The need for high-quality oocyte mitochondria at extreme ploidy dictates mammalian germline development
Figures
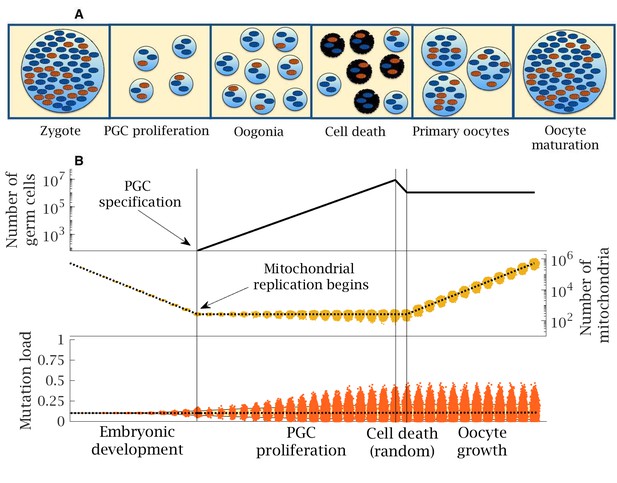
Stages in female germline development.
(A) Timeline of human oocyte development showing the main stages modeled, with wildtype (blue) and mutant mitochondria (orange). (B) Numerical simulation of the base model. Top panel: number of germ cells from specification of the 32 primordial germ cells (PGCs) after 12 cell divisions; proliferation to form 8 million oogonia; random cell death reducing to 1 million primary oocytes; quiescent period (not shown) and finally oocyte maturation at puberty. Middle panel: copy number of mitochondria (i.e. mtDNA); from zygote with ~500,000 copies, which are partitioned at cell division during early embryo development until replication begins (first vertical line) during PGC proliferation; copy number is amplified during oocyte maturation back to ~500,000 copies; dotted line shows the mean mitochondria copy number, with the distribution across oocytes shown in yellow. Note, skew reflects the log-scale. Bottom panel: mean (dotted line) and distribution of mutation load through development. The yellow shaded area shows the 90% quantile. Other parameter values , .
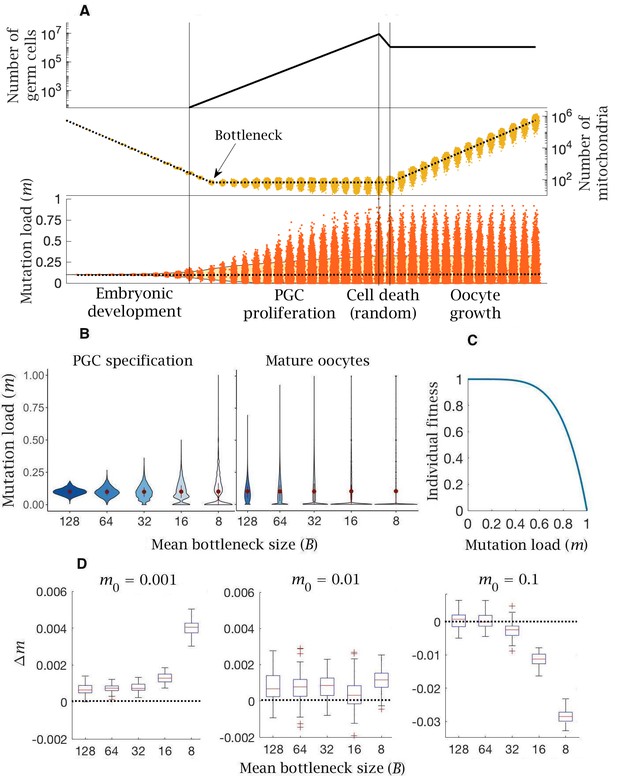
Model of germline bottleneck and individual selection.
