The mitochondrial iron transporter ABCB7 is required for B cell development, proliferation, and class switch recombination in mice
Figures
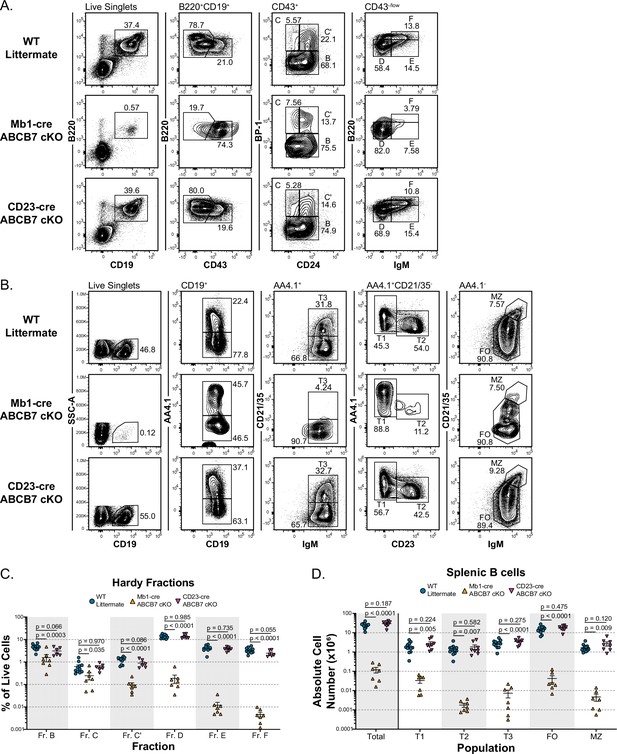
ABCB7 is required for pro-B cell development but not peripheral B cell homeostasis.
(A) Flow cytometry analysis of B cell development in bone marrow from wild-type (WT), Mb1-cre ABCB7 conditional knockout (cKO), and CD23-cre ABCB7 cKO mice. Pro-B cells were divided into Hardy fractions as follows: Fr. B (B220+ CD19+ CD43+ BP-1-), Fr. C (B220+ CD19+ CD43+ CD24lo BP-1+), and Fr. C’ (B220+ CD19+ CD43+ CD24hi BP-1+), Fr. D (B220+ CD19+ CD43-/low sIgM-), Fr. E (B220+ CD19+ CD43-/low sIgM+), and Fr. F (B220hi CD19+ CD43-/low sIgM+). Contour plots are representative of six independent experiments (total of 6–11 mice/group). (B) Flow cytometry analysis of splenic B cell populations in WT, Mb1-cre ABCB7 cKO, and CD23-cre ABCB7 cKO mice. Populations were identified by gating on CD19+ splenocytes: transitional type 1 (T1; AA4.1+CD21/35- IgM+ CD23-), transitional type 2 (T2; AA4.1+ CD21/35- IgM+ CD23+), transitional type 3 (T3; AA4.1+CD21/35+IgM+), follicular (FO; AA4.1- CD21/35+ IgM+), and marginal zone (MZ; AA4.1- CD21/35hi IgMhi). Contour plots are representative of seven independent experiments (total of 7–12 mice/group). (C) Graph showing the percentage of total live bone marrow cells for each Hardy fraction in (A). (D) Graph showing absolute cell numbers of splenic B cell populations in (B). (C, D) Lines represent the mean ± SEM. Statistics were obtained by using a one-way ANOVA with Dunnett’s test for multiple comparisons.
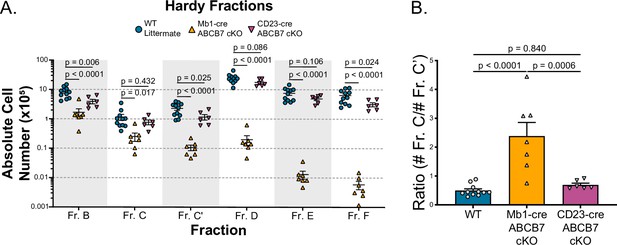
Analysis of pro-B cell block in Mb1-cre ABCB7 conditional knockout (cKO) mice.
