High potency of sequential therapy with only β-lactam antibiotics
Figures
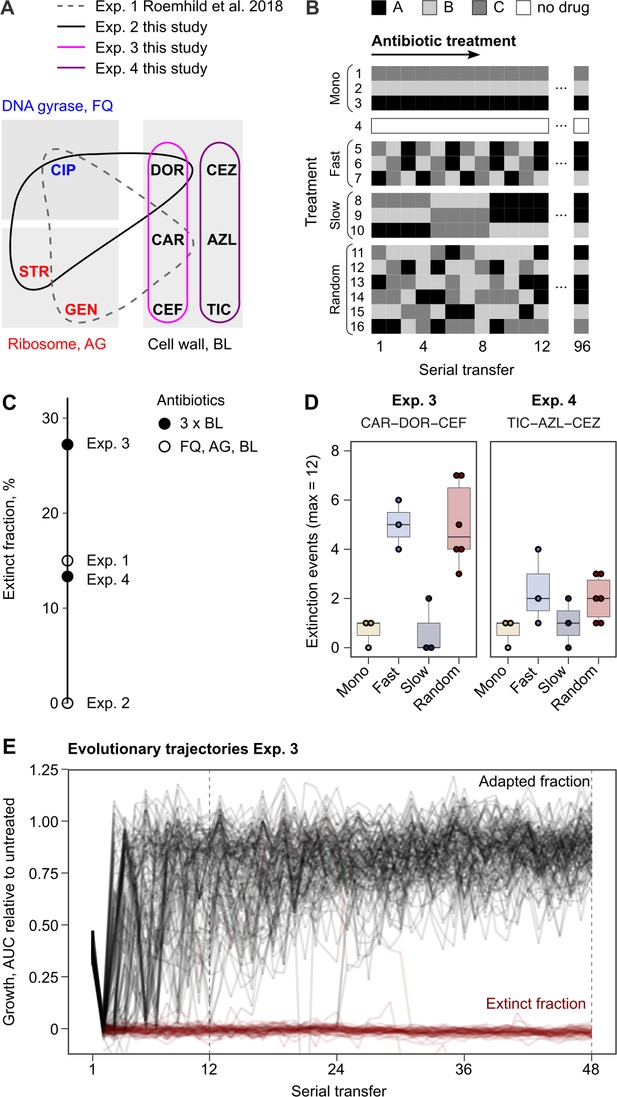
Probability of evolutionary rescue depends on drug triplets and treatment type.
(A) The evaluated antibiotic combinations comprise different types of antibiotic targets. Fluoroquinolone antibiotics (FQ) target DNA gyrase, aminoglycosides (AG) inhibit translation, and β-lactams (BL) inhibit cell-wall synthesis. (B) The evaluated treatment protocols test the effects of switching rate and temporal regularity. (C) A fraction of lineages is eradicated by the sublethal dosage sequential treatments. Lineage extinction is high for combinations of cell-wall targeting β-lactams. (D) Variation in extinction for the β-lactam combinations by treatment type (n = 3–6 protocols per treatment type). (E) The distribution of evolutionary trajectories for Exp. 3 with CAR-DOR-CEF shows that the majority of extinction events occur within the first 12 serial transfers (n = 180 lineages). Growth of evolving lineages is quantified relative to untreated reference populations using the relative area under the growth curve (AUC). AZL: azlocillin; CAR: carbenicillin; CEF: cefsulodin; CEZ: ceftazidime; CIP: ciprofloxacin; DOR: doripenem; GEN: gentamicin; STR: streptomycin; TIC: ticarcillin. The following supplementary material is available for Figure 1: Figure 1—figure supplement 1, Figure 1—source data 1, Figure 1—figure supplement 1—source data 1, Supplementary file 1A.
-
Figure 1—source data 1
Source data for the panels of Figure 1.
- https://cdn.elifesciences.org/articles/68876/elife-68876-fig1-data1-v2.xlsx
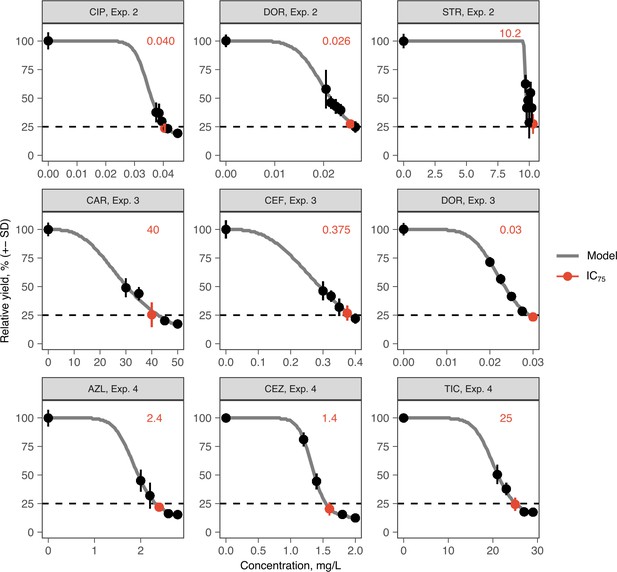
Antibiotic dose-response curves for PA14 (mean ± s.d.; n = 6 biological replicates).
