 | 17 | 11.526 | N/A | 2 |
| 1M SP DN #3 | 1018 | 17 | 17.306 | N/A | 2 |
| 4M Sp DN #1 | 653 | 17 | 11.101 | 9.3 | 2.2 |
| 4M SP DN #2 | 692 | 17 | 11.764 | 9.6 | 2.3 |
| 4M SP DN #3 | 1028 | 17 | 17.476 | 9.4 | 2.1 |
| 1M mLN Trms | 78 | 17 | 1.326 | 9.2 | 1.8 |
| 1M mLN Trms | 139 | 23 | 3.197 | 7.4 | 1.8 |
| 4M mLN Trms | 1200 | 17 | 20.4 | 9.1 | 1.8 |
| 4M mLN Trms | 818 | 17 | 13.906 | 8.7 | 1.8 |
RNAseq heatmaps and GSEA full data.
RNAseq expression related to Figure 6.
| Gene | Sp1M-1 | Sp1M-2 | Sp1M-3 | Sp4M-1 | Sp4M-2 | Sp4M-3 | ||
|---|---|---|---|---|---|---|---|---|
| Neurl3 | −1.448372 | −0.744367 | 1.413726 | 0.029106 | 0.558649 | 0.191258 | ||
| Ctla4 | −1.472581 | −0.893646 | −0.085631 | 0.764674 | 0.93726 | 0.749924 | ||
| Fn1 | −0.4761 | −0.784447 | 0.313751 | −0.4761 | 1.898997 | −0.4761 | ||
| Arl4c | −0.219927 | −0.637316 | −0.954088 | −0.368204 | 1.839574 | 0.33996 | ||
| Pdcd1 | −0.974857 | −1.521079 | 0.271857 | 0.647515 | 0.79075 | 0.785815 | ||
| Bcl2 | −1.381925 | −0.653217 | 1.527735 | −0.080661 | 0.019993 | 0.568075 | ||
| Rgs2 | −1.527485 | −0.983776 | 0.344262 | 0.818042 | 0.6478 | 0.701157 | ||
| Rgs16 | −1.15537 | −1.037752 | −0.469227 | 0.887162 | 0.836029 | 0.939158 | ||
| Fcgr2b | −0.959014 | −0.670621 | −1.068859 | 0.853631 | 0.752644 | 1.092219 | ||
| Cd244 | −0.4632 | −0.776816 | −1.37756 | 0.884462 | 0.876998 | 0.856116 | ||
| Slamf7 | −1.139613 | −1.165538 | −0.294917 | 0.903817 | 0.860608 | 0.835643 | ||
| Atf3 | −0.779708 | −0.779708 | −0.779708 | 0.517244 | 0.120688 | 1.701193 | ||
| Il2ra | −1.322889 | −1.127548 | 0.021861 | 0.733615 | 0.889219 | 0.805742 | ||
| Zeb2 | −1.316736 | −1.060834 | −0.124986 | 0.843957 | 0.692407 | 0.966192 | ||
| Nr4a2 | −1.166233 | −1.064171 | −0.391073 | 0.70188 | 0.861206 | 1.058391 | ||
| Itga6 | 0.103598 | −1.354866 | 1.208944 | −1.045011 | 0.477086 | 0.610248 | ||
| Chn1 | −0.408248 | −0.408248 | −0.408248 | 2.041241 | −0.408248 | −0.408248 | ||
| Itga4 | −1.533817 | −0.935003 | 0.956091 | 0.590266 | 0.254859 | 0.667605 | ||
| Cd44 | −0.687106 | −0.641904 | −1.152441 | 0.138632 | 1.061634 | 1.281186 | ||
| Sema6d | −1.095775 | −1.095775 | −0.277219 | 1.304133 | 0.315154 | 0.849482 | ||
| Dusp2 | −1.284804 | −1.248205 | 0.363333 | 0.516972 | 0.711724 | 0.94098 | ||
| Bcl2l1 | −1.599476 | −0.920734 | 0.650549 | 0.563217 | 0.660348 | 0.646096 | ||
| Fgf13 | −1.184461 | −1.352896 | 0.33382 | 0.580607 | 0.836326 | 0.786603 | ||
| Cxcr3 | −1.558265 | −0.934718 | 0.89392 | 0.368323 | 0.670138 | 0.560601 | ||
| Fabp5 | −0.937223 | −0.580515 | −1.161553 | 0.966239 | 0.987902 | 0.72515 | ||