(A) Graph showing the absolute cell numbers of bone marrow Hardy fractions from wild-type (WT), Mb1-cre ABCB7 cKO, and CD23-cre ABCB7 cKO mice from Figure 1A. (B) Graph showing the ratio of absolute numbers of Fr. C cells (B220+ CD19+ CD43+ CD24lo BP-1+) over absolute numbers of Fr. C’ cells (B220+ CD19+ CD43+ CD24hi BP-1+) in WT, Mb1-cre ABCB7 cKO, and CD23-cre ABCB7 cKO mice from (A). (A, B) Error bars represent SEM, and p-values are indicated above the data. Results were collected from six independent experiments (total of 6–11 mice/group). Statistics were obtained by using a one-way ANOVA. Dunnett’s multiple comparisons test was used for (A) while Tukey’s test was used for (B).
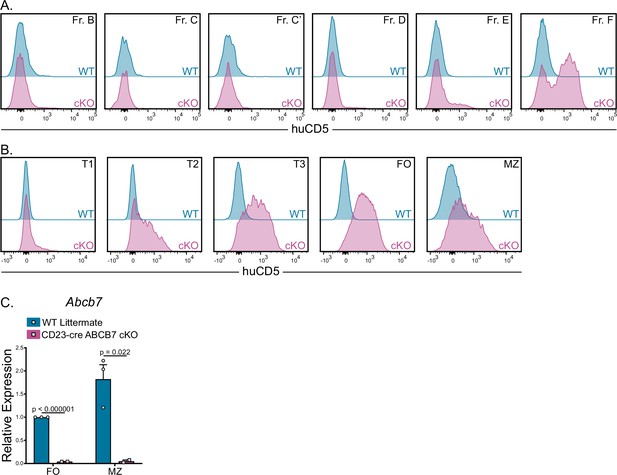
ABCB7 deletion efficiency in CD23-cre ABCB7 conditional knockout (cKO) peripheral B cells.
Flow cytometry analysis of huCD5 reporter expression in BM (A) and splenic (B) B cell populations from wild-type (WT) and CD23-cre ABCB7 cKO mice.
Offset histograms are representative of three independent experiments. (C) Quantitative real-time PCR analysis of Abcb7 expression in sorted follicular (FO) and marginal zone (MZ) B cells from WT and CD23-cre ABCB7 cKO mice. 18S rRNA was used as an endogenous control, and relative expression values were normalized to expression in WT FO B cells. Results were obtained from three independent experiments (total of 2–3 mice/group). Error bars represent SEM, and p-values are indicated above the data. Statistics were obtained by using an unpaired Student’s t-test.
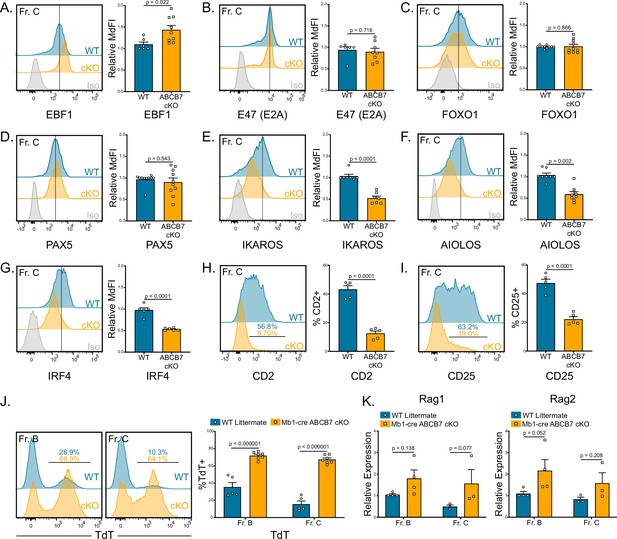
Gene expression changes confirm absence of pre-B cells in Mb1-cre ABCB7 conditional knockout (cKO) mice.