Red text and points indicate IC75 inhibitory concentrations as applied in the evolution experiments. Gray line indicates Nelder–Mead dose-response model (R package drc). The source data is provided in Figure 1—figure supplement 1—source data 1.
-
Figure 1—figure supplement 1—source data 1
Source data for Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/68876/elife-68876-fig1-figsupp1-data1-v2.xlsx
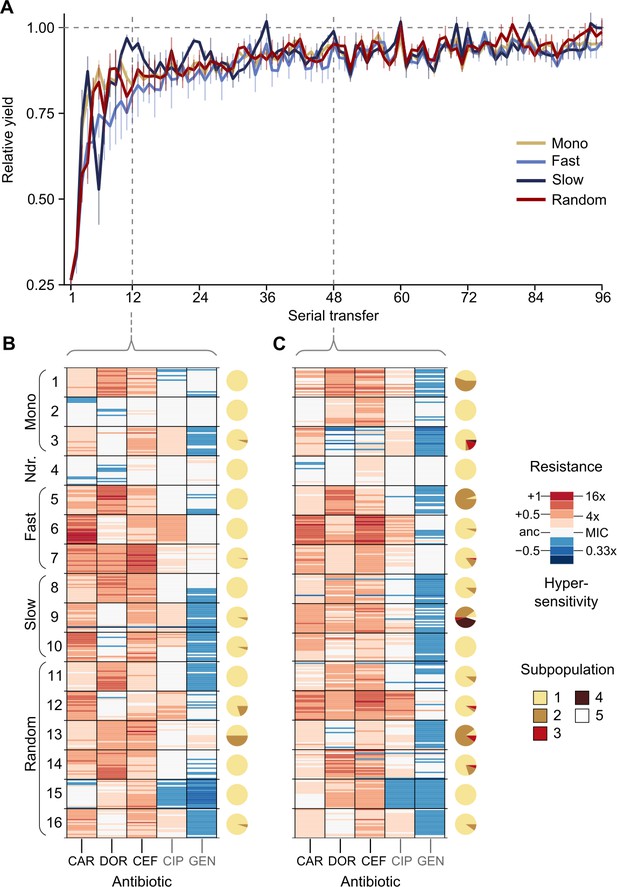
Resistance to doripenem is constrained in the CAR-CEF-DOR triple β-lactam experiment.
(A) Rapid adaptive increase of biomass yields relative to the untreated reference populations (mean ± CI95; n = 3–6 protocols per treatment type and 12 biological replicates per sequence; extinct lineages excluded). Vertical dotted lines separate the three growth phases. Evolved changes in the susceptibility to the treatment antibiotics CAR, DOR, and CEF and the non-treatment antibiotics CIP and GEN after transfer 12 (B) or transfer 48 (C), evaluated with 20 isolates each for the 16 representative adapting populations at each time point. Mono 1 is monotherapy with CAR, mono 2 is monotherapy with DOR, and mono 3 is monotherapy with CEF. The evolution of resistance and hypersensitivity is indicated by red and blue colors, respectively, given for the considered isolates as horizontal lines (total of 640 isolates), sorted according to evolution treatment (main rows in the figures) and tested antibiotics (main columns; antibiotics given at the bottom). Pie charts on the right show phenotypic within-population diversity, where different colors indicate subpopulations inferred from hierarchical clustering of resistance phenotypes. CAR: carbenicillin; CEF: cefsulodin; CIP: ciprofloxacin; DOR: doripenem; GEN: gentamicin. The following supplementary material is available for Figure 2: Figure 2—figure supplement 1, Figure 2—figure supplement 2, Figure 2—figure supplement 3, Figure 2—figure supplement 4, Figure 2—figure supplement 5, Figure 2—source data 1, Figure 2—figure supplement 1—source data 1, Figure 2—figure supplement 2—source data 1, Figure 2—figure supplement 3—source data 1, Figure 2—figure supplement 5—source data 1, Supplementary file 1B.
