Exosome component 1 cleaves single-stranded DNA and sensitizes human kidney renal clear cell carcinoma cells to poly(ADP-ribose) polymerase inhibitor
Figures
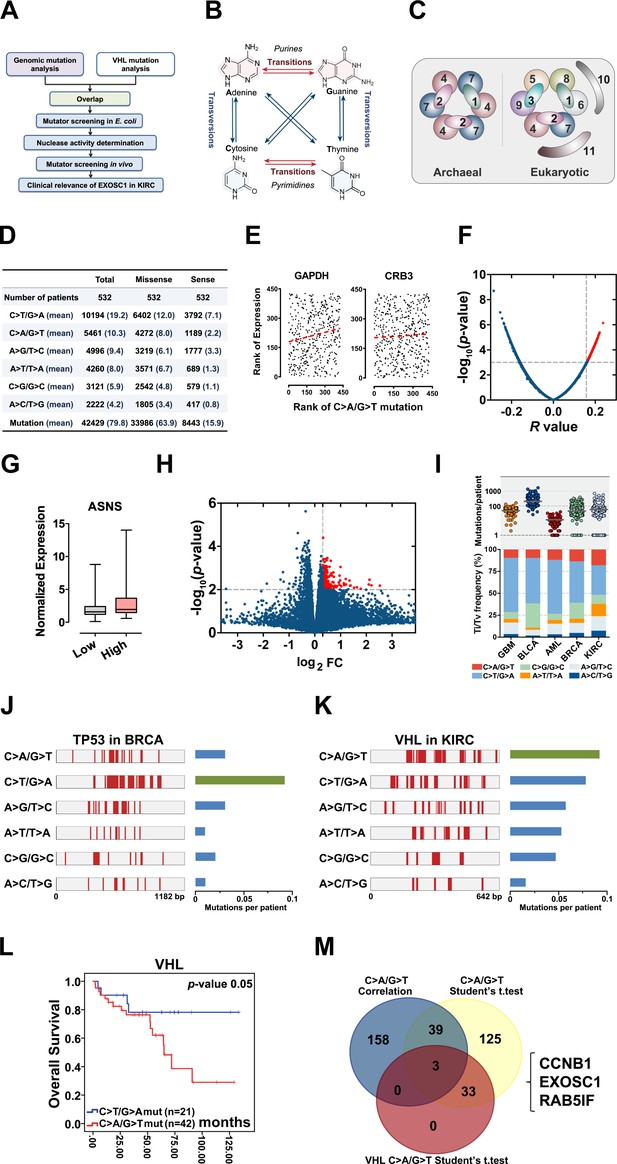
Identification of candidate ESMs in KIRC by statistical analyses.
(A) Schematic of this study. (B) Illustration of base substitutions. (C) Schematic showing the archaeal and eukaryotic exosome complexes viewed from the top. (D) Summary statistics for the six types of c-substitutions in KIRC. (E) Scatter plots showing the correlation between the rank of mutation and gene expression. Each plot represents one KIRC sample. The orange dashed line shows the best fit for visualization. P values were calculated by Spearman’s rank correlation. (F) Volcano plots of p and r values calculated by Spearman's correlation analyses. Each plot represents one gene. The top 1% of genes were taken as candidates and marked in red. (G) Box plots showing ASNS expression in the high and low C>A/G>T mutation groups. The expression was normalized to TBP. (H) Volcano plots showing the p and fold change (FC) values calculated by the two-tailed Student’s t-test. Each plot represents one gene. FC was calculated by the formula: FC=the mean gene expression in the high group/that in the low group. The top 1% of genes were taken as candidates and marked in red. (I) C-substitution mutation frequencies in five types of major cancers. (J, K) Mutation spectra of the TP53 gene in BRCA (J) and VHL gene in KIRC (L). (L) Kaplan-Meier (KM) analyses of OS between VHL C>A/G>T and C>T/G>A mutation groups. The median OSs in the C>A/G>T and C>T/G>A groups were 72.95 and 108.91 months, respectively. The p value was obtained from the log-rank test. (M) Venn diagram showing the overlap of the candidate genes identified by three types of statistical analyses as noted. ESM, endogenous source of mutation; KIRC, kidney renal clear cell carcinoma; OS, overall survival.
-
Figure 1—source data 1
Identification of candidate ESMs in KIRC by statistical analyses.