| Tnfsf10 | −0.757581 | −1.3349 | −0.529128 | 0.782834 | 0.745086 | 1.093689 | ||
| Rorc | −0.833666 | −0.833666 | 0.249621 | −0.833666 | 0.726727 | 1.52465 | ||
| Cd101 | −1.953973 | −0.122477 | 0.328694 | 0.424373 | 0.621766 | 0.701617 | ||
| Csf1 | −1.089549 | −0.89601 | −0.735068 | 0.905082 | 0.872774 | 0.94277 | ||
| S1pr1 | −1.414071 | −1.10549 | 0.261792 | 0.695359 | 0.834468 | 0.727942 | ||
| Usp33 | −1.470917 | −1.045478 | 0.725238 | 0.869896 | 0.325855 | 0.595405 | ||
| Bach2 | −1.176816 | −0.99054 | 1.523135 | 0.417138 | 0.352504 | −0.125421 | ||
| Aqp3 | −1.330326 | −1.216594 | 0.833799 | 0.371489 | 0.762789 | 0.578843 | ||
| Coro2a | −1.303957 | −1.114023 | −0.062495 | 0.784956 | 0.872198 | 0.823321 | ||
| Tnfsf8 | −1.655898 | −0.080202 | 1.478025 | 0.013587 | 0.258145 | −0.013656 | ||
| Dmrta1 | −0.845576 | −1.149819 | 1.698186 | −0.055975 | 0.094192 | 0.258992 | ||
| Jun | −1.376227 | −1.162485 | 0.313126 | 0.681802 | 0.765077 | 0.778706 | ||
| Zfp683 | −1.593545 | −0.902161 | 0.462586 | 0.734325 | 0.491177 | 0.807619 | ||
| Runx3 | −1.150841 | −1.327161 | 0.987223 | 0.185512 | 0.48935 | 0.815917 | ||
| Tnfrsf1b | −1.135037 | −1.426812 | 0.622179 | 0.498893 | 0.688943 | 0.751834 | ||
| Abcb1a | −1.375583 | −1.001921 | −0.109472 | 0.691716 | 0.866081 | 0.929179 | ||
| Cd36 | −1.132023 | −1.132023 | 0.647012 | 1.350724 | 0.414674 | −0.148364 | ||
| Fosl2 | 0.574051 | −1.743753 | −0.617194 | 0.260588 | 0.853012 | 0.673296 | ||
| Cd38 | −1.726006 | −0.546715 | 0.169224 | 0.342229 | 0.768355 | 0.992914 | ||
| Klf3 | −1.580977 | −0.948517 | 0.612343 | 0.634295 | 0.623061 | 0.659795 | ||
| Cxcl9 | −0.779708 | −0.779708 | 0.517244 | 1.701193 | 0.120688 | −0.779708 | ||
| Tgfbr3 | −1.496104 | −1.003156 | 0.272229 | 0.611554 | 0.842089 | 0.773388 | ||
| Dtx1 | −1.323911 | −0.532375 | 1.651323 | −0.271103 | 0.079677 | 0.39639 | ||
| Vps37b | −1.327024 | −1.248228 | 0.731573 | 0.523888 | 0.673187 | 0.646604 | ||
| Hsph1 | −0.89509 | −1.269058 | −0.482019 | 0.763431 | 0.921505 | 0.961231 | ||
| Chn2 | −1.469041 | −1.039823 | 0.366715 | 0.565777 | 0.966551 | 0.609821 | ||
| Il12rb2 | −1.512436 | −0.863636 | −0.044159 | 0.717179 | 0.841954 | 0.861097 | ||
| Cd8b1 | −0.897526 | −1.057029 | −0.74355 | 0.650528 | 1.07277 | 0.974808 | ||
| Cd8a | −1.369955 | −0.899035 | −0.293171 | 0.691408 | 0.958922 | 0.91183 | ||
| Sema4f | −0.711384 | −1.581909 | −0.130766 | 0.707607 | 0.841139 | 0.875313 | ||
| Mxd1 | −1.636166 | −0.835304 | 0.820257 | 0.420078 | 0.520451 | 0.710683 | ||
| Bhlhe40 | −1.377693 | −0.960477 | −0.194366 | 0.794975 | 0.876639 | 0.860922 | ||