-
Figure 2—source data 1
Source data for the panels of Figure 2.
- https://cdn.elifesciences.org/articles/68876/elife-68876-fig2-data1-v2.xlsx
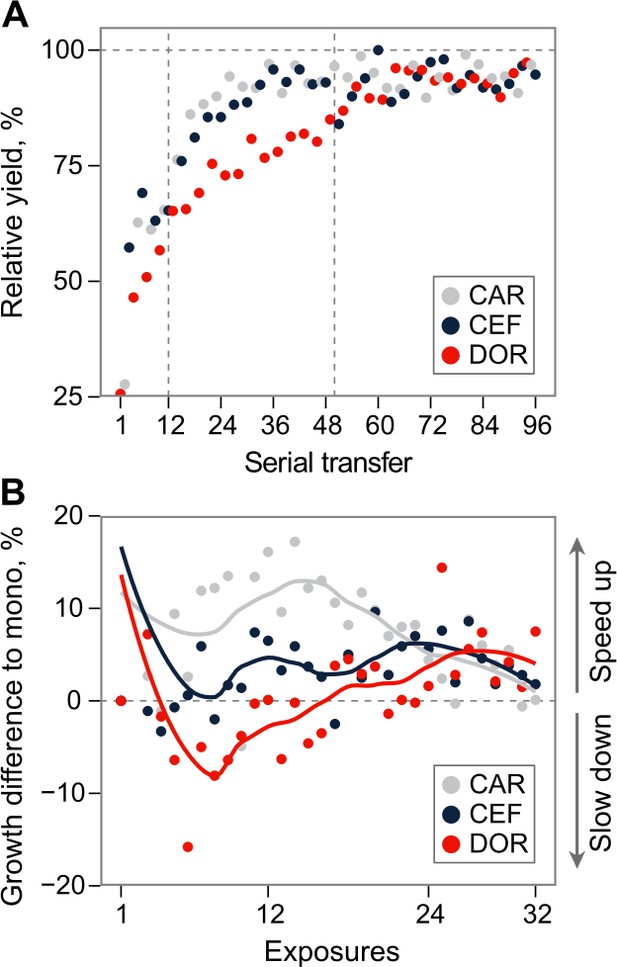
Growth dynamics in fast sequential protocols.
(A) Evolutionary growth improvements for fast protocol #6 (mean of the seven surviving lineages). Relative growth increased to the antibiotics at different rates, demonstrating the consecutive evolution of resistance, and thus coexistence of genetic subpopulations. The resulting clonal interference may explain the drop of growth on CEF around transfer 50, and the subsequent growth oscillations during CEF. (B) Mean difference of growth during exposures to particular antibiotics in fast sequential protocols #5–7 compared to growth in monotherapies after the same number of exposures to that drug. X-axis denotes exposures to a particular antibiotic, and thus goes to 96/3 = 32. Multidrug exposure accelerated adaptation compared to monotherapy against CAR and CEF, but slowed down adaptation against DOR. The source data is provided in Figure 2—figure supplement 1—source data 1.CAR: carbenicillin; CEF: cefsulodin; DOR: doripenem.
-
Figure 2—figure supplement 1—source data 1
Source data for Figure 2—figure supplement 1.
- https://cdn.elifesciences.org/articles/68876/elife-68876-fig2-figsupp1-data1-v2.xlsx
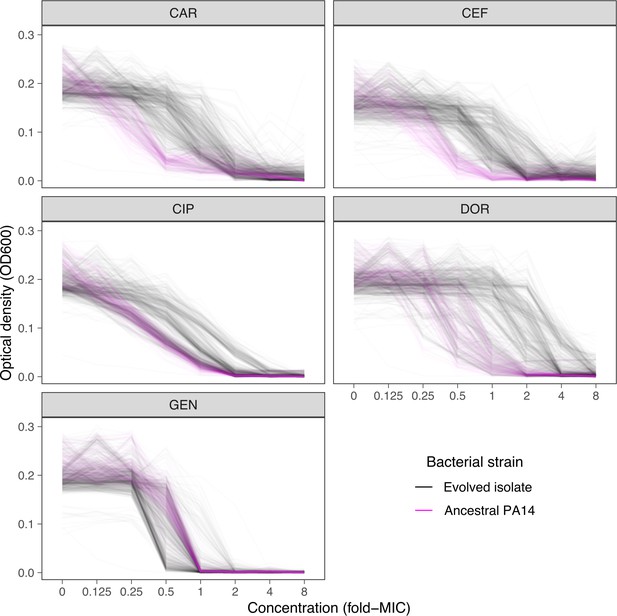
Dose-response curve distributions for Exp. 3 with CAR-DOR-CEF, underlying Figure 2B and C.