- https://cdn.elifesciences.org/articles/69454/elife-69454-fig1-data1-v2.xlsx
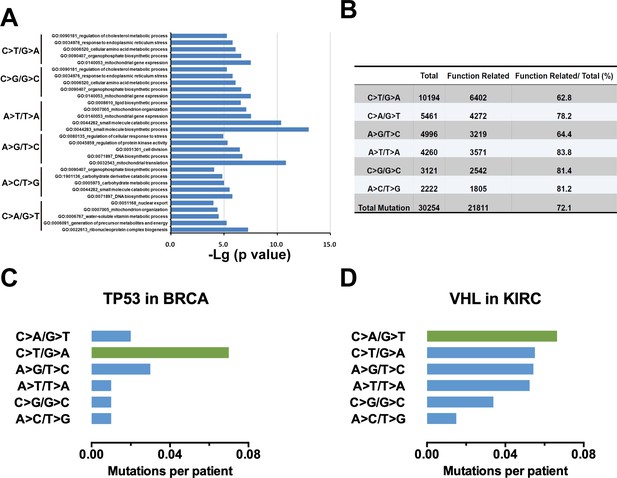
Identification of candidate ESMs in KIRC by statistical analyses.
(A) The top-ranked 200 genes of each c-substitution type (Supplementary file 1) were applied for Gene Ontology (GO) enrichment analyses by Metascape (metascape.org). (B) Summary statistics for the information of function-related mutations in KIRC. (C, D) Mutation spectra of TP53 (C) and VHL (D) normalized by the nucleotide abundance. ESM, endogenous source of mutation.
-
Figure 1—figure supplement 1—source data 1
Identification of candidate ESMs in KIRC by statistical analyses.
- https://cdn.elifesciences.org/articles/69454/elife-69454-fig1-figsupp1-data1-v2.xlsx
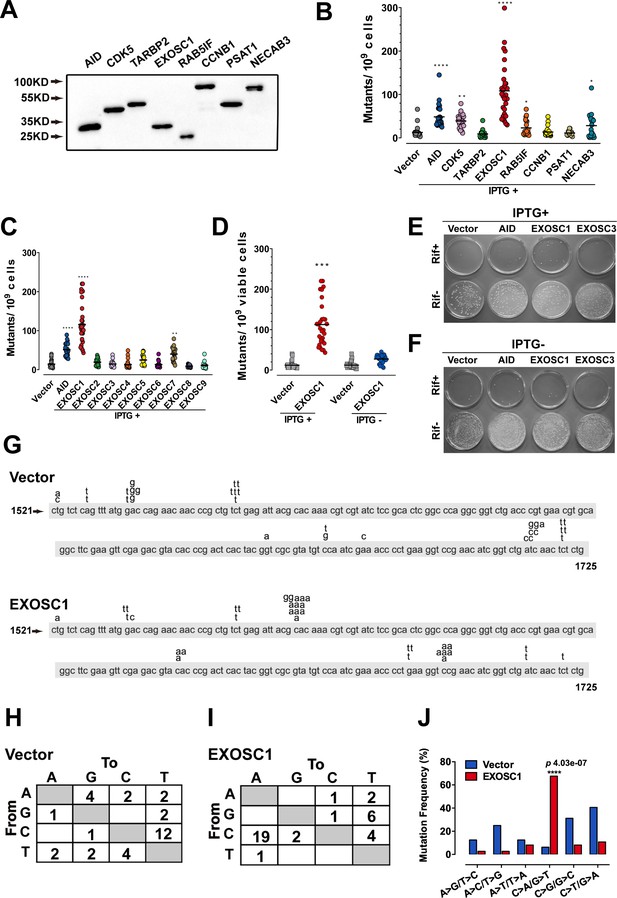
EXOSC1 promotes mutations in Escherichia coli.
(A) Western blot showing His-tagged protein levels in E. coli. (B, C) The frequencies of the RifR mutation in the E. coli cells expressing candidate ESMs (B) or exosome family members (C). The vector and AID were used as negative and positive controls, respectively. Each plot represents the mutational frequency of an independent overnight culture (n=30). Median mutational frequency of the gene is noted. (D) Frequencies of the RifR mutation in the E. coli cells treated with and without IPTG (n=30). (E, F) Representative images of E. coli cells treated with (E) and without (F) IPTG. (G) The mutational distribution in 25 independent RifR colonies transformed by vector or EXOSC1. (H, I) Summary of the c-substitutions in RifR colonies transformed by vector (H) and EXOSC1 (I). (J) The mutational frequencies of each c-substitution in RifR colonies. The p value was calculated by Fisher's exact test.
-
Figure 2—source data 1
EXOSC1 promotes mutations in E. coli.