Analysis of critical transcription factors in wild-type (WT) and Mb1-cre ABCB7 cKO Fr. C cells (B220+CD19+CD43+BP-1+). (A–G) Intracellular flow cytometry analysis of EBF1 (A), E47 (E2A) (B), FOXO1 (C), PAX5 (D), IKAROS (E), AIOLOS (F), and IRF4 (G) expression. Quantification of MdFI is shown on the right of each plot. Isotype controls are shown in gray. Offset histograms are representative of at least three independent experiments (total of 6–10 mice/group). (H, I) Flow cytometry analysis of CD2 (H) and CD25 (I) expression. Indicated values are the proportion of Fr. C cells positive for either marker, and quantifications are shown on the right of each plot. Offset histograms are representative of three independent experiments (total of five mice/group). (J) Intracellular flow cytometry analysis of TdT expression in Fr. B and Fr. C cells. Indicated values are the proportion of cells positive for TdT expression, and quantifications are shown on the right. Offset histograms are representative of three independent experiments (total of 5–7 mice/group). (K) Quantitative real-time PCR analysis of Rag1 and Rag2 expression in sorted Fr. B and Fr. C cells. 18S rRNA was used as an endogenous control, and relative expression values were normalized to expression in WT Fr. B cells. Results were obtained from three independent experiments (total of 3–4 mice/group). (A–K) Error bars represent SEM, and p-values are indicated above the data. Statistics were obtained by using an unpaired Student’s t-test.

Failure to generate pre-B cell colony-forming unit (CFU) from Mb1-cre ABCB7 conditional knockout (cKO) mice.
P re-B cell CFU assay to examine pre-B cell colony-forming potential of wild-type (WT) and Mb1-cre ABCB7 cKO bone marrow cells. Total bone marrow was used as input in a MethoCult M3606 pre-B CFU kit and was seeded with 2 × 105 cells, according to the manufacturer’s instructions. Colonies were counted after 8 days of incubation. Reported colony numbers are averages across triplicate plates. Results were obtained from two independent experiments (total of four mice/group). Error bars represent SEM, and p-values are indicated above the data. Statistics were obtained by using an unpaired Student’s t-test.
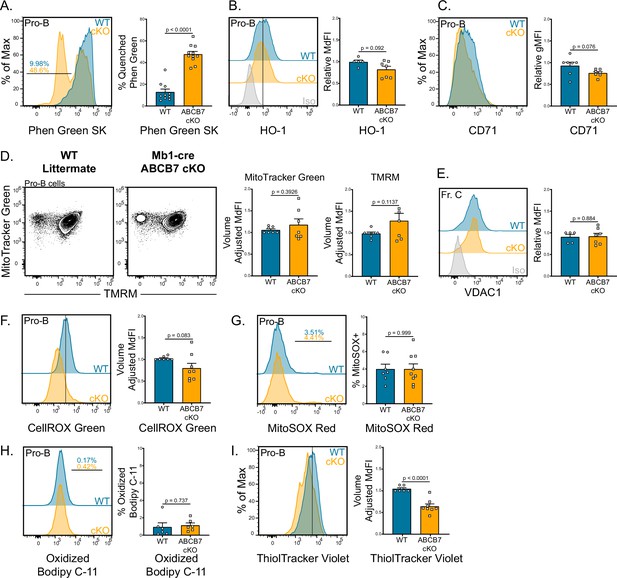
Iron accumulation in ABCB7-deficient pro-B cells. Analysis of mitochondria, iron accumulation, and reactive oxygen species (ROS) in wild-type (WT) and Mb1-cre ABCB7 conditional knockout (cKO) pro-B cells (B220+ CD19+ CD43+).