Gray lines show data from evolved isolates, magenta lines show repeated measurements of the PA14 ancestor. The source data is provided in Figure 2—figure supplement 2—source data 1. CAR: carbenicillin; CEF: cefsulodin; DOR: doripenem.
-
Figure 2—figure supplement 2—source data 1
Source data for Figure 2—figure supplement 2.
- https://cdn.elifesciences.org/articles/68876/elife-68876-fig2-figsupp2-data1-v2.xlsx
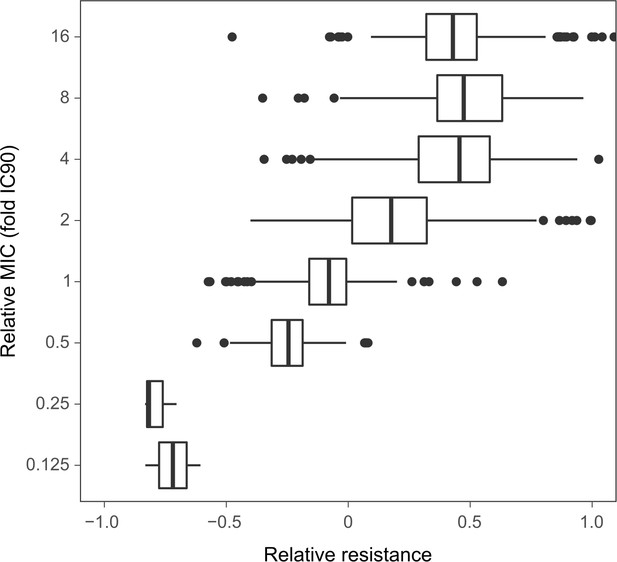
Relation between the resistance values and the fold-change of the minimal inhibitory concentrations (MIC) approximated by IC90.
The resistance values are depicted in Figure 2B, C. The boxes show interquartile range (25th to 75th percentile), the thick line indicates the median. Whiskers cover data range but are capped at maximum 1.5× the interquartile range. The source data is provided in Figure 2—figure supplement 3—source data 1.
-
Figure 2—figure supplement 3—source data 1
Source data for Figure 2—figure supplement 3.
- https://cdn.elifesciences.org/articles/68876/elife-68876-fig2-figsupp3-data1-v2.xlsx
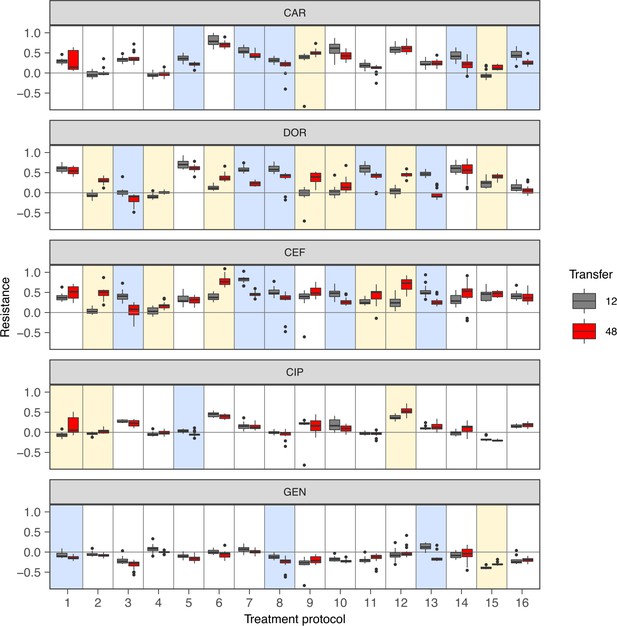
Change of population antibiotic resistance between transfer 12 (indicated in gray) and transfer 48 (indicated in red).
The resistance values are depicted in Figure 2B, C. The boxes show interquartile range (25th to 75th percentile), the thick line indicates the median. Whiskers cover data range but are capped at maximum 1.5× the interquartile range. Statistical difference between time points was assessed using Wilcoxon rank-sum test as described in Supplementary file 1F. A blue shading of the background indicates significant decrease of resistance, and a yellow background shading indicates a significant increase of resistance. p-values were adjusted by Bonferroni correction. The figure is a different representation of the data shown in the heatmaps of Figure 2B, C. The source data is accordingly provided in Figure 2—source data 1. The results of the statistical analysis are provided in Supplementary file 1F.
