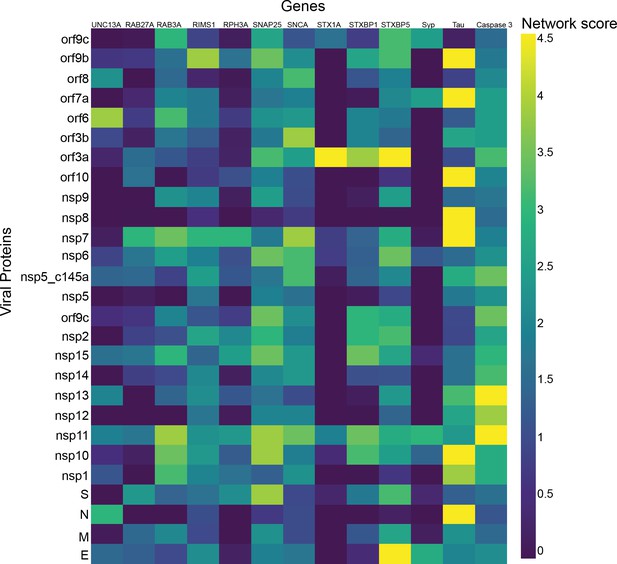
Effect of SARS-CoV-2 proteins on vascular permeability
Figures
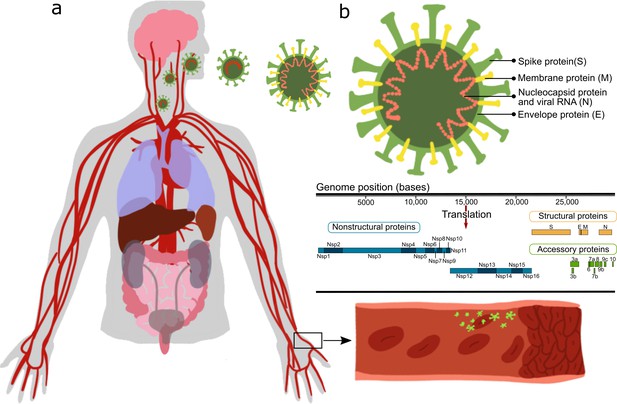
Effect of severe acute respiratory syndrome (SARS)-CoV-2 proteins on endothelial cells.
(a) Sketch representing the main organs affected by SARS-CoV-2; (b) structure and gene composition of SARS-CoV-2.
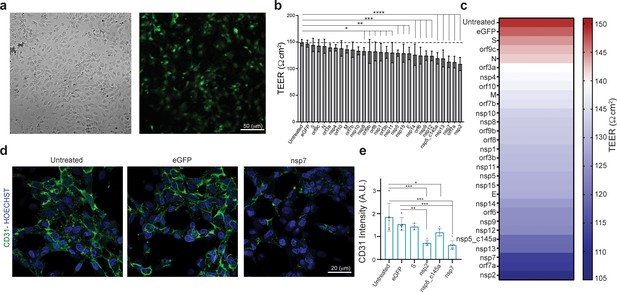
Effect of severe acute respiratory syndrome (SARS)-CoV-2 proteins on human umbilical vein endothelial cell (HUVEC) at day 3.
(a) Bright-field and fluorescent image of infected eGFP HUVEC, scale bar: 50 µm; (b) changes in barrier functions as a result of SARS-CoV-2 proteins were assessed by trans-epithelial-endothelial electrical resistance (TEER) measurement. Note the statistical differences compared to the untreated control condition, assessed by F-statistic with two-way ANOVA test, followed by the Holm–Sidak test for multiple comparisons; (c) color map showing a gradual decrease in TEER values compared to the untreated condition at day 3; (d) immunocytochemistry (ICC) for CD31 (green) and Hoechst (blue) for the three specified conditions, scale bar: 20 µm; (e) analysis of CD31 expression levels.