| Klrg1 | −0.649531 | −0.92849 | −1.127976 | 0.880401 | 0.945822 | 0.879774 | ||
| Lag3 | −1.314079 | −1.250661 | 0.47627 | 0.562176 | 0.736842 | 0.789452 | ||
| Klre1 | −0.817609 | −0.924946 | −0.991491 | 0.900879 | 0.893107 | 0.940061 | ||
| Klrd1 | −0.82806 | −1.237129 | −0.605683 | 0.779021 | 0.84455 | 1.0473 | ||
| Klrc2 | −1.060961 | −0.22419 | −1.157839 | 1.312817 | 0.318738 | 0.811434 | ||
| Klrc1 | −0.456844 | −0.628525 | −1.48154 | 0.799308 | 0.874063 | 0.893538 | ||
| Dusp16 | −1.549192 | −0.590124 | −0.304361 | 0.482539 | 0.940286 | 1.020853 | ||
| Emp1 | −0.820283 | −1.020728 | −0.88602 | 0.790046 | 0.961756 | 0.975229 | ||
| Fosb | −1.192929 | −1.192929 | −0.104719 | 0.817943 | 0.642677 | 1.029956 | ||
| Nkg7 | 0.856662 | 0.552759 | −1.93607 | 0.002826 | 0.455585 | 0.068237 | ||
| Ppp1r15a | −1.743276 | −0.690768 | 0.651869 | 0.583382 | 0.59713 | 0.601662 | ||
| Swap70 | −1.185293 | −1.273627 | 0.02884 | 0.760466 | 0.836827 | 0.832787 | ||
| Il4ra | −1.240991 | −1.270277 | 1.021941 | 0.40852 | 0.380698 | 0.70011 | ||
| Il21r | −1.376141 | −1.176848 | 0.858231 | 0.430021 | 0.630542 | 0.634194 | ||
| Itgal | −1.41421 | −1.139199 | 0.44835 | 0.575817 | 0.77812 | 0.751121 | ||
| Itgax | −1.299963 | −1.141223 | −0.005996 | 0.730109 | 0.843686 | 0.873388 | ||
| Bag3 | −1.261842 | −1.302585 | 0.579948 | 0.471619 | 0.819237 | 0.693622 | ||
| Adam8 | −1.448605 | −0.760408 | −0.355088 | 0.802297 | 0.908979 | 0.852826 | ||
| Ifitm2 | −1.931568 | 0.697426 | −0.221955 | 0.354391 | 0.572163 | 0.529542 | ||
| Ifitm1 | −0.116103 | −0.604934 | −1.639024 | 0.602024 | 0.9939 | 0.764137 | ||
| Ifitm3 | −0.399411 | −1.471071 | −0.006047 | −0.164996 | 1.552387 | 0.489138 | ||
| Ifitm10 | −1.619476 | −0.810367 | 0.198443 | 0.630475 | 0.833945 | 0.76698 | ||
| Ifngr1 | −1.689527 | −0.724236 | 0.542852 | 0.31725 | 0.873566 | 0.680095 | ||
| Prdm1 | −0.763724 | −0.527892 | −1.266607 | 1.19839 | 0.974068 | 0.385764 | ||
| Prf1 | −1.450717 | −1.022122 | 0.129833 | 0.732409 | 0.819384 | 0.791213 | ||
| Gzmm | −0.148129 | −1.768864 | 0.896369 | −0.297885 | 0.868697 | 0.449812 | ||
| Gadd45b | −1.732358 | −0.68227 | 0.405453 | 0.592053 | 0.623696 | 0.793427 | ||
| Phlda1 | −1.336075 | −1.197598 | 0.286457 | 0.783069 | 0.814378 | 0.649768 | ||
| Ifng | −1.270643 | −0.812118 | −0.587444 | 0.872087 | 0.942033 | 0.856084 | ||
| A430078g2 | −0.768864 | −1.672411 | 0.352995 | 0.762701 | 0.546624 | 0.778955 | ||
| Rasa3 | −1.298438 | −1.268276 | 0.456223 | 0.612102 | 0.740735 | 0.757654 | ||
| Dusp4 | −0.013625 | −1.524651 | −0.248373 | −0.013625 | 1.604724 | 0.19555 | ||