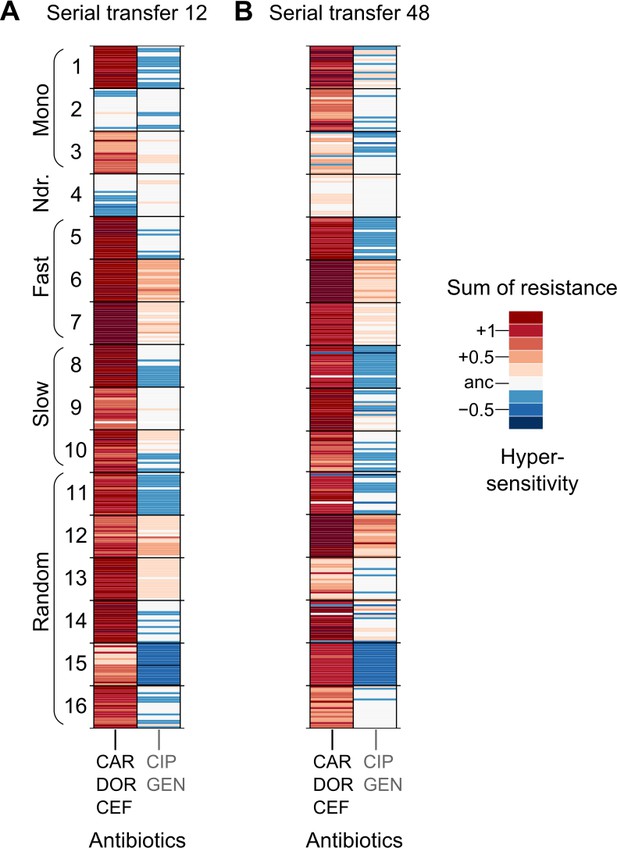
Population multidrug resistance after (A) transfer 12 and (B) transfer 48.
The left column indicates the sum of resistance scores for the β-lactam antibiotics CAR, DOR, and CEF, which were used for the evolution experiment. The right column indicates the sum of collateral resistance to the antibiotics CIP and GEN, which were not used in the evolution experiment. Clones are depicted in the same order as in Figure 2B, C, and on the same color scale. The source data is provided in Figure 2—figure supplement 5—source data 1. CAR: carbenicillin; CEF: cefsulodin; CIP: ciprofloxacin; DOR: doripenem; GEN: gentamicin.
-
Figure 2—figure supplement 5—source data 1
Source data for Figure 2—figure supplement 5.
- https://cdn.elifesciences.org/articles/68876/elife-68876-fig2-figsupp5-data1-v2.xlsx
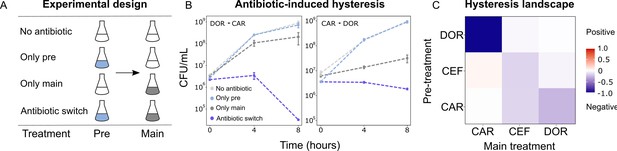
Negative hysteresis is common among the tested β-lactam antibiotics.
(A) Hysteresis effects were measured using the previously established experimental approach (see Materials and methods). (B) Bacterial counts were plotted over time after the pretreatment to obtain time-kill curves (mean ± sem, n = 3). Level of hysteresis was quantified as the difference between the antibiotic switch and the only main curves. Negative values indicate negative hysteresis and positive values indicate positive hysteresis. (C) Heatmap of hysteresis levels between all nine combinations of the three β-lactams. DOR and CAR show asymmetric bidirectional negative hysteresis. Negative hysteresis is also observed in switches from CEF to CEF and CAR to CEF. Weak positive hysteresis is found for the switch from CEF to CAR. The following supplementary material is available for Figure 3: Figure 3—figure supplement 1, Figure 3—figure supplement 2, Figure 3—source data 1, Figure 3—figure supplement 1—source data 1. CAR: carbenicillin; CEF: cefsulodin; DOR: doripenem.
-
Figure 3—source data 1
Source data for the panels of Figure 3.