- https://cdn.elifesciences.org/articles/69454/elife-69454-fig2-data1-v2.xlsx
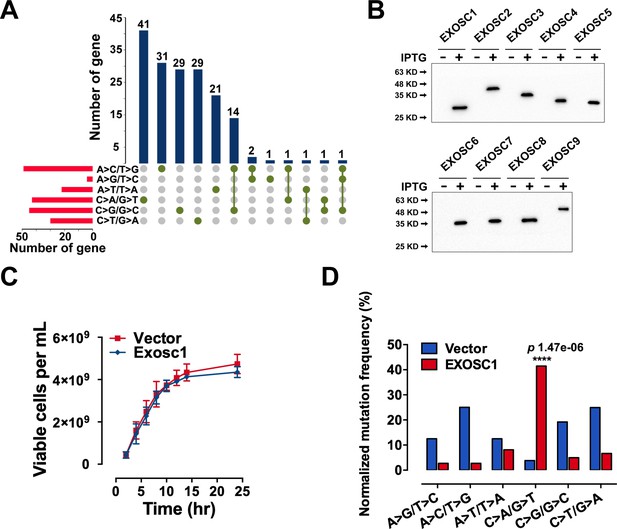
EXOSC1 promotes mutations in Escherichia coli.
(A) Setup plot showing the overlap numbers of the candidate genes of the six types of c-substitutions. More details can be found in Supplementary file 4. (B) Western blot showing the His-tagged protein levels of the exosome complex members in the E. coli cells. (C) Growth rates of control and EXOSC1-transformed cells in the presence of IPTG (n=5). (D) Frequencies of c-substitutions in RifR colonies were normalized to the nucleotide abundances. The p value was calculated by Fisher's exact test.
-
Figure 2—figure supplement 1—source data 1
EXOSC1 promotes mutations in E. coli.
- https://cdn.elifesciences.org/articles/69454/elife-69454-fig2-figsupp1-data1-v2.xlsx
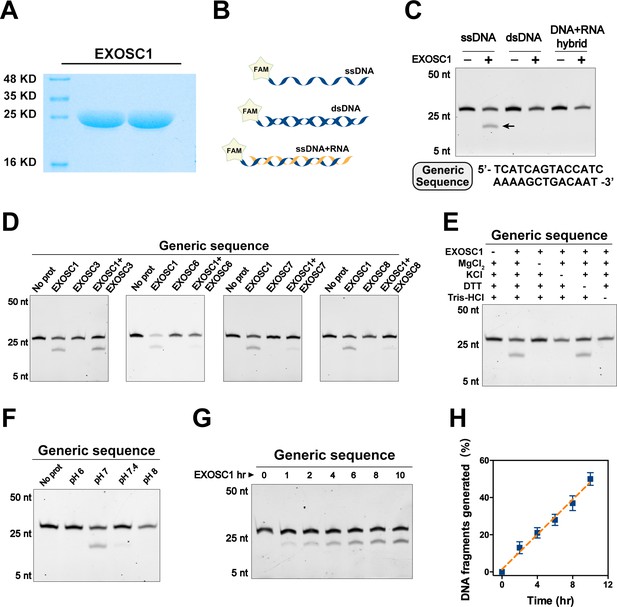
EXOSC1 cleaves ssDNA.
(A) Coomassie blue staining of in vitro purified EXOSC1 protein. (B) Schematic of synthetic DNA substrates. (C) In vitro cleavage assays of EXOSC1 using generic ssDNA, dsDNA, and DNA-RNA hybrid as substrates. The assays were performed in the presence of 1 μM oligonucleotides, 1 μM EXOSC1, 70 mM KCl, 700 μM MgCl2, 1 mM DTT, and 20 mM Tris–HCl pH 7.0. After incubation at 37°C for 4 hr, resultant samples were analyzed by 15% polyacrylamide TBE-urea gels. (D) Cleavage assays of EXOSC1 in the presence or absence of EXOSC3, EXOSC6, EXOSC7, and EXOSC8 using generic ssDNA as substrates. (E) Cleavage assays in the presence of the components as noted. (F) Cleavage assays of EXOSC1 at the pH as noted. (G) Time course cleavage assays of EXOSC1 using generic ssDNA as substrates. (H) Rate curve of EXOSC1 cleavage at 37°C and pH 7.0. dsDNA, double-stranded DNA; ssDNA, single-stranded DNA.
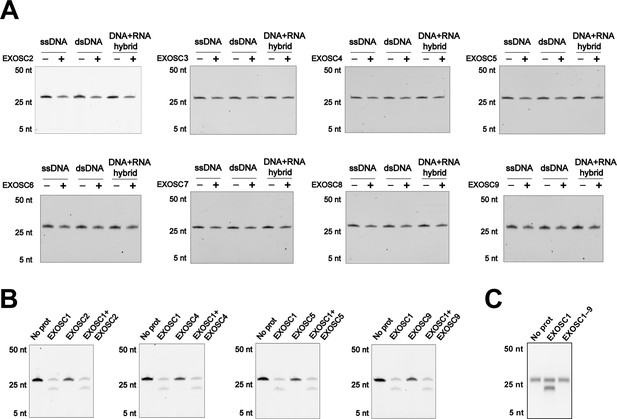
EXOSC1 cleaves ssDNA.