| Lpl | −0.751462 | −0.751462 | −0.751462 | −0.058456 | 0.596117 | 1.716724 | ||
| Klf2 | −0.890686 | −1.058738 | −0.622839 | 0.304069 | 1.26258 | 1.005613 | ||
| Inpp4b | −1.411831 | −1.122956 | 0.917949 | 0.548702 | 0.408487 | 0.65965 | ||
| Dnajb1 | −1.351654 | −1.187033 | 0.50308 | 0.456015 | 0.956211 | 0.623381 | ||
| Junb | −1.248952 | −1.324103 | 0.75357 | 0.508582 | 0.679045 | 0.631859 | ||
| Cdh1 | −1.503072 | −0.854578 | 1.239026 | 0.498073 | 0.17862 | 0.441931 | ||
| Itgb1 | −1.070625 | −1.246026 | −0.245061 | 0.669811 | 0.908323 | 0.98358 | ||
| Fut11 | −1.566705 | −0.907343 | 0.264437 | 0.640465 | 0.710643 | 0.858503 | ||
| Gzmc | −1.129587 | −1.129587 | 1.393745 | 0.701502 | 0.081963 | 0.081963 | ||
| Gzmb | −1.019828 | −0.852547 | −0.860309 | 0.860605 | 0.936848 | 0.935231 | ||
| Tmem123 | −0.486668 | −0.568492 | −1.383266 | 0.289273 | 1.332877 | 0.816276 | ||
| Icam1 | −1.395117 | −1.177656 | 0.622649 | 0.592831 | 0.735718 | 0.621575 | ||
| S1pr5 | −1.401126 | −1.070617 | 0.107256 | 0.68738 | 0.826563 | 0.850543 | ||
| Gm10080 | −0.187622 | −0.228053 | −1.797739 | 0.584955 | 0.734187 | 0.894272 | ||
| Smad3 | −1.296473 | −1.254074 | 0.436554 | 0.494756 | 0.817683 | 0.801554 | ||
| Anxa2 | −1.161681 | −1.045596 | −0.444054 | 0.777762 | 0.935755 | 0.937813 | ||
| Cx3cr1 | −1.267574 | −1.170718 | −0.023068 | 0.778225 | 0.846035 | 0.837099 | ||
| Ccr8 | −1.030745 | −1.030745 | −0.649317 | 0.859499 | 0.902093 | 0.949215 | ||
| Crr9 | −1.163685 | −1.163685 | 0.518173 | 0.059333 | 1.369219 | 0.380645 | ||
| Cxcr6 | −1.008609 | −0.713182 | −0.998656 | 0.85504 | 0.986542 | 0.878867 | ||
| Ccr1 | −0.959366 | −0.959366 | −0.044992 | −0.460137 | 1.148955 | 1.274906 | ||
| Ccr2 | −1.244353 | −0.427864 | −0.835107 | 0.647598 | 0.470274 | 1.389451 | ||
| Ccr5 | −1.267395 | −1.197781 | 0.084813 | 0.600219 | 0.830419 | 0.949725 | ||
| Adam19 | −1.693324 | −0.353517 | 1.307494 | 0.407904 | 0.361716 | −0.030273 | ||
| Havcr2 | −0.267637 | −1.661471 | −0.479217 | 0.747014 | 0.819872 | 0.841438 | ||
| Itgae | 0.265873 | −0.131851 | −1.890634 | 0.287854 | 1.030981 | 0.437777 | ||
| Traf4 | −1.26168 | −1.317019 | 0.664003 | 0.656493 | 0.700534 | 0.557669 | ||
| Ccl5 | 0.717805 | 0.069708 | −1.965756 | 0.155463 | 0.696447 | 0.326333 | ||
| Ccl9 | −0.096406 | −1.204224 | −1.176934 | 0.685658 | 1.095686 | 0.69622 | ||
| Ccl3 | 0.321741 | 0.441288 | −2.033821 | 0.347205 | 0.557351 | 0.366236 | ||
| Ccl4 | −0.900786 | −1.071795 | −0.743744 | 0.770811 | 0.976086 | 0.969428 | ||
| Wfikkn2 | −1.521007 | −0.464233 | 1.525421 | −0.037938 | 0.151938 | 0.345819 | ||