- https://cdn.elifesciences.org/articles/68876/elife-68876-fig3-data1-v2.xlsx
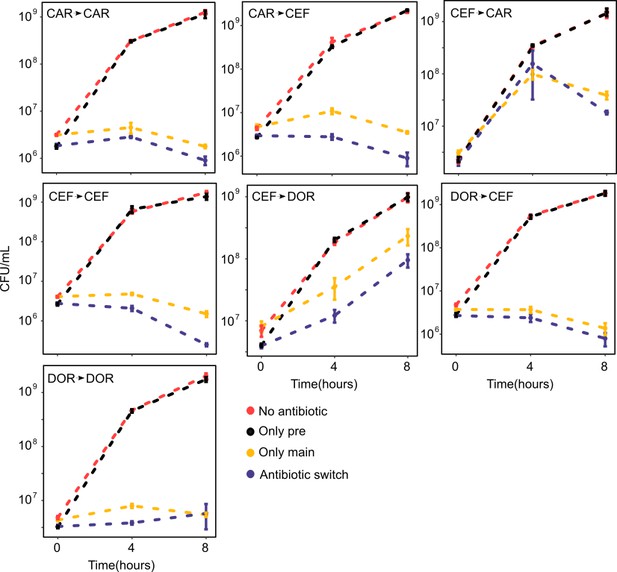
Time-kill curves of hysteresis experiments for the combinations not presented in Figure 3 (mean ± sem, n = 3).
The source data is provided in Figure 3—figure supplement 1—source data 1.
-
Figure 3—figure supplement 1—source data 1
Source data for Figure 3—figure supplement 1.
- https://cdn.elifesciences.org/articles/68876/elife-68876-fig3-figsupp1-data1-v2.xlsx
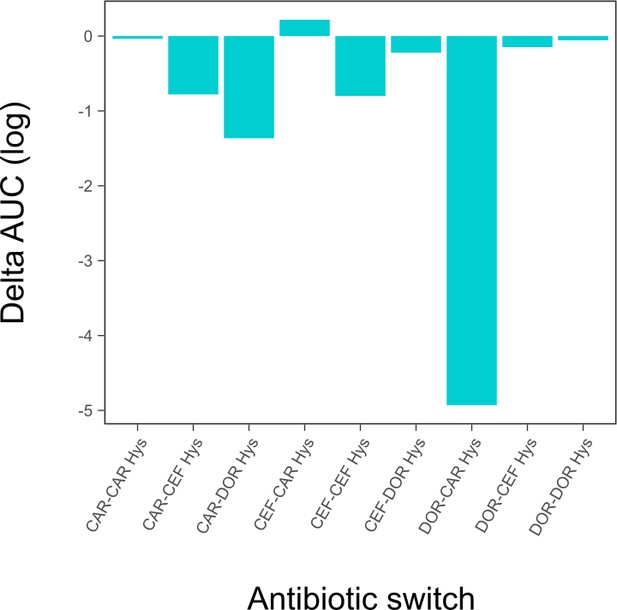
Hysteresis effects quantified as area under the curve (AUC) difference between the ‘only main’ and ‘antibiotic switch’ curves from the time-kill dynamics.
This figure is a different representation of the same data shown in the heatmap of Figure 3C. The source data for this figure is accordingly provided in Figure 3—source data 1.
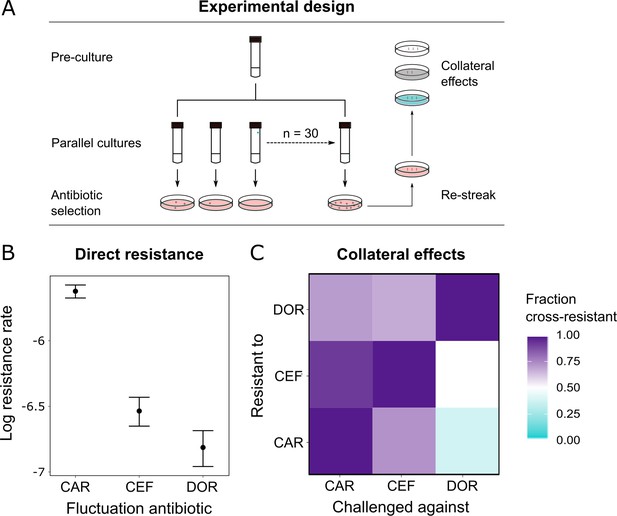
Doripenem has the lowest rates of direct and indirect resistance.
(A) Schematic of the experimental protocol to determine spontaneous rates of resistance on each of the three β-lactams and the resulting collateral landscape. Briefly, an overnight culture was taken and split into 30 parallel cultures where bacteria were allowed to divide in the absence of an antibiotic and any other constraint. Spontaneous resistant mutants were selected on minimal inhibitory concentration (MIC) plates and restreaked to ensure genetic resistance. These mutants were then patched on MIC plates of the other two β-lactams to test for cross-resistance. (B) Comparison of rates of spontaneous resistance on the three β-lactams on a Log10 scale. Error bars depict CI95. All comparisons were found to be significantly different from each other (likelihood ratio test; CAR vs. CEF p<0.0001, CAR vs. DOR p<0.0001, and DOR vs. CEF p<0.01). (C) Landscape of collateral effects between the three β-lactams. Fraction of cross-resistant mutants per antibiotic combination is plotted. DOR has the least cases of cross-resistance of the three. A total of 60 mutants per antibiotic were used for collateral effect testing. The following supplementary material is available for Figure 4: Figure 4—source data 1, Supplementary file 1G–I. CAR: carbenicillin; CEF: cefsulodin; DOR: doripenem.