(A) Flow cytometry analysis of Phen Green SK fluorescence quenching by heavy metal atoms. Indicated values are the proportion of cells with quenched fluorescence, and quantification is shown on the right. Overlaid histogram is representative of five independent experiments (total of 10–11 mice/group). (B) Intracellular flow cytometry analysis of HO-1 expression. Quantification of HO-1 MdFI is shown on the right. Offset histogram is representative of three independent experiments (total of 5–7 mice/group). (C) Flow cytometry analysis of CD71 expression. Quantification of CD71 gMFI is shown on the right. Overlaid histogram is representative of three independent experiments (total of seven mice/group). (D) Flow cytometry analysis of mitochondria abundance (MitoTracker Green) and membrane potential (tetramethylrhodamine methyl ester [TMRM]). Quantification of MitoTracker Green volume-adjusted MdFI and TMRM volume-adjusted MdFI is shown on the right. Contour plots are representative of four independent experiments (total of seven mice/group). (E) Intracellular flow cytometry analysis of VDAC1 expression in Fr. C cells (B220+ CD19+ CD43+ BP-1+). Quantification of VDAC1 MdFI is shown on the right. Offset histogram is representative of three independent experiments (total of 5–7 mice/group). (F) Flow cytometry analysis of CellROX Green ROS detection probe. Quantification of CellROX Green volume-adjusted MdFI is shown on the right. Offset histogram is representative of five independent experiments (total of 7–8 mice/group). (G) Flow cytometry analysis of MitoSOX Red mitochondrial ROS detection probe. Indicated values are the proportion of cells positive for MitoSOX Red dye, and quantification is shown on the right. Offset histogram is representative of four independent experiments (total of 7–9 mice/group). (H) Flow cytometry analysis of Bodipy C-11 lipid peroxidation probe. Indicated values are the proportion of cells positive for oxidized Bodipy C-11, and quantification is shown on the right. Offset histogram is representative of three independent experiments (total of six mice/group). (I) Flow cytometry analysis of ThiolTracker Violet glutathione detection agent. Quantification of ThiolTracker Violet volume-adjusted MdFI is shown on the right. Overlaid histogram is representative of four independent experiments (total of 7–8 mice/group). (A–H) Error bars represent SEM, and p-values are indicated above the data. Statistics were obtained by using an unpaired Student’s t-test.
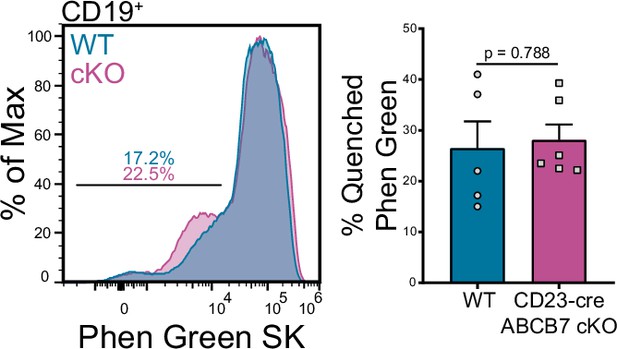
Splenic B cells in CD23-cre ABCB7 conditional knockout (cKO) do not have iron accumulation.
Flow cytometry analysis of Phen Green SK fluorescence quenching by heavy metal atoms in CD19+ cells from wild-type (WT) and CD23-cre ABCB7 cKO mice. Indicated values are the proportion of cells with quenched fluorescence, and quantification is shown on the right. Overlaid histogram is representative of three independent experiments (total of 5–6 mice/group).
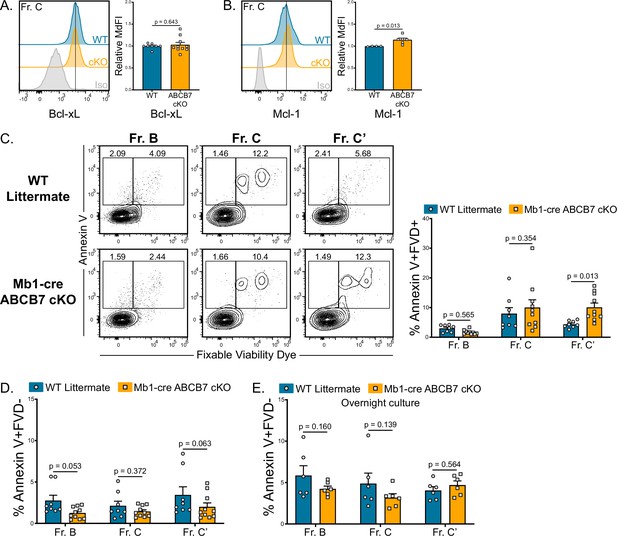
Analysis of apoptosis in ABCB7-deficient pro-B cells.