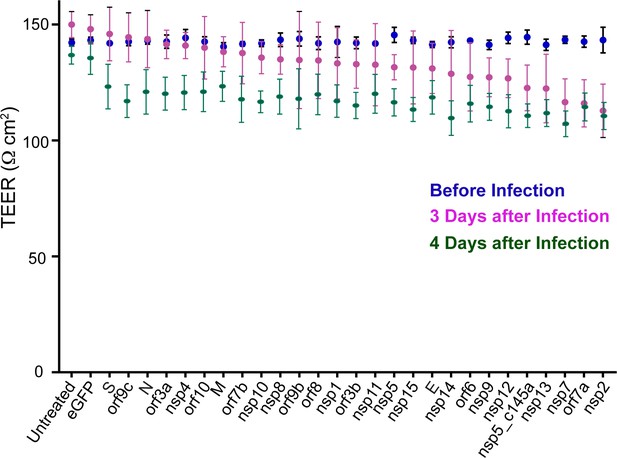
Effect of severe acute respiratory syndrome (SARS)-CoV-2 proteins on human umbilical vein endothelial cell (HUVEC) functionality.
Trans-epithelial-endothelial electrical resistance (TEER) values at three different time points: before the infection (blue dots), 3 and 4 days after the infections (pink and green dots).
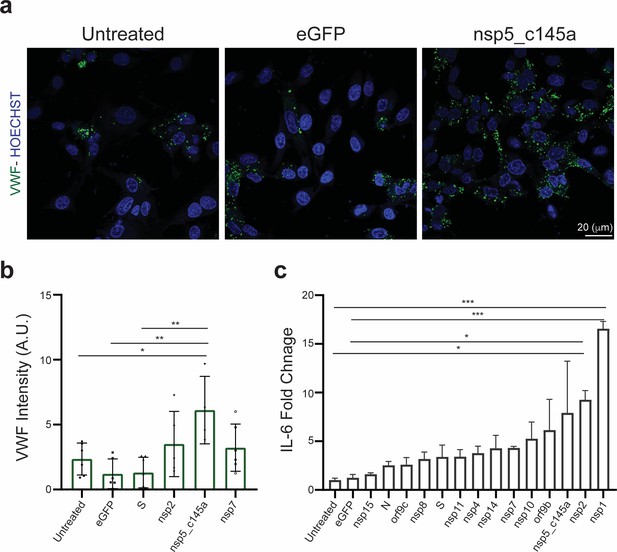
Human umbilical vein endothelial cell (HUVEC) response to specific proteins.
(a) Confocal reconstructions of HUVEC stained for von Willebrand factor (VWF) (green) and Hoechst (blue) for three conditions: control (untreated), eGFP, and nsp5_c145a, scale bar: 20 µm; (b) analysis of VWF expression levels; (c) fold change of interleukin (IL)-6 in response to the different proteins.
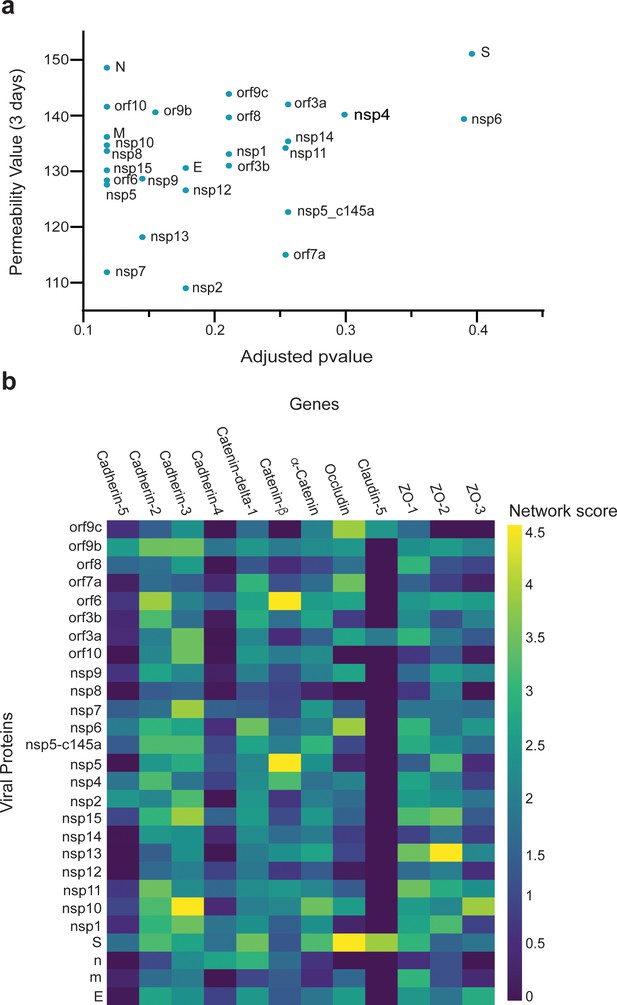
Correlation between viral protein effect on permeability and proximity to permeability-related proteins in a protein–protein interaction (PPI) network.