| Tbx21 | −1.188999 | −1.37588 | 0.475345 | 0.610345 | 0.759099 | 0.720091 | ||
| Arl5c | −1.071685 | −1.481244 | 0.622113 | 0.521195 | 0.755916 | 0.653704 | ||
| Stat3 | −1.33085 | −1.244827 | 0.568421 | 0.575872 | 0.722103 | 0.709281 | ||
| Icam2 | −1.427743 | −1.074286 | 0.307995 | 0.501199 | 0.965903 | 0.726932 | ||
| Cmah | −1.619268 | −0.820081 | 1.012438 | 0.481339 | 0.443919 | 0.501653 | ||
| Fam65b | −1.435265 | −1.087651 | 0.278384 | 0.756549 | 0.73623 | 0.751753 | ||
| Irf4 | −0.942848 | −1.402093 | −0.166421 | 0.737652 | 0.874145 | 0.899565 | ||
| Ly86 | −1.176696 | −1.176696 | 0.271478 | 0.702255 | 0.093094 | 1.286564 | ||
| Nfil3 | −1.352986 | −1.210674 | 0.442696 | 0.664949 | 0.780773 | 0.675242 | ||
| Cdc14b | −1.660352 | −0.625526 | −0.018991 | 0.682479 | 0.624463 | 0.997927 | ||
| Naip3 | −0.645497 | −0.645497 | −0.645497 | −0.645497 | 1.290994 | 1.290994 | ||
| Elovl7 | −0.792302 | −0.420558 | −1.350578 | 1.107228 | 0.934382 | 0.521828 | ||
| Gzma | −0.686737 | −0.814558 | −1.197182 | 0.848737 | 0.941036 | 0.908703 | ||
| Gzmk | −1.406303 | −0.870165 | −0.278402 | 0.764809 | 0.91127 | 0.878791 | ||
| Itga1 | −1.285811 | −1.102208 | −0.113217 | 0.834949 | 0.703502 | 0.962785 | ||
| Rhob | −0.230141 | −1.869004 | 0.130554 | 0.363198 | 0.71253 | 0.892863 | ||
| Id2 | −1.046589 | −0.909419 | −0.760447 | 0.760224 | 0.914986 | 1.041244 | ||
| Adam4 | 0.587047 | −1.985993 | 0.480645 | −0.035246 | 0.587047 | 0.3665 | ||
| Fos | −0.879435 | −1.290058 | −0.474026 | 0.882188 | 0.81645 | 0.94488 | ||
| Il7r | −0.170366 | −0.087313 | −1.825959 | 0.423786 | 0.64009 | 1.019763 | ||
| Ly6c1 | −0.753065 | −1.398044 | −0.214637 | 0.174751 | 1.067836 | 1.123158 | ||
| Ly6c2 | −1.276681 | −1.209469 | 0.15969 | 0.548136 | 0.896153 | 0.882171 | ||
| Il2rb | −1.273008 | −1.30208 | 0.705662 | 0.505892 | 0.665374 | 0.698162 | ||
| Bin2 | −1.441078 | −1.122664 | 0.546309 | 0.600436 | 0.709577 | 0.70742 | ||
| Nr4a1 | −1.212606 | −1.312252 | 1.027233 | 0.554366 | 0.462063 | 0.481197 | ||
| Itga5 | −0.995646 | −0.995646 | 1.538635 | −0.396988 | 0.681175 | 0.16847 | ||
| Litaf | −1.67839 | −0.692126 | 0.153189 | 0.817433 | 0.827332 | 0.572561 | ||
| Klhl6 | −1.527286 | −0.908352 | 0.137948 | 0.561523 | 0.888302 | 0.847865 | ||
| Bcl6 | −1.042258 | −1.315998 | −0.140006 | 0.851054 | 1.025231 | 0.621977 | ||
| Tigit | −0.636695 | −0.492403 | −1.452925 | 0.80108 | 0.972809 | 0.808135 | ||
| Sidt1 | −1.416356 | −0.917945 | 1.352363 | 0.320931 | 0.308518 | 0.352489 | ||
| Ccr6 | −0.408248 | −0.408248 | −0.408248 | −0.408248 | 2.041241 | −0.408248 | ||