(A, B) Intracellular flow cytometry analysis of Bcl-xL (B) and Mcl-1 (C) expression in Fr. C cells (B220+ CD19+ CD43+ BP-1+) from wild-type (WT) and Mb1-cre ABCB7 conditional knockout (cKO) mice. Quantification of MdFI is shown on the right of each plot. Isotype controls are shown in gray. Offset histograms are representative of at least three independent experiments (total of 4–10 mice/group). (C) Flow cytometry analysis of Annexin V binding and fixable viability dye (FVD) labeling of Fr. B-C’ cells from WT and Mb1-cre ABCB7 cKO mice. Quantification of the proportion of each fraction that are dead (Annexin V+ FVD+) is shown on the right. Contour plots are representative of four independent experiments (total of 8–10 mice/group). (D) Quantification of the proportion of each fraction that are apoptotic (Annexin V+ FVD-) from (C). (E) Quantification of the proportion of each fraction that are apoptotic (Annexin V+ FVD-) after 16 hr in culture. Data represent three independent experiments (total of six mice/group). (A–E) Error bars represent SEM, and p-values are indicated above the data. Statistics were obtained by using an unpaired Student’s t-test.

Bcl2 expression was undetected in pro-B cells.
Intracellular flow cytometry analysis of Bcl2 expression in Fr. C cells (B220+CD19+CD43+BP-1+) from wild-type (WT) and Mb1-cre ABCB7 conditional knockout (cKO) mice. WT Fr. A cells (B220+CD19-) were used as a positive control for Bcl2 expression in the bone marrow. Isotype control is shown in gray. Offset histogram is representative of three independent experiments.
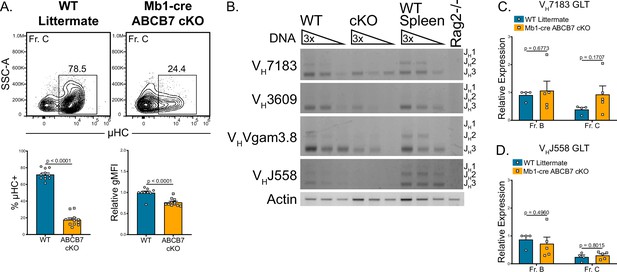
Reduced heavy chain recombination in ABCB7-deficient pro-B cells.
(A) Intracellular flow cytometry analysis of μ heavy chain (μHC) expression in Fr. C cells (B220+ CD19+ CD43+ BP-1+) from wild-type (WT) and Mb1-cre ABCB7 conditional knockout (cKO) mice. Quantification of the proportion of μHC+ cells is shown on the left graph. Quantification of μHC gMFI is shown on the right graph. Contour plots are representative of six independent experiments (total of 12–15 mice/group). (B) Semiquantitative PCR analysis of heavy chain locus recombination. DNA was purified from magnetically enriched pro-B cells. DNA from magnetically enriched CD19+ WT splenocytes was used as a positive control, while DNA from a Rag2-deficient cell line was used as a negative control. DNA was adjusted to an equivalent concentration and subjected to threefold serial dilutions. Recombined VH gene segments were amplified using the indicated family-specific forward primer and a reverse primer specific to JH3. Three bands corresponding to the usage of JH1, JH2, and JH3 were expected for each VH gene amplified (JH1 band is underrepresented due to product length). Results are ordered from proximal (VH7183) to distal (VHJ558) VH gene families. Actin was used as a loading control. Results are representative of four independent experiments. Image contrast and brightness were adjusted and colors were inverted for the final image. Source images are provided in Figure 5—source data 1. (C, D) Quantitative real-time PCR analysis of sterile VH7183 (C) and VHJ558 (D) germline transcript (GLT) expression in FACS sorted Fr. B (B220+ CD19+ CD43+ BP-1-) and Fr. C cells. Hprt1 was used as an endogenous control, and relative expression values were normalized to expression in WT Fr. B cells. Results were obtained from three independent experiments (total of 4–5 mice/group). (A, C, D) Error bars represent SEM, and p-values are indicated above the data. Statistics were obtained using an unpaired Student’s t-test.
-
Figure 5—source data 1
Source images.
This zip archive contains all raw gel images taken for semiquantitative PCR data shown in Figure 5B. Gels were photographed using an Omega Lum G gel imager, which saved the raw image files provided here. Individual files were named based on the VH gene family that was analyzed, and images were saved as full-resolution, 16-bit grayscale TIFF files. In addition to the unedited gel images, a labeled image is provided (named as ‘labeled’) for each gel.