(a) Correlation of adjusted p-value versus permeability (Pearson = 0.295); (b) proximity between vascular proteins and the viral proteins.
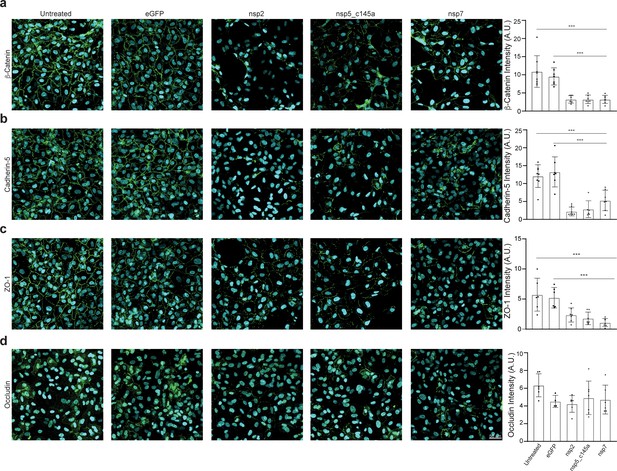
Tight-junctions impairment by severe acute respiratory syndrome (SARS)-CoV-2 proteins.
Immunocytochemistry (ICC) for (a) β-catenin (green), (b) cadherin-5 (green), (c) ZO-1 (green), (d) occludin (green), and Hoechst (blue) for five specified conditions: untreated, eGFP, nsp2, nsp5-c1451, and nsp7. Relative quantification is shown for each tight junction (TJ) protein for all five conditions on the right. Scale bar: 50 µm.
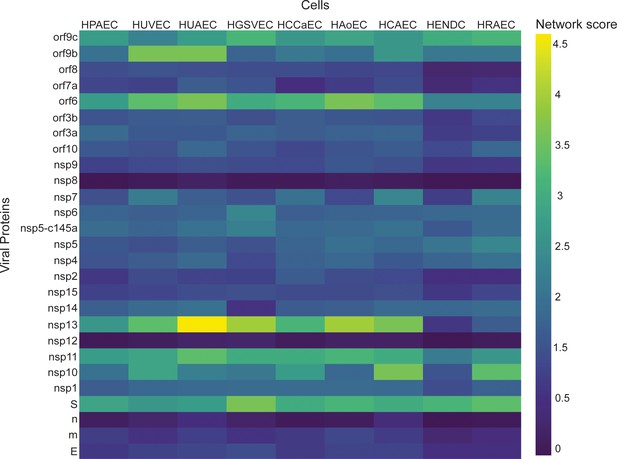
Correlation between viral proteins and different types of vascular endothelial cells.
Correlation of adjusted p-value between vascular proteins identified in vascular endothelial cells and the viral proteins.