(A) In vitro cleavage assays of EXOSC1 homologs (EXOSC2–EXOSC9) using generic ssDNA, dsDNA, and DNA-RNA hybrid as substrates. (B) Cleavage assays of EXOSC1 in the presence/absence of EXOSC2, EXOSC4, EXOSC5, or EXOSC9. (C) Cleavage assays of exosome complex (EXOSC1–9) using generic ssDNA. dsDNA, double-stranded DNA; ssDNA, single-stranded DNA.
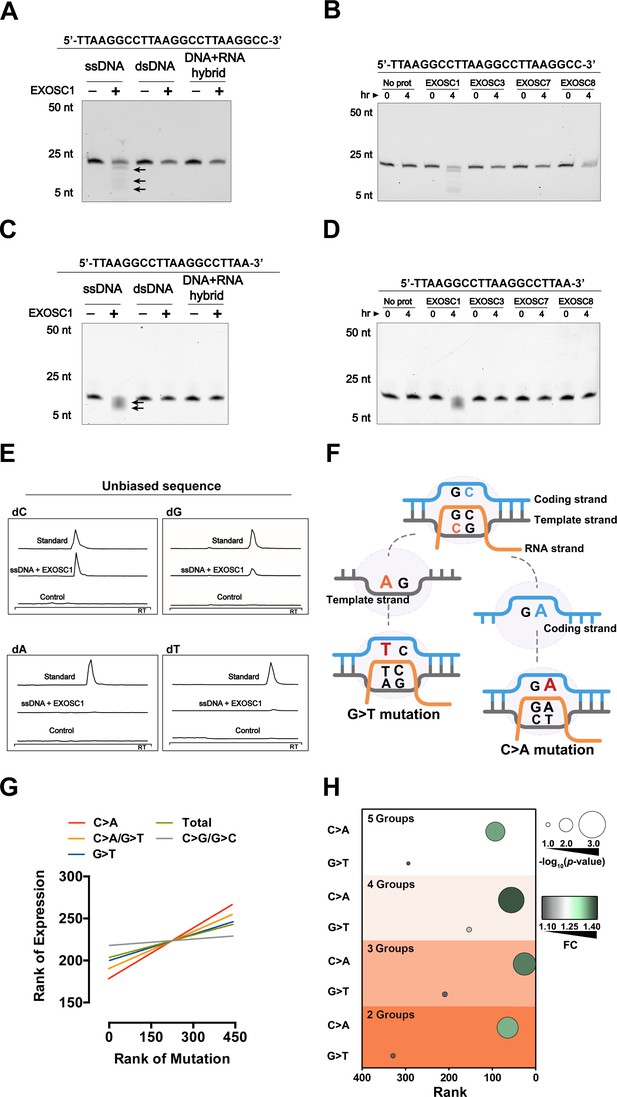
EXOSC1 prefers to cleave C sites in ssDNA.
(A, B) Cleavage assay of EXOSC1 using unbiased DNA, dsDNA, and DNA-RNA hybrid as substrates. (C, D) Cleavage assays of EXOSC1, EXOSC3, EXOSC7 and EXOSC8 using unbiased ssDNA as substrates. (E) Mass spectrometry (MS) analyses of the resultant mixtures described in (B). C, G, A, and T were detected by MS using nucleoside to base ion mass transitions of 228.1–112.2 (C), 268.1–152.1 (G), 252.2–136.1 (A), and 243.1–127.2 (T), respectively (Figure 3—figure supplement 1E). Standard curves were generated by a serial dilution of C, G, A, and T (Figure 3—figure supplement 1F). Free C, G, A, and T contained in the reaction mixtures were quantified by standard curves. (F) Schematic showing that the consequence of C>A mutation in the coding strand is distinct from that in the template strand. (G) Correlation between EXOSC1 expression and the c-substitution mutation as noted in KIRC. Each line represents one best fit for visualization. P values were from Spearman’s rank correlation. P and r values of C>A mutations were 0.0001 and 0.19, respectively; C>A/G>T: p=0.0006, r=0.17; G>T: p=0.0271, r=0.10; total mutations: p=0.0594, r=0.09; C/G>G/C: p=0.5730, r=0.03. (H) Student’s t-test analyses of the expression difference of EXOSC1 between the high and low mutation groups. 2, 3, 4, and 5 represent the group numbers. For example, 4: the KIRC patients were grouped into four groups and the expression difference of EXOSC1 between the high and low mutation groups was analyzed by Student’s t-tests. FC = mean gene expression in the high group/that in the low group. dsDNA, double-stranded DNA; KIRC, kidney renal clear cell carcinoma; ssDNA, single-stranded DNA.