- https://cdn.elifesciences.org/articles/69621/elife-69621-fig5-data1-v1.zip
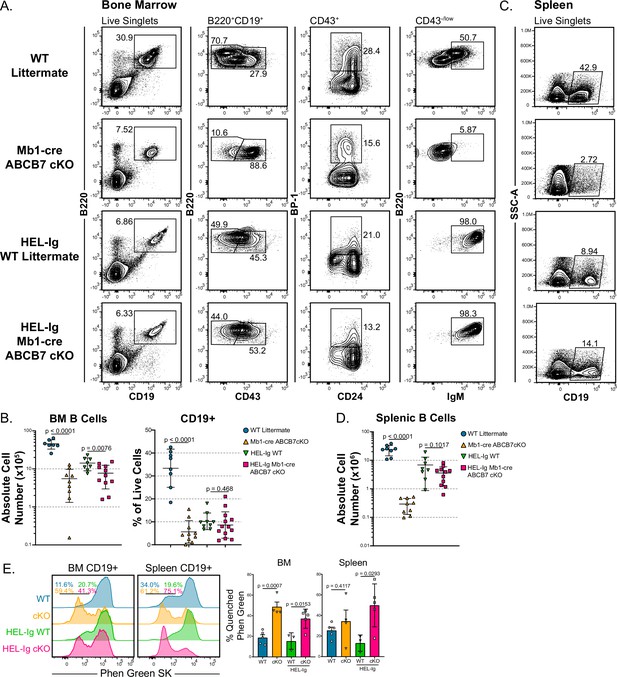
MD4 HEL-Ig transgenic B cell receptor (BCR) normalizes bone marrow B cell populations and restores splenic B cells in Mb1-cre ABCB7 conditional knockout (cKO) mice.
(A) Flow cytometry analysis of B cell development in bone marrow from wild-type (WT) and Mb1-cre ABCB7 cKO mice in the presence or absence of a fully rearranged transgenic BCR specific for hen egg lysozyme (HEL-Ig). B cell populations were identified by gating on B220+ CD19+ cells: pro-B cells (CD43+), BP-1+ pro-B cells, and Fr. E/F cells (CD43-/low IgM+). Contour plots are representative of seven independent experiments (total of 9–13 mice/group). (B) Graphs showing CD19+ absolute cell numbers (left) and percentage of live cells (right) in the bone marrow of mice analyzed in (A). (C) Flow cytometry analysis of splenic CD19+ cells in mice from (A). Contour plots are representative of seven independent experiments (total of 9–13 mice/group). (D) Graph showing absolute cell numbers of CD19+ cells in the spleen of mice analyzed in (C). (E) Flow cytometry analysis of Phen Green SK fluorescence quenching by heavy metal ions in bone marrow and splenic CD19+ cells from WT and Mb1-cre ABCB7 cKO mice. Indicated values are the proportion of cells with quenched fluorescence, and quantification is shown on the right. Offset histograms are representative of three independent experiments (total of 3–5 mice/group). (B, D, E) Error bars represent SEM, and p-values are indicated above the data. Statistics were obtained by using an unpaired Student’s t-test.
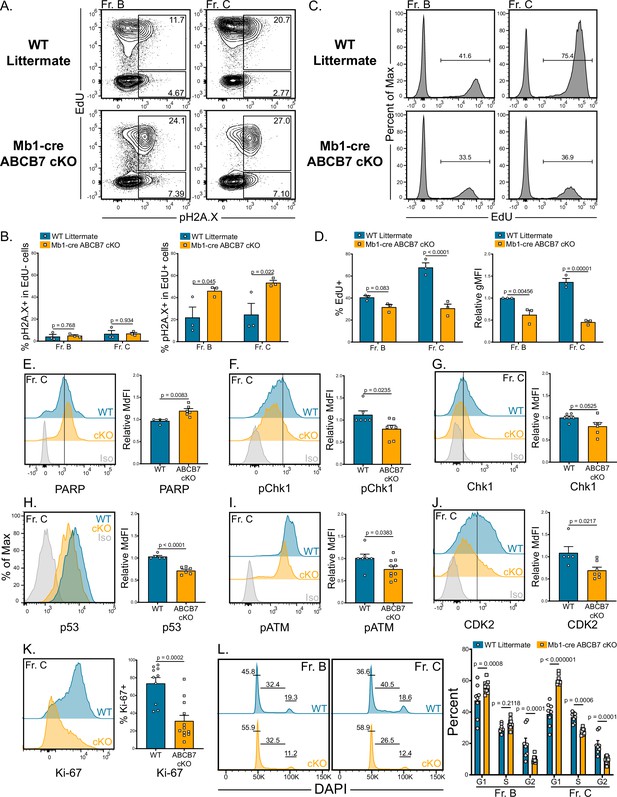
Reduced proliferation and evidence of DNA damage in ABCB7-deficient pro-B cells.