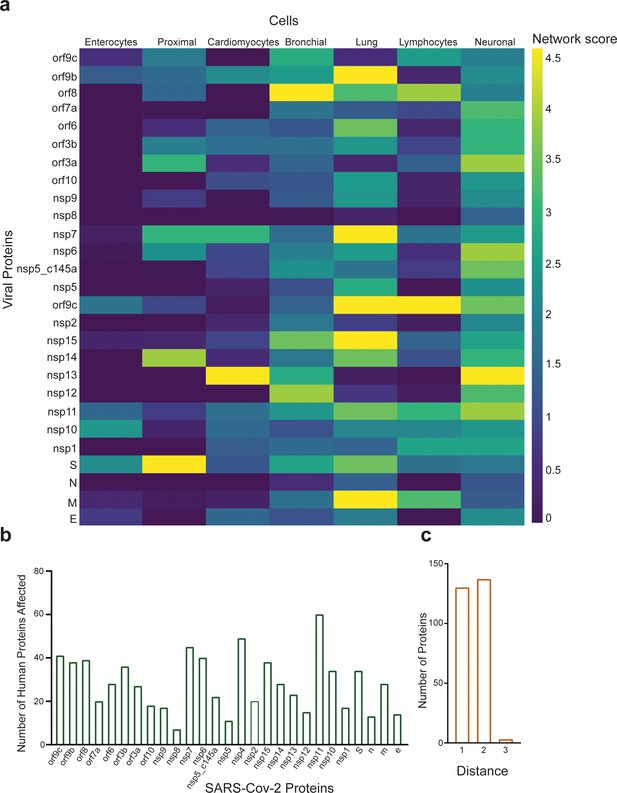
Protein identification using protein–protein interaction (PPI).
(a) PPI results for the severe acute respiratory syndrome (SARS)-CoV-2 proteins that have a significant effect on the proteins presented in SI Table 1 for each system; (b) number of proteins affected by each SARS-CoV-2 protein, as calculated by PPI; (c) number of proteins with a specific distance factor from the viral proteins (also shown in SI Table 1).
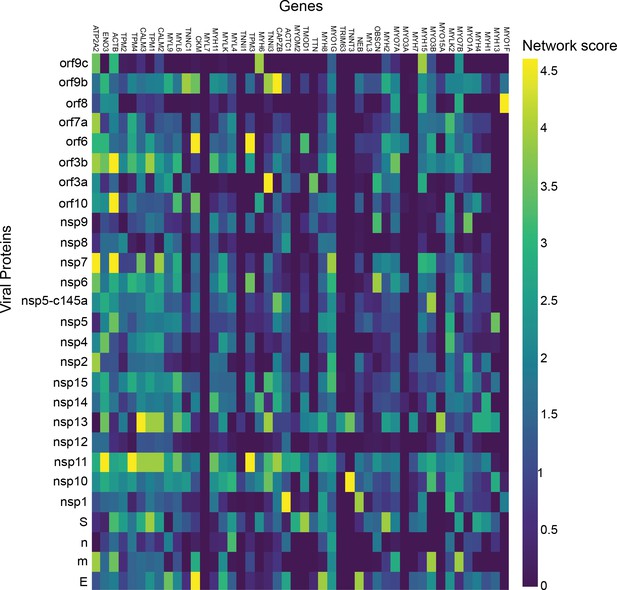
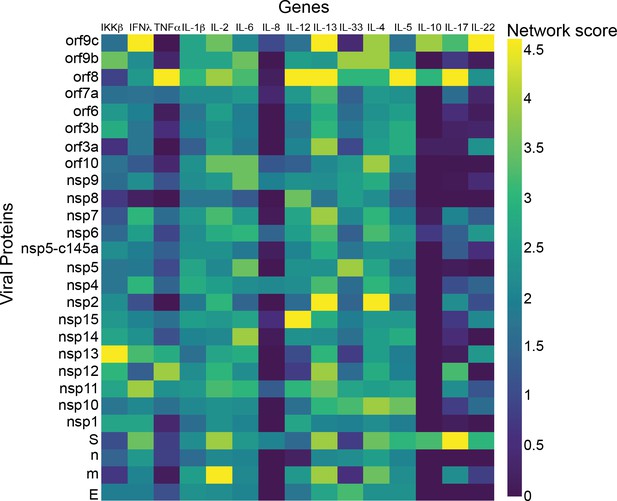
Cardiomyocytes.
p-Value correlation of a target protein with a specific viral protein, calculated empirically using 100 random samples.