-
Figure 4—source data 1
Source data for Figure 4.
- https://cdn.elifesciences.org/articles/68876/elife-68876-fig4-data1-v2.xlsx
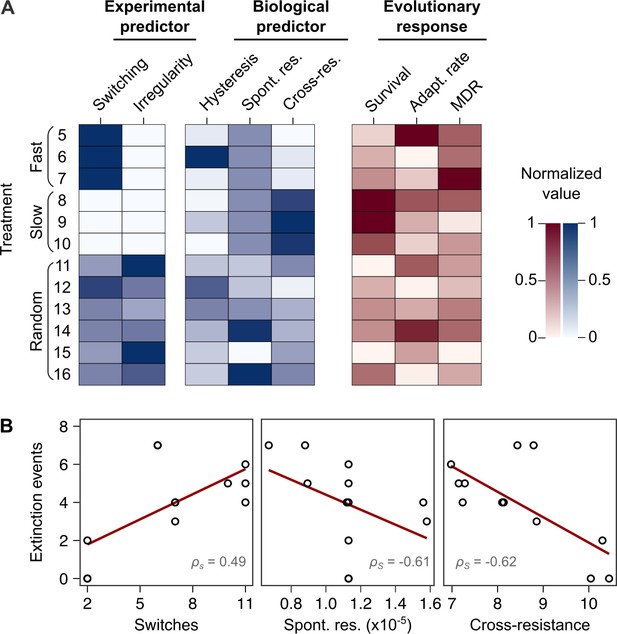
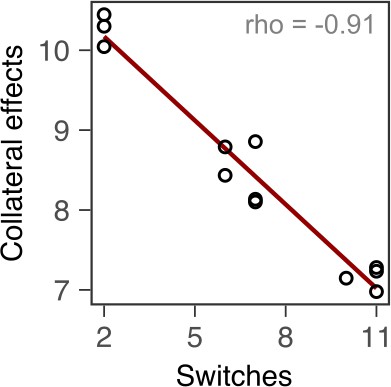
Bacterial extinction is correlated to switching rate, spontaneous rate of resistance, and spontaneous cross-resistance.
(A) Variation in experimental parameters, potential biological predictors, and the measured traits up to transfer 12. The experimental parameters include switching rate and regularity of change (high irregularity in dark). Potential biological predictors are cumulative levels of hysteresis (dark indicates protective effects), cumulative probabilities of spontaneous resistance (Spont. res., dark indicates higher probability), and cumulative level of collateral effects (Cross-res., dark indicates high fraction of cross-resistance). The evolutionary response was measured for population survival (max = 12), adaptation rate (Adapt. rate, n ≤ 12, extinct lineages excluded), evolved multidrug resistance (MDR) to treatment antibiotics CAR, DOR, and CEF (MDR, n = 16). (B) Variation in extinction was best explained by collateral effects between the antibiotics (for illustrative purposes, the red line depicts linear regression and ρS the Spearman’s rank correlation coefficient). The following supplementary material is available for Figure 5: Figure 5—figure supplement 1, Figure 5—source data 1, Supplementary file 1J–O. CAR: carbenicillin; CEF: cefsulodin; DOR: doripenem.
-
Figure 5—source data 1
Source data for Figure 5.
- https://cdn.elifesciences.org/articles/68876/elife-68876-fig5-data1-v2.xlsx
Tables
Evolved genetic changes inferred from whole-genome sequencing.
| Treatment type | ID* | AA change† | Gene name | Annotation | Freq‡ |
|---|---|---|---|---|---|
| Monotherapy | 1 | V471G | ftsI | Peptidoglycan synthesis | 3/3 |
| 2§ | N242S | ftsI | Peptidoglycan synthesis | 3/3 | |
| 3 | T157P | pepA | Virulence | 3/3 | |
| Fast-rgular | 5 | V471G | ftsI | Peptidoglycan synthesis | 3/3 |
| 6 | K26 | nalD | Efflux | 3/3 | |
| S379ISR | rmcA | Biofilm maintenance | 1/3 | ||
| 7 | R220C | phoQ | Two-component | 3/3 | |
| - | PA14_55631 | 23srRNA, translation | 1/3 | ||
| Slow-rgular | 8 | V471G | ftsI | Peptidoglycan synthesis | 3/3 |
| 9 | D357N | pepA | Virulence | 3/3 | |
| 10 | T157P | pepA | Virulence | 3/3 | |
| E115VAAWIPK | PA14_21540 | Lipid metabolism (3-exoacyl ACP synthase) | 1/3 | ||
| Q117AEEQ | PA14_21540 | Lipid metabolism (3-exoacyl ACP synthase) | 1/3 | ||
| R178C | zipA | Cell division | 2/3 | ||
| P483PEP | dnaX | Cell division | 1/3 |
-
* Individual treatment of evolution experiment.