(A) A bottleneck with two extra rounds of cell division without replication (cell division 13 and 14; after the first vertical line), reducing mitochondria copy number per PGC (by a quarter on average). Two extra rounds of mitochondrial replication are required to regenerate the copy number in mature oocytes. Compared to the base model (Figure 1), mean mutation load (dotted line, bottom panel) is slightly higher and variation in load is substantially greater (yellow shaded area, 90% quantile). Parameter values , . (B) Violin plots of the distribution of mutations (mean ± SD shown in red) at two developmental stages, PGC specification and mature oocytes, given 5 mean bottleneck sizes () when . (C) Strength of selection on individual fitness, with a concave fitness function based on clinical data from mitochondrial diseases. (D) Change in mutation load () across a single generation for three initial mutation loads (), given 5 mean bottleneck sizes (), showing the median (red line) and distribution (box plot IQR with min/max whiskers and outliers).
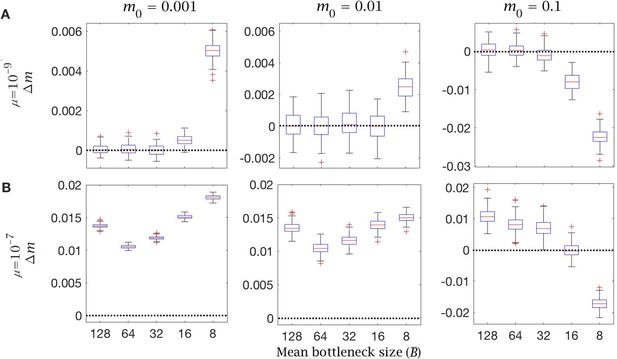
Bottleneck change in mutation load with different mutation rates.
Change in mutation load () across a single generation after individual selection with variable mean bottleneck size (). This is shown with (A) low () and (B) high () mutation rate, for individuals with low (), medium (), and high () initial mutation loads. Box plots show the median (red line) and distribution (box plot IQR with min/max whiskers and outliers).
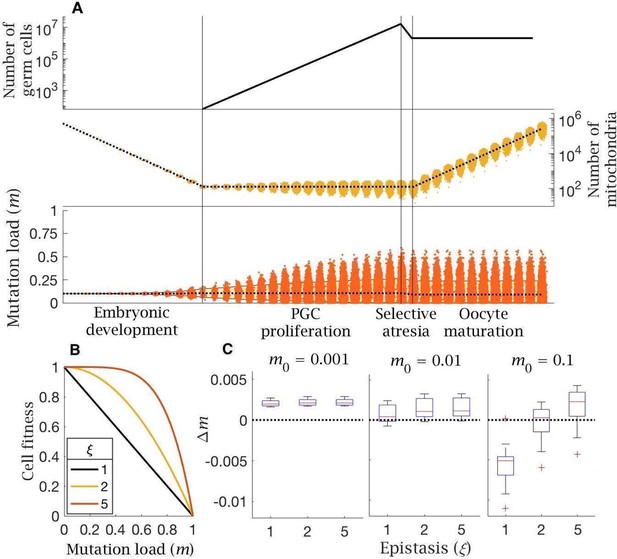
Model of follicular atresia and cell selection.
(A) After PGC proliferation, follicular atresia occurs through selective apoptosis of oogonia. (B) Cell fitness is assumed to be linear () or follow negative epistasis ( in which mutations are more deleterious in combination. (C) Change in mutation load, , across a single generation after cell selection, at an intermediate mutation rate (), for individuals with low (), medium () and high () initial mutation loads, for variable levels of epistasis (median (red line) and distribution (box plot IQR with min/max whiskers and outliers)).
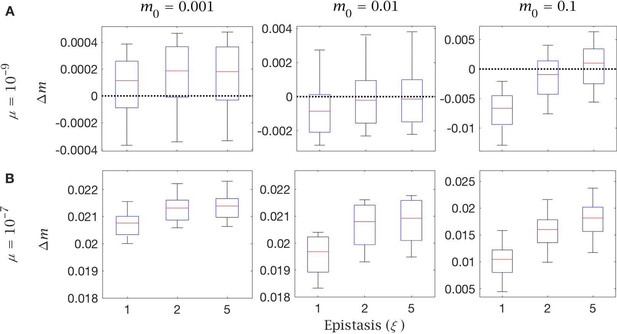
Follicular atresia and cell selection change in mutation load with different mutation rates.