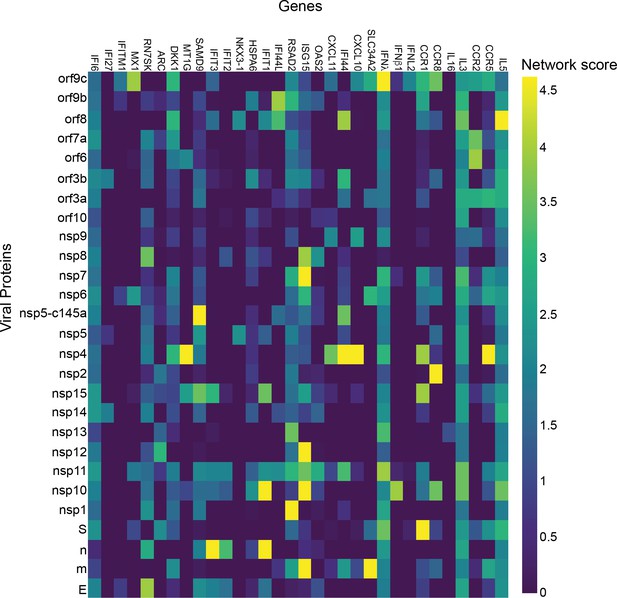
Enterocytes.
p-Value correlation of a target protein with a specific viral protein, calculated empirically using 100 random samples.
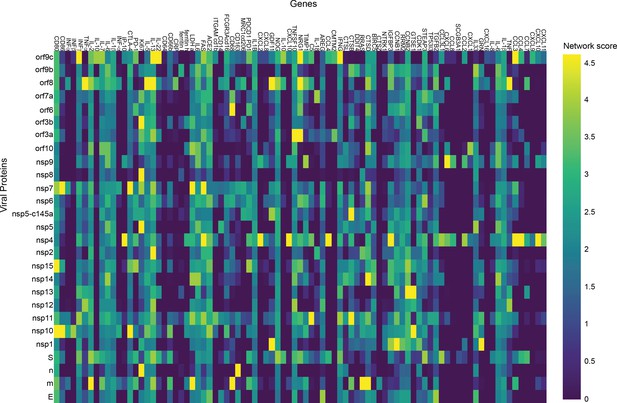
Lymphocytes.
p-Value correlation of a target protein with a specific viral protein, calculated empirically using 100 random samples.
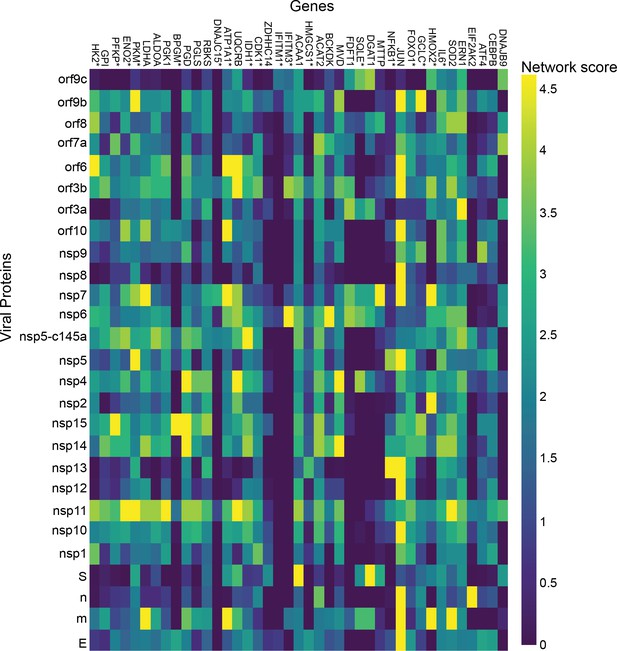
Lung.
p-Value correlation of a target protein with a specific viral protein, calculated empirically using 100 random samples.