-
Figure 4—source data 1
EXOSC1 prefers to cleave C sites in ssDNA.
- https://cdn.elifesciences.org/articles/69454/elife-69454-fig4-data1-v2.xlsx
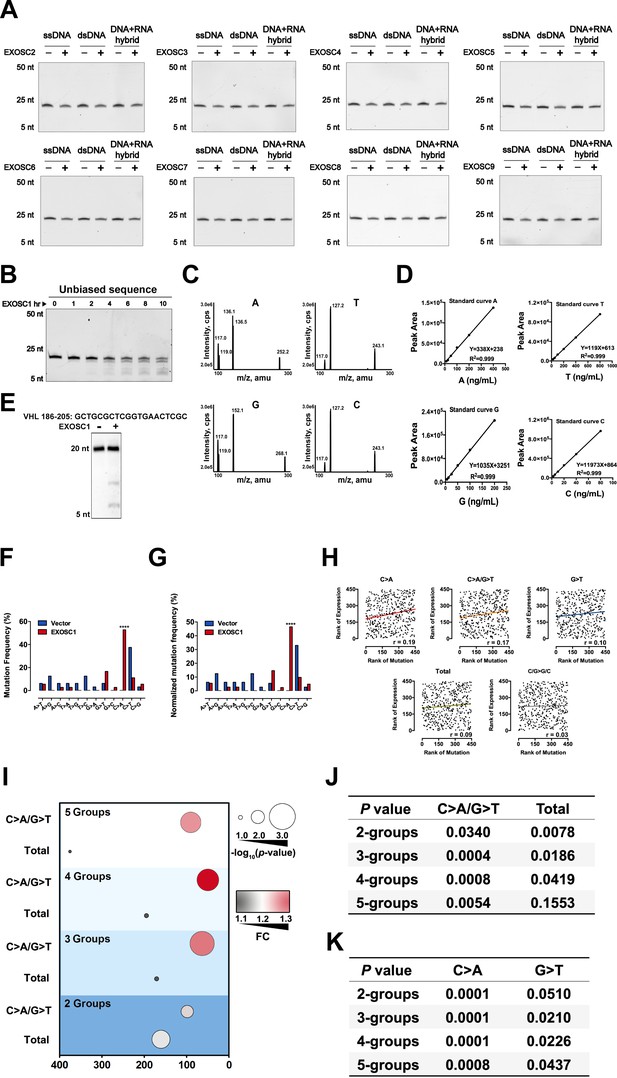
EXOSC1 prefers to cleave C sites in ssDNA.
(A) Cleavage assays of EXOSC2–EXOSC9 using unbiased ssDNA, dsDNA, and DNA-RNA hybrid as substrates. (B) Time course cleavage assays of EXOSC1 using unbiased ssDNA as substrates. (C) Base ion mass transitions for LC-MS/MS analyses of C, G, A, and T. Free nucleosides were detected using nucleoside to base ion mass transitions of 228.1–112.2 (C), 268.1–152.1 (G), 252.2–136.1 (A), and 243.1–127.2 (T). (D) LC-MS/MS standard curves of nucleoside as noted. (E) Cleavage assays of EXOSC1 using a 20 nt ssDNA containing hot spot of VHL mutation (186–205) as substrate. (F, G) Frequencies (F) and normalized frequencies (G) of 12 types of substitutions in RifR colonies. P values were calculated by Fisher's exact test. (H) Correlation between EXOSC1 expression and the mutation as noted in KIRC. Each plot represents one KIRC sample. (I) Student’s t-test analyses of EXOSC1 expression difference between the high and low mutation groups. The 532 KIRC patients were grouped into two, three, four, or five groups to evaluate the impact of group number. (J, K) Summary statistics for the mutation as noted in KIRC. P values were calculated by the two-tailed Student’s t-test. dsDNA, double-stranded DNA; KIRC, kidney renal clear cell carcinoma; ssDNA, single-stranded DNA.
-
Figure 4—figure supplement 1—source data 1
EXOSC1 prefers to cleave C sites in ssDNA.