| Hspa1a | −1.489338 | −1.05789 | 0.546575 | 0.59207 | 0.600862 | 0.807722 | ||
| Tnf | −1.932303 | −0.226197 | 0.37222 | 0.508253 | 0.666342 | 0.611686 | ||
| Tnfsf9 | −0.721049 | −1.122053 | −0.869307 | 0.871738 | 0.99594 | 0.84473 | ||
| Qpct | −0.972332 | 0.746934 | −0.972332 | −0.773832 | 0.919129 | 1.052433 | ||
| Slc3a2 | −1.412175 | −1.03627 | 0.068812 | 0.641712 | 0.919915 | 0.818006 | ||
| Dtx4 | −1.895821 | 0.031087 | 0.985432 | 0.206382 | 0.623526 | 0.049393 | ||
| Dusp5 | −1.204462 | −1.231643 | −0.009555 | 0.655736 | 0.90792 | 0.882003 | ||
| Aff3 | 0.334158 | 0.934136 | 1.302955 | −0.695519 | −1.131655 | −0.744075 | ||
| Icos | 1.471137 | 0.841393 | 0.145304 | −0.792242 | −1.047218 | −0.618376 | ||
| Ikzf2 | −0.165671 | 1.704978 | 0.658251 | −0.793835 | −0.609887 | −0.793835 | ||
| Cxcr4 | 0.869115 | 0.919333 | 0.906884 | −0.886976 | −1.182769 | −0.625587 | ||
| Cd55 | 0.694467 | 0.444026 | 1.397141 | −0.459842 | −1.074231 | −1.001562 | ||
| Rgs1 | 0.282992 | −0.028889 | 1.320663 | 0.669655 | −1.444527 | −0.799896 | ||
| Sell | 0.343005 | 0.577399 | 1.57097 | −0.879322 | −0.741027 | −0.871025 | ||
| Xcl1 | 1.158069 | 0.726765 | 0.823087 | −0.933921 | −0.948108 | −0.825893 | ||
| Slamf6 | 0.878115 | 0.648026 | 1.158166 | −0.969274 | −0.688009 | −1.027024 | ||
| Dapl1 | 0.95876 | 0.953171 | 0.822988 | −0.894936 | −0.945047 | −0.894936 | ||
| Mal | 0.278549 | −1.166388 | 0.89838 | −0.024742 | 1.180589 | −1.166388 | ||
| Pmepa1 | 1.000943 | 1.530105 | −0.429028 | −0.700673 | −0.700673 | −0.700673 | ||
| Fabp4 | −0.408248 | 2.041241 | −0.408248 | −0.408248 | −0.408248 | −0.408248 | ||
| Skil | 1.47811 | 1.07101 | −0.815616 | −0.553424 | −0.577221 | −0.602859 | ||
| Il2 | 0.843097 | 1.08096 | 0.786459 | −0.913213 | −0.735725 | −1.061578 | ||
| Foxo1 | 1.398004 | 1.047696 | −0.261312 | −1.167157 | −0.508615 | −0.508615 | ||
| Il6ra | 0.192477 | 0.123263 | 1.813008 | −0.551343 | −0.551343 | −1.026062 | ||
| Npr1 | −0.408248 | 2.041241 | −0.408248 | −0.408248 | −0.408248 | −0.408248 | ||
| Lef1 | −0.577413 | −0.122582 | 2.008562 | −0.513026 | −0.534901 | −0.26064 | ||
| Lpar3 | −0.600795 | −0.600795 | 1.779753 | −0.600795 | −0.600795 | 0.623426 | ||
| Ifi44 | −1.522188 | 1.370539 | 0.77727 | −0.414406 | −0.162093 | −0.049122 | ||
| Nr4a3 | 1.658887 | −1.434777 | 0.298688 | −0.213395 | −0.096008 | −0.213395 | ||
| Tlr1 | 1.087014 | −1.759701 | 0.693949 | −0.332077 | −0.04672 | 0.357535 | ||
| Cxcl10 | 0.819849 | −1.553617 | 1.217093 | 0.268993 | −0.179362 | −0.572955 | ||
| Cd27 | −0.812702 | 0.736012 | 1.686726 | −0.76644 | −0.35287 | −0.490726 | ||