Bronchial.
p-Value correlation of a target protein with a specific viral protein, calculated empirically using 100 random samples.
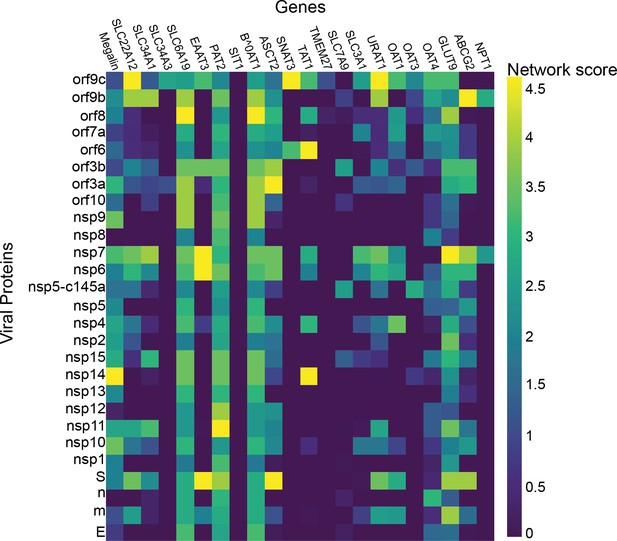
Proximal tubule cells.
p-Value correlation of a target protein with a specific viral protein, calculated empirically using 100 random samples.
Tables
Severe acute respiratory syndrome (SARS)-CoV-2 proteins.
| SARS-CoV-2 proteins | General impact |
|---|---|
| Structural proteins | |
| S (spike) | Spike protein, mediates binding to ACE2, fusion with host membrane Surface glycoprotein, needs to be processed by cellular protease TMPRSS2 (Gordon et al., 2020) |
| M (membrane) | Membrane glycoprotein, the predominant component of the envelope A major driver for virus assembly and budding (Gordon et al., 2020) |
| E (envelope) | Envelope protein, involved in virus morphogenesis and assembly Coexpression of M and E is sufficient for virus-like particle formation and release (Gordon et al., 2020) |
| N (nucleocapsid) | Nucleocapsid phosphoprotein binds to RNA genome (Gordon et al., 2020) |
| Nonstructural proteins | |
| nsp1 | Leader sequence, suppresses host antiviral response Antagonizes interferon induction to suppress host antiviral response (Gordon et al., 2020) |
| nsp2 | Interferes with host cell signaling, including cell cycle, cell-death pathways, and cell differentiation May serve as an adaptor for nsp3 Not essential for virus replication, but deletion of nsp2 diminishes viral growth and RNA synthesis (Gordon et al., 2020; Procko, 2020) |
| nsp3 | nsp3–nsp4–nsp6 complex involved in viral replication Functions as papain-like protease (Gordon et al., 2020) |
| nsp4 | nsp3–nsp4–nsp6 complex involved in viral replication (Gordon et al., 2020) The complex is predicted to nucleate and anchor viral replication complexes on double-membrane vesicles in the cytoplasm (mitochondria) |
| nsp5 | Inhibits interferon I signaling processes by intervening in the NF-κB process and breaking down STAT one transcription factor Functions as 3-chymotrypsin-like protease, cleaves the viral polyprotein (Gordon et al., 2020) |
| nsp5_c145a | Catalytic dead mutant of nsp5 (Gordon et al., 2020) |
| nsp6 | nsp3–nsp4–nsp6 complex involved in viral replication Limits autophagosome expansion Components of the mitochondrial complex V (the complex regenerates ATP from ADP) copurify with nsp6 (Gordon et al., 2020) |
| nsp7 | Cofactor of nsp12 nsp7–nsp8 complex in part of RNA polymerase (nsp7, 8, 12 – replication complex)Affects electron transport, GPCR signaling, and membrane trafficking (Gordon et al., 2020; Peng et al., 2020b; Romano et al., 2020; Hillen et al., 2020) |
| nsp8 | Cofactor of nsp12 nsp7–nsp8 complex in part of RNA polymerase. Affects the signal recognition particle and mitochondrial ribosome (Gordon et al., 2020; Peng et al., 2020b; Romano et al., 2020; Chi et al., 2003) |
| nsp9 | ssRNA binding protein (can bind both DNA and RNA, but prefers ssRNA) Interacts with the replication complex (nsp7, 8, 12) (Cornillez-Ty et al., 2009) |