- https://cdn.elifesciences.org/articles/69454/elife-69454-fig4-figsupp1-data1-v2.xlsx
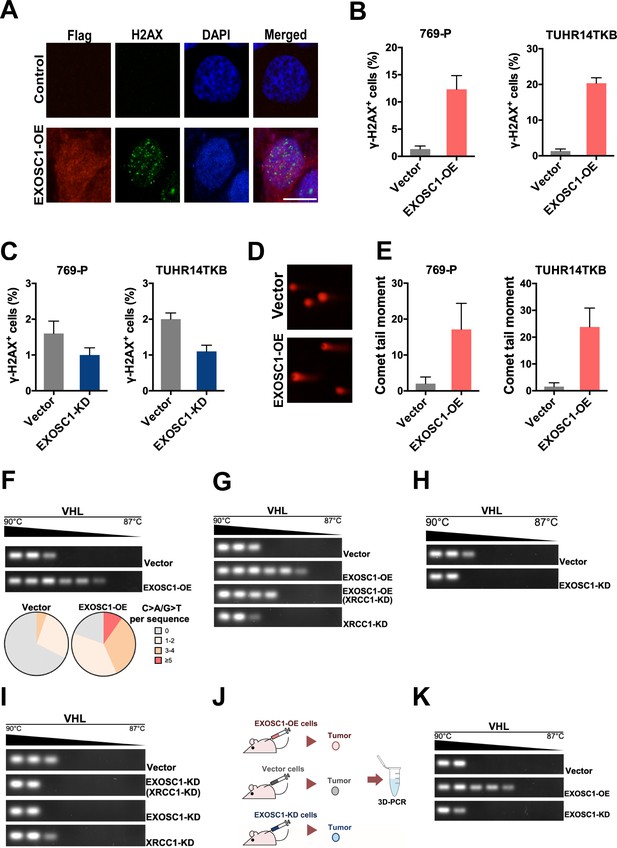
EXOSC1 enhances DNA damages and mutations in KIRC Cells.
(A) Representative fluorescent images of γ-H2AX foci in 769 P cells transfected with control (pCDH, empty vector) or pCDH-Flag EXOSC1 plasmids for 2 days. Scale bar=10 μm. (B) Percentage of cells with more than 20 γ-H2AX foci in the KIRC cells transfected with control or pCDH-Flag EXOSC1 plasmids for 2 days. (C) Percentage of cells with more than 20 γ-H2AX foci in the KIRC cells infected with lentivirus encoding shRNAi control (pLKO scramble) or pLKO sh-EXOSC1 for 2 days. (D) Representative images of DNA comets in 769 P cells transfected with control or pCDH-Flag EXOSC1 plasmids for 2 days. (E) Comet tail moment of the 769 P and TUHR14TKB cells transfected with control or pCDH-Flag EXOSC1 plasmids for 2 days. (F) 3D-PCR and subsequent sequencing analyses of the VHL mutations in the TUHR14TKB cells stably expressing control (vector) or EXOSC1 (pCDH-Flag EXOSC1, EXOSC1-OE). (G) 3D-PCR analyses of VHL in TUHR14TKB cells stably expressing control (pLKO.1 vector) or shRNA against EXOSC1 (pLKO shEXOSC1-1, EXOSC1-KD). (H) 3D-PCR analyses of VHL in TUHR14TKB cells stably expressing control, EXOSC1-OE and/or shRNA against XRCC1 (pMKO.1 shXRCC1-1, XRCC1-KD). (I) 3D-PCR analyses of VHL in TUHR14TKB cells stably expressing control (pLKO.1 vector), shRNA against EXOSC1 (pLKO shEXOSC1-1, EXOSC1-KD) and/or shRNA against XRCC1 (pMKO.1 shXRCC1-1, XRCC1-KD). (J) Schematic showing the subcutaneous xenograft tumor models. The 5 × 106 control, EXOSC1-OE and EXOSC1-KD 769 P cells were implanted subcutaneously. After 2 weeks, the resultant tumors were analyzed by 3D-PCR. (K) 3D-PCR analyses of VHL in the tumors described in (J). KIRC, kidney renal clear cell carcinoma.
-
Figure 5—source data 1
EXOSC1 enhances DNA damages and mutations in KIRC cells.
- https://cdn.elifesciences.org/articles/69454/elife-69454-fig5-data1-v2.xlsx
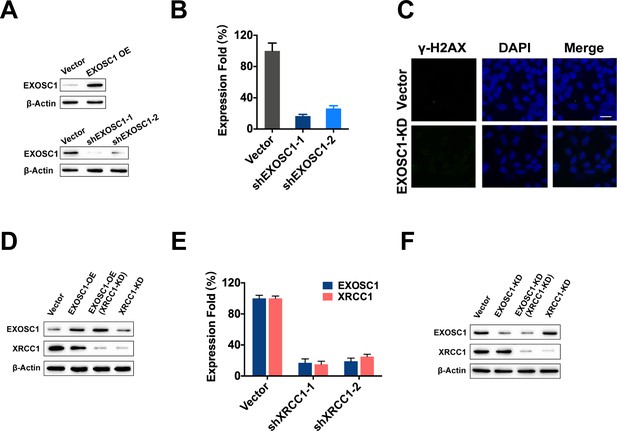
EXOSC1 enhances DNA damages and mutations in KIRC cells.