| Cd9 | 1.139853 | 0.963004 | 0.584072 | −0.83021 | −0.83021 | −1.02651 | ||
| Cd69 | 0.624643 | 0.93042 | 1.125309 | −0.853084 | −0.712926 | −1.114363 | ||
| Isg20 | 1.393478 | 0.934687 | 0.197602 | −0.930029 | −0.737091 | −0.858648 | ||
| Rgs10 | 0.96634 | 1.037646 | 0.595869 | −0.585759 | −0.644411 | −1.369686 | ||
| Themis | −0.659209 | 1.957135 | −0.786652 | −0.207908 | −0.035484 | −0.267882 | ||
| Fyn | 1.066584 | 1.346547 | 0.054683 | −0.726098 | −0.837588 | −0.904128 | ||
| Egr2 | 1.036188 | −1.636837 | 1.073379 | −0.057847 | −0.260015 | −0.154868 | ||
| Timp3 | −0.408248 | 2.041241 | −0.408248 | −0.408248 | −0.408248 | −0.408248 | ||
| Dusp6 | −0.205991 | 0.085139 | 1.646718 | −1.49606 | −0.014903 | −0.014903 | ||
| Crtam | 1.079203 | 1.14537 | 0.408387 | −1.043006 | −0.745838 | −0.844116 | ||
| Cxcr5 | −1.475245 | 0.994979 | 1.204565 | 0.116051 | −0.50998 | −0.330371 | ||
| Eomes | −0.005112 | 0.362201 | 1.517082 | −1.559944 | 0.048375 | −0.362601 | ||
| Tcf7 | −0.562937 | 0.066607 | 1.970539 | −0.522208 | −0.289205 | −0.662796 | ||
| Ccr7 | −0.893954 | −0.163325 | 1.969748 | −0.395389 | −0.21246 | −0.30462 | ||
| Cd86 | 1.380865 | 1.183911 | −0.546108 | −0.52407 | −0.777066 | −0.717531 | ||
| Btla | 0.971706 | 0.874718 | 0.860138 | −0.76399 | −1.171717 | −0.770855 | ||
| Sik1 | 0.195164 | 0.195164 | 0.195164 | 1.116809 | 0.195164 | −1.897467 | ||
| Epcam | 1.063844 | 0.498488 | 0.93633 | −1.165034 | −1.165034 | −0.168594 | ||
| Egr1 | −0.013398 | −0.699301 | 1.896141 | −0.699301 | 0.151141 | −0.635282 | ||
| Ifit3 | 1.114624 | 0.9678 | 0.615644 | −0.896586 | −0.812924 | −0.988558 | ||
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Mus musculus) | C57BL6/J | Jackson Laboratory | Stock No: 000664 (RRID:IMSR_JAX:000664) | |
| Strain, strain background (Mus musculus) | B6.PL(84NS)/Cy | Jackson Laboratory | Stock No: 000983 (RRID:IMSR_JAX:000406) | C57BL6/J Thy1.1 |
| Strain, strain background (Mus musculus) | B6.Cg-Tcratm1MomTg(TcrLCMV)327Sdz (P14) | Jackson Laboratory | Stock No: 37394-JAX (RRID:IMSR_TAC:4138) | |
| Strain, strain background (Mus musculus) | Thy1.1/1.1- B6.Cg-Tcratm1Mom Tg(TcrLCMV)327Sdz | This paper | Thy1.1/1.1 P14 | Can be acquired through lab contact or breeding of above commercially available strains |
| Strain, strain background (Mus musculus) | Thy1.1/1.2- B6.Cg-Tcratm1Mom Tg(TcrLCMV)327Sdz | This paper | Thy1.1/1.2 P14 | Can be acquired through lab contact or breeding of above commercially available strains |