| nsp10 | Cofactor of nsp16 and nsp14 (Romano et al., 2020) Essential for nsp16 methyltransferase activity (stimulator of nsp16) Zinc finger protein essential for replication (Gordon et al., 2020; Peng et al., 2020b) |
| nsp11 | Unknown function |
| nsp12 | Functions as an RNA-direct RNA polymerase, the catalytic subunitAffects the spliceosome (Gordon et al., 2020; Peng et al., 2020b; Romano et al., 2020; Hillen et al., 2020) |
| nsp13 | Has helicase and 5’ triphosphatase activity Initiates the first step in viral mRNA capping nsp13,14,16 installs the cap structure onto viral mRNA in the cytoplasm instead of in the nucleus, where the host mRNA is capped (Gordon et al., 2020; Peng et al., 2020b; Romano et al., 2020; Ivanov et al., 2004) |
| nsp14 | In addition to the capping function of the methyltransferase, nsp14 is also an endonuclease (3’–5’ exoribonuclease) that corrects mutations during genome replication (Gordon et al., 2020; Peng et al., 2020b; Romano et al., 2020) |
| nsp15 | Endoribonuclease has uridine-specific endonuclease activity, essential for viral RNA synthesis (Gordon et al., 2020; Romano et al., 2020) |
| nsp16 | May involve complexation with nsp10 and nsp14, for stabilization of homoenzyme, for capping the mRNA (Gordon et al., 2020; Peng et al., 2020b; Romano et al., 2020) |
| Open reading frame (accessory factors) | |
| orf3a | Packaging into virions Mediates trafficking of spike protein by providing ER/golgi retention signals Induces IL-6b, activates NF-κB, activates the NLRP3 inflammasome (Gordon et al., 2020) |
| orf3b | Interferon antagonist and involved in pathogenesis (Gordon et al., 2020) |
| orf6 | Type I interferon antagonist, suppresses the induction of interferon, and interferon signaling pathways (Gordon et al., 2020) |
| orf7a | May be related to viral-induced apoptosis (Gordon et al., 2020) |
| orf7b | Unknown function |
| orf8 | Recombination hotspot Induces ER stress and activates NLRP3 inflammasomes Low similarity to SAR-CoV (Gordon et al., 2020) |
| orf9b | Suppresses host antiviral response Targets the mitochondrion-associated adaptor molecules MAVS and limits host cell interferon responses (Gordon et al., 2020) |
| orf9c | No evidence that this protein is expressed during SARS-CoV-2 infection (Gordon et al., 2020) |
| orf10 | No evidence that this protein is expressed during SARS-CoV-2 infection (Gordon et al., 2020) |
Comparison of tight junction (TJ) proteins expression among different types of vascular endothelial cells.
Additional files
-
Supplementary file 1
Protein change by SARS-Cov-2.
(A) Documented change by severe acute respiratory syndrome (SARS)-Cov-2. (B) Significant target for each viral protein.
- https://cdn.elifesciences.org/articles/69314/elife-69314-supp1-v1.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/69314/elife-69314-transrepform1-v1.docx
-
Source data 1
The source data file contains all the data that was used for Figure 2, Figure 3, Figure 5, and Figure 2—figure supplement 1b.
- https://cdn.elifesciences.org/articles/69314/elife-69314-supp2-v1.xlsx