(A) Western blot analyses of EXOSC1 in stable control, enhanced EXOSC1, and EXOSC1 knockdown cells. (B) Quantitative PCR analyses of EXOSC1 mRNA in the 769 P cells stably expressing shRNA (pLKO EXOSC1-1 or pLKO EXOSC1-2) against EXOSC1. (C) Representative fluorescence images of γ-H2AX foci in the 769 P cells stably expressing shRNA against control (empty vector) or EXOSC1 (pLKO EXOSC1-1, EXOSC1-KD). Scale bar=10 μm. (D, F) Western blot analyses of EXOSC1 and XRCC1 protein levels in the stable cells as noted. (E) Quantitative PCR analyses of the mRNA in the cells stably expressing shRNA against EXOSC1 and/or XRCC1. KIRC, kidney renal clear cell carcinoma.
-
Figure 5—figure supplement 1—source data 1
EXOSC1 enhances DNA damages and mutations in KIRC cells.
- https://cdn.elifesciences.org/articles/69454/elife-69454-fig5-figsupp1-data1-v2.xlsx
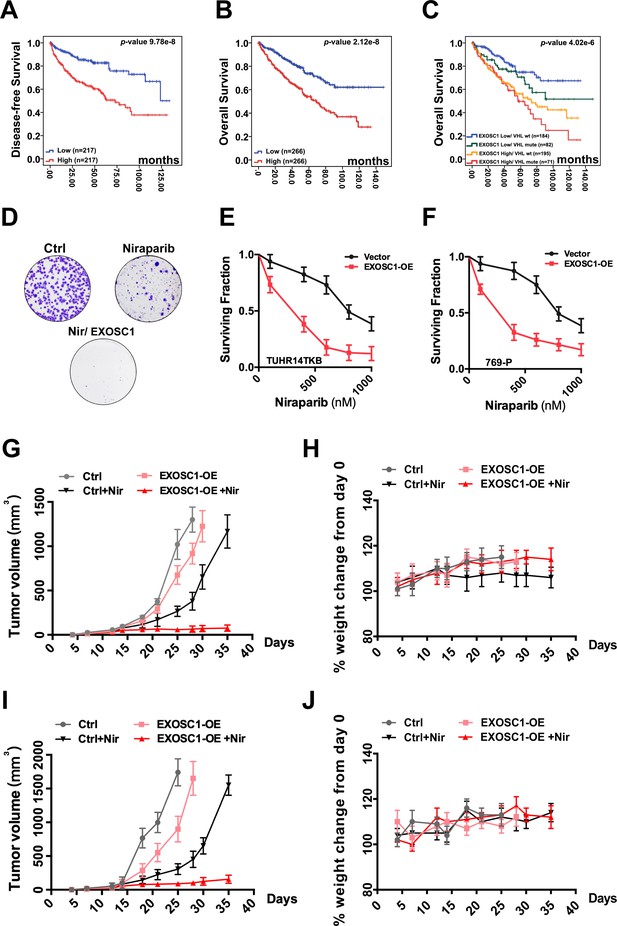
EXOSC1 sensitizes KIRC cells to PARP inhibitor.
(A, B) KM analyses of DFS (A) and OS (B) in KIRC patients with different EXOSC1 levels. P values were obtained from the log-rank test. (C) KM analysis of OS in KIRC patients with different EXOSC1 expression levels and VHL mutations. (D) Colony formation of control and EXOSC1-OE TUHR14TKB cells treated with 600 nM niraparib. (E, F) Clonogenic survival of control and EXOSC1-OE TUHR14TKB (E) and 769 P (F) cells treated with niraparib. (G, I) Tumor volumes of 769 P (G) and Caki-2 (I) xenografts treated with or without niraparib (n=4 groups; n=6 mice/group; ± SEM). (H, J) Body weight change percentage of 769 P (H) and Caki-2 (J) xenografts treated as described in (G). DFS, disease-free survival; KIRC, kidney renal clear cell carcinoma; KM, Kaplan-Meier; OS, overall survival.
-
Figure 6—source data 1
EXOSC1 sensitizes KIRC cells to PARP inhibitor.