| Strain, strain background (Influenza A virus) | Recombinant influenza A/PR/8/34 expressing (H1N1) GP33-41 | Laidlaw et al. Cooperativity Between CD8+ T Cells, Non- Neutralizing Antibodies, and Alveolar Macrophages Is Important for Heterosubtypic Influenza Virus Immunity. Plos Pathog. 9(3) e1003207 (2013). | PR8-GP33 | Can be acquired through lab contact. |
| Strain, strain background (Influenza A virus) | Recombinant influenza A/X-31 (H3N2) expressing GP33-41 | Laidlaw et al. Cooperativity Between CD8+ T Cells, Non- Neutralizing Antibodies, and Alveolar Macrophages Is Important for Heterosubtypic Influenza Virus Immunity. Plos Pathog. 9(3) e1003207 (2013). | X31-GP33 | Can be acquired through lab contact. |
| Peptide, recombinant protein | GP33-44 | AnaSpec | Catalog #: AS-61296 | |
| Antibody | CD11a (rat monoclonal) | Biolegend | M17/4 (AB_312776) | FACs (1:100) |
| Antibody | IFNγ (rat monoclonal) | eBioscience | XMG1.2 (AB_465410) | FACs (1:100) |
| Antibody | CD8a (rat monoclonal) | eBioscience | 53–6.7 (AB_1853141) | FACs (1:100) |
| Antibody | Thy1.1 (mouse monoclonal) | eBioscience | OX-7 (AB_2201314) | FACs (1:100) |
| Antibody | Thy1.2 (rat monoclonal) | eBioscience | 30-H12 (AB_1853152) | FACs (1:100) |
| Antibody | CD45.2 (mouse monoclonal) | eBioscience | 104 (AB_469724) | FACs (1:100) |
| Antibody | CD103 (hamster monoclonal) | Biolegend | 2E7 (AB_469040) | FACs (1:100) |
| Antibody | CD69 (hamster monoclonal) | Biolegend | H1.2F3 (AB_1853105) | FACs (1:100) |
| Antibody | CD44 (rat monoclonal) | Biolegend | 1M7 (AB_223593) | FACs (1:100) |
| Antibody | CD62L (rat monoclonal) | Biolegend | MEL-14 (AB_1853103) | FACs (1:100) |
| Antibody | KLRG1 (mouse monoclonal) | eBioscience | 2F1 (AB_540279) | FACs (1:100) |
| Antibody | CX3CR1 (mouse monoclonal) | eBioscience | SA011F11 (AB_2565701) | FACs (1:100) |
| Antibody | CXCR3 (Armenian hamster monoclonal) | eBioscience | CXCR3-173 (AB_1210593) | FACs (1:100) |
| Antibody | Eomesodermin (rat monoclonal) | eBioscience | Dan11mag (AB_11042577) | FACs (1:100) |
| Antibody | TNF (rat monoclonal) | eBioscience | MP6-XT22 (AB_465416) | FACs (1:100) |
| Antibody | IL-2 (rat monoclonal) | Biolegend | JES6-5H4 (AB_315298) | FACs (1:100) |
| Antibody | Granzyme A (mMouse monoclonal) | Biolegend | 3G8.5 (AB_2565308) | FACs (1:100) |
| Antibody | Granzyme B (rat monoclonal) | Biolegend | 12F9B65 (AB_2564373) | FACs (1:100) |
| Antibody | BrdU (mouse monoclonal) | Biolegend | Bu20a (AB_1595472) | FACs (1:100) |
| Commercial assay or kit | Foxp3/Transcription Factor Staining Buffer Set | Invitrogen | 00-5523-00 | |
| Software, algorithm | GraphPad Prism | GraphPad Prism 8 | Version 8.4.2 (464) (RRID:SCR_002798) |