(A) Intracellular flow cytometry analysis of EdU incorporation and pH2A.X expression in Fr. B (B220+ CD19+ CD43+ BP-1-) and Fr. C (B220+ CD19+ CD43+ BP-1+) cells from wild-type (WT) and Mb1-cre ABCB7 conditional knockout (cKO) mice. Cells were pulsed with EdU for 3 hr in culture. Contour plots are representative of three independent experiments (total of three mice/group). (B) Quantification of the proportion of EdU- cells (left graph) and EdU+ cells (right graph) that were positive for pH2A.X expression. (C) Flow cytometric analysis of the proportion of Fr. B and Fr. C cells (A) that incorporated EdU. Histograms are representative of three independent experiments (total of three mice/group). (D) Quantification of the proportion of cells that incorporated EdU (left plot) and EdU gMFI (right graph) in Fr. B and Fr. C cells from (C). gMFI was normalized to WT Fr. B cells. (E–K) Intracellular flow cytometry analysis of PARP (E), pChk1 (F), total Chk1 (G), p53 (H), pATM (I), and CDK2 (J), and Ki-67 (K) expression in Fr. C cells from WT and Mb1-cre ABCB7 cKO mice. Quantification of the MdFI or percent positive is shown on the right of each plot. Isotype controls are shown in gray. Offset and overlaid histograms are representative of at least three independent experiments (total of 4–10 mice/group). (L) Analysis of cell cycle status using intracellular DAPI staining in Fr. B and Fr. C cells from WT and Mb1-cre ABCB7 cKO mice. Leftmost gate marks cells in G1, middle gate marks cells in S phase, and rightmost gate marks cells in G2/M phases. Values shown above gates were derived from the FlowJo cell cycle analysis modeling tool. Quantification of the proportion of cells in G1, S, and G2/M phases is shown on the right of the plot. Proportions were determined by using the FlowJo cell cycle analysis modeling tool. Offset histograms are representative of six independent experiments (total of 8–10 mice/group). (B, D–L) Error bars represent SEM, and p-values are indicated above the data. Statistics were obtained by using an unpaired Student’s t-test.
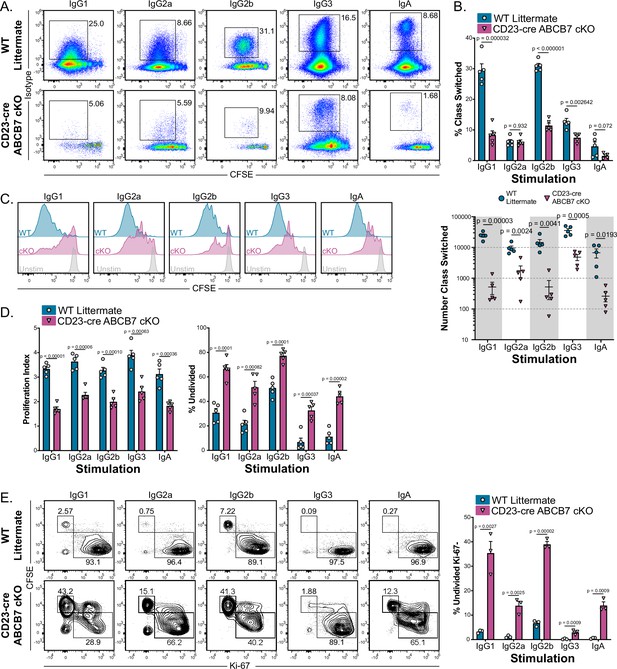
ABCB7 is required for peripheral B cell proliferation and class switching.