- https://cdn.elifesciences.org/articles/69454/elife-69454-fig6-data1-v2.xlsx
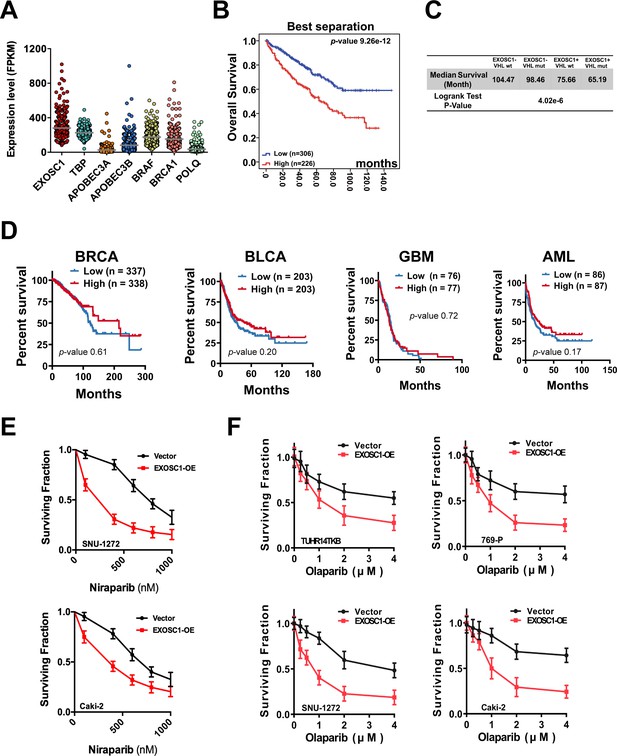
EXOSC1 sensitizes KIRC cells to PARP inhibitor.
(A) The fragments per kilobase per million mapped reads (FPKMs) of the noted gene in 532 KIRC patients. Each dot represents one KIRC patient. (B) Best separation KM analyses of OS in KIRC patients with different EXOSC1 levels. The median OS (low vs. high EXOSC1 group)=106.40 vs. 71.91 months. (C) Summary statistics for Figure 6C. (D) Median separation KM analyses of OS in the noted cancer patients with different EXOSC1 levels. (E) Clonogenic survival of control/EXOSC1-OE SNU-1272 and Caki-2 cells treated with niraparib. (F) Clonogenic survival of control/EXOSC1-OE KIRC cells treated with olaparib. KIRC, kidney renal clear cell carcinoma; KM, Kaplan-Meier; OS, overall survival.
-
Figure 6—figure supplement 1—source data 1
EXOSC1 sensitizes KIRC cells to PARP inhibitor.
- https://cdn.elifesciences.org/articles/69454/elife-69454-fig6-figsupp1-data1-v2.xlsx
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Anti-Flag (rabbit polyclonal) | Sigma-Aldrich | Cat. number F7425 (RRID:AB_439687) | Western blot (1:1000) |
| Antibody | Anti-His (rabbit polyclonal) | Sigma-Aldrich | Cat. number SAB1306085 | Western blot (1:1000) |
| Antibody | Anti-phospho-γ-H2AX (Ser139) (mouse monoclonal) | Millipore | Cat. number 05–636 (RRID:AB_309864) | IF (1:200) |
| Antibody | Anti-XRCC1 (rabbit polyclonal) | Millipore | Cat. number ABE559 | Western blot (1:1000) |
| Antibody | Anti-EXOSC1 (rabbit monoclonal) | Abcam | Cat. number EPR13526 | Western blot (1:1000) |
| Chemical compound, drug | Niraparib | MedChem Express | Cat. number HY-10619 | |
| Chemical compound, drug | Olaparib | MedChem Express | Cat. number HY-10162 | |
| Chemical compound, drug | Rifampicin | Sigma-Aldrich | Cat. number R3501 | |
| Software, algorithm | Prism | GraphPad | Version 8 | data analyses |
| Other | Lipofectamine 2000 | Thermo Fisher Scientific Inc | Cat. number 11668019 |
Additional files
-
Supplementary file 1
Table for the candidates identified by Spearman’s rank analyses at whole genomic level.
- https://cdn.elifesciences.org/articles/69454/elife-69454-supp1-v2.xlsx
-
Supplementary file 2
Table for the candidates identified by Student’s t-test analyses at whole genomic level.
- https://cdn.elifesciences.org/articles/69454/elife-69454-supp2-v2.xlsx
-
Supplementary file 3
Table for the genes showing significant association with VHL.
- https://cdn.elifesciences.org/articles/69454/elife-69454-supp3-v2.xlsx
-
Supplementary file 4
Table for the overlap numbers of the candidate genes of the six types of c-substitutions.
- https://cdn.elifesciences.org/articles/69454/elife-69454-supp4-v2.xlsx
-
Supplementary file 5
Table for primer sequence.
- https://cdn.elifesciences.org/articles/69454/elife-69454-supp5-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/69454/elife-69454-transrepform-v2.docx