Change in mutation load () across a single generation after cell selection with variable levels of epistasis (). This is shown with (A) low () and (B) high () mutation rate, for individuals with low (), medium () and high () initial mutation loads. Box plots show the median (red line) and distribution (box plot IQR with min/max whiskers and outliers).
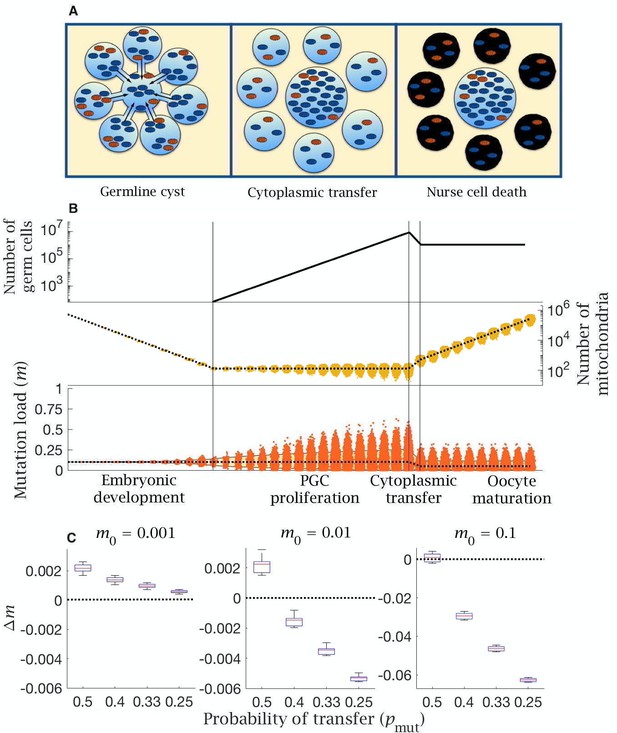
Model of cytoplasmic transfer and mitochondria selection.
(A) Cytoplasmic bridges form among oogonia in the germline cyst, leading to selective transfer of wild-type mitochondria (blue) to the primary oocyte, leaving mutant mitochondria (red) in nurse cells that then undergo apoptosis. (B) Cytoplasmic transfer which selectively pools of mtDNA from eight germline cyst cells into a single primary oocyte causes a large increase in the number of mitochondria (middle panel) and a large reduction in the mean (dotted line, bottom panel) and distribution of mutation load (yellow shaded area shows the 90% quantile, bottom panel), which persists during oocyte maturation. Pooling of mtDNA requires two fewer rounds of mtDNA replication to regenerate copy number in mature oocytes. Parameter values , . (C) Change in mutation load () across a single generation (median (red line) and distribution (box plot IQR with min/max whiskers and outliers)), for individuals with low (), medium (), and high () initial mutation loads, with variable strengths of selective transfer (). Parameter value , .
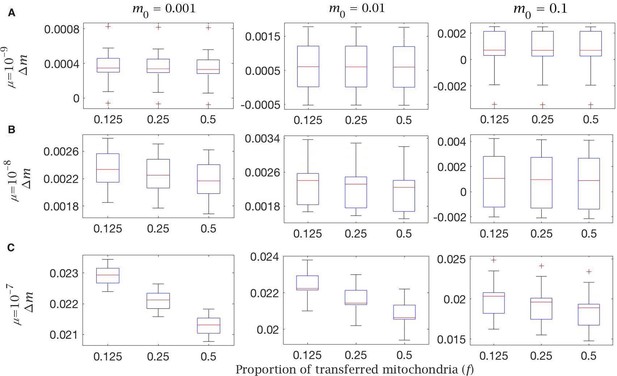
Cytoplasmic transfer and mitochondria selection change in mutation load with different mutation rates and proportion of transferred mitochondria ().