(A) Flow cytometry analysis of IgG1, IgG2a, IgG2b, IgG3, and IgA expression on enriched B220+ CD19+ B cells from wild-type (WT) and CD23-cre ABCB7 conditional knockout (cKO) mice after 4 days in culture conditions that induce class switching to the indicated isotypes. Pseudocolor dot plots are representative of five independent experiments (total of five mice/group). (B) Quantification of the proportion (top) and number (bottom) of cells from (A) that class switched to the indicated antibody isotypes. The reported cell number was derived from flow cytometry live CD19+ cells during analysis. (C) Flow cytometry analysis of carboxyfluorescein diacetate succinimidyl diester (CFSE) dilution in cells from (A). Offset histograms are representative of five independent experiments (total of five mice/group). (D) FlowJo proliferation modeling tool was used to quantify the proliferation index (left) and percentage of undivided cells (right) in cells from (C). (E) Intracellular flow cytometric analysis of Ki-67 expression in proliferating B220+ CD19+ cells after 4 days in culture conditions that induce class switching. Contour plots are representative of three independent experiments (total of three mice/group). Quantification of the percentage of undivided, Ki-67- cells is shown on the graph on the right. (B, D, E) Error bars represent SEM, and p-values are indicated above the data. Statistics were obtained by using an unpaired Student’s t-test.
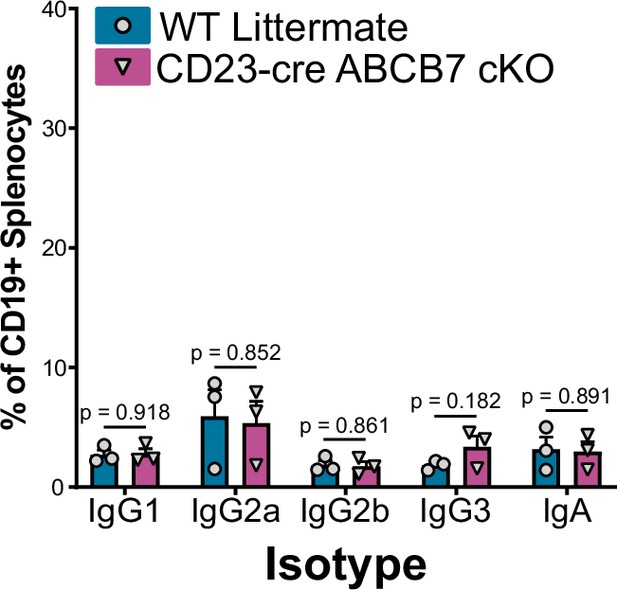
Class-switched B cells in the spleens of CD23-cre ABCB7 conditional knockout (cKO) mice.
Quantification of the proportion of IgG1+, IgG2a+ IgG2b+, IgG3+, and IgA+ class-switched B220+ CD19+ B cells in the spleens of unchallenged mice. Analyzed by flow cytometry in three independent experiments (total of three mice/group). Error bars represent SEM, and p values are indicated above the data. Statistics were obtained by using an unpaired Student’s t-test.
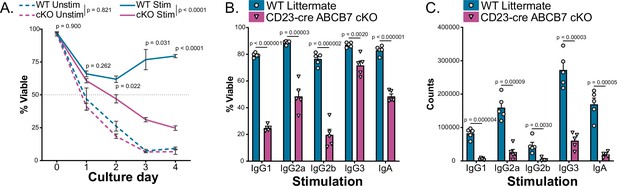
Cell viability in class switch assay.
(A) Quantification of cell viability over time in IgG1-stimulating culture conditions. Cells were analyzed for fixable viability dye (FVD) staining by flow cytometry. Enriched B220+CD19+ B cells from wild-type (WT) and CD23-cre ABCB7 conditional knockout (cKO) mice were cultured for 4 days (see Figure 8). Data were obtained from three independent experiments (total of three mice/group). Error bars represent SEM, and adjusted p-values on the line graph represent comparison between stimulated WT and CD23-cre ABCB