† Amino acid change.
-
‡ Occurrence frequency of the identified variant (before slash) out of the total number of isolates sequenced (behind slash).
§ Mutations listed are from isolates obtained from the populations frozen at transfer 48, no variants were found in the isolates from transfer 12.
-
Table 1—source data 1
Source data for the summary of the genome sequencing analysis shown in Table 1.
- https://cdn.elifesciences.org/articles/68876/elife-68876-table1-data1-v2.xlsx
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Pseudomonas aeruginosa) | PA14 | https://doi.org/10.1126/science.7604262 | UCBPP-PA14 | |
| Chemical compound, drug | AZL (azlocillin) | Sigma | A7926-1G | |
| Chemical compound, drug | CAR (carbenicillin) | Carl Roth | 6344.2 | |
| Chemical compound, drug | CIP (ciprofloxacin) | Sigma | 17850-5 G-F | |
| Chemical compound, drug | CEF (cefsulodin) | Carl Roth | 4014.2 | |
| Chemical compound, drug | CEZ (ceftazidime) | Sigma | C3809.1G | |
| Chemical compound, drug | DOR (doripenem) | Sigma | 32138-25 MG | |
| Chemical compound, drug | GEN (gentamicin) | Carl Roth | 2475.1 | |
| Chemical compound, drug | STR (streptomycin) | Sigma | S6501-5 | |
| Chemical compound, drug | TIC (ticarcillin) | Sigma | T5639-1G | |
| Software, algorithm | R: A language and environment for statistical computing | https://www.R-project.org/ |
Additional files
-
Supplementary file 1
Tables with information on antibiotics used and summaries of the statistical analyses.
(A) List of antibiotics used for the evolution experiments. (B) Statistical analysis of main evolution treatments for the evolutionary dynamics shown in Figure 2A. (C) Statistical analysis of main evolution treatments for the multidrug β-lactam resistance after transfer 12 in Figure 2B. (D) Statistical analysis of main evolution treatments for the multidrug β-lactam resistance after transfer 48 in Figure 2C. (E) Statistical analysis of main evolution treatments for Shannon diversity in Figure 2B, C. (F) Comparison of resistance profiles between transfer 12 and transfer 48 for Figure 2—figure supplement 4 using Wilcoxon test and Bonferroni correctiona. (G) Minimum inhibitory concentrations (MICs) as determined by agar dilution for the three β-lactams and as used for the fluctuation assays shown in Figure 4. (H) Likelihood ratio test to assess pairwise variation in spontaneous resistance rates for the three β-lactam antibiotics shown in Figure 4B. (I) Post hoc comparisons based on the false discovery rate for phenotype of cross-resistance on secondary antibiotic as shown in Figure 4C. (J) Analysis of variance of the consequences of the main treatment type on the three measured responses for transfer 12 of the triple β-lactam experiment as summarized in Figure 5. (K) Post hoc comparison based on the false discovery rate of the effect of the main treatment types on extinction for the triple β-lactam experiment as summarized in Figure 5. (L) General linear model analysis of the consequences of the experimental predictors switching rate and irregularity on the three measured responses for transfer 12 of the triple β-lactam experiment as summarized in Figure 5. (M) Main effect tests for the consequences of switching rate and irregularity on extinction and multidrug resistance (MDR) for transfer 12 of the triple β-lactam evolution experiment as summarized in Figure 5. (N) General linear model analysis of the consequences of the three considered biological predictors, cumulative probability of spontaneous resistance, cross-resistance, and hysteresis on the three measured responses for transfer 12 of the triple β-lactam experiment as summarized in Figure 5. (O) Main effect tests for the consequences of the three considered biological predictors, cumulative probability of spontaneous resistance, cross-resistance, and hysteresis on extinction of the triple β-lactam evolution experiment as summarized in Figure 5.
- https://cdn.elifesciences.org/articles/68876/elife-68876-supp1-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/68876/elife-68876-transrepform-v2.docx