Change in mutation load () across a single generation, given a variable proportion of transferred mitochondria () to the Balbiani body when transfer is non-selective (). This is shown with (A) low (), (B), standard () and (C) high () mutation rate, for individuals with low (), medium (), and high () initial mutation loads. Box plots show the median (red line) and distribution (box plot IQR with min/max whiskers and outliers).
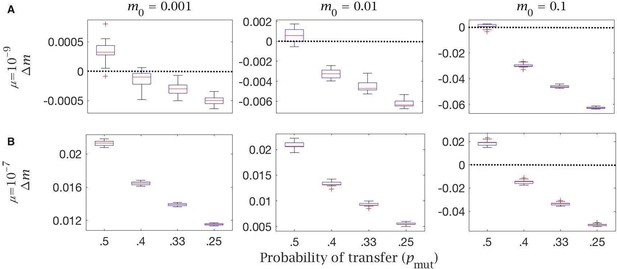
Cytoplasmic transfer and mitochondria selection change in mutation load with different mutation rates and probability of mutant transfer ().
Change in mutation load () across a single generation, for individuals undergoing cytoplasmic transfer the Balbiani body with variable strength of selection, given a fixed probability of transfer of wild-type mitochondria () and a decreasing probability of transfer of mutant mitochondria (). Note the null case is when (). This is shown with (A) low () and (B) high () mutation rate, for individuals with low (), medium (), and high () initial mutation loads, and a fixed proportion of transferred mitochondria (). Box plots show the median (red line) and distribution (box plot IQR with min/max whiskers and outliers).
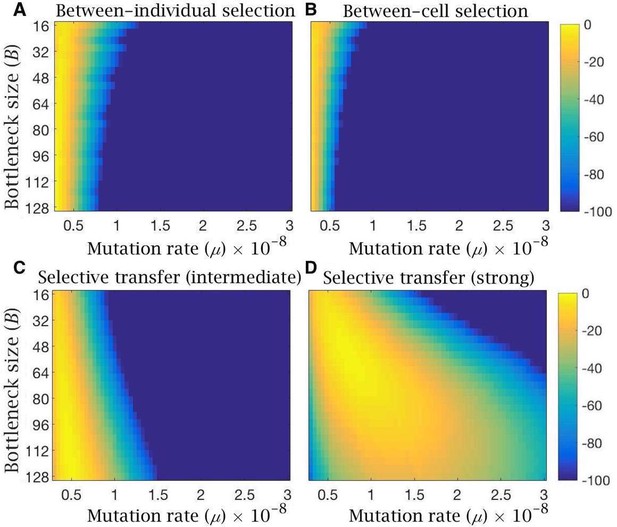
Log-likelihood of the models reproducing clinical data of mitochondria mutation load and disease frequency.
Heatmaps showing log-likelihood of reproducing the observed mutation load and disease frequency in humans, for equilibrium conditions under the evolutionary model with (A) bottleneck and selection on individuals, (B) follicular atresia and selection on cells (), (C) cytoplasmic transfer with intermediate (, ,) or (D) strong (, , ) selective transfer of wild-type mitochondria. Yellow depicts high likelihood; blue, low likelihood. All models are shown for variable bottleneck size (the minimum mitochondria population size at which replication commences) and variable mutation rates.
Tables
Parameter and variable symbols and values.
| Maximum number of germ cells | |
| mtDNA number in mature oocytes | |
| Minimum mtDNA ploidy | |
| Final number of germ cells | |
| Initial mutation load | |
| Mutation rate per bp per cell division | μ |
| Strength of epistatic interactions | |
| Transfer probability of mutant mtDNA | |
| Transfer probability of wildtype mtDNA | |
| Human mitochondrial genome size | bp (Palca, 1990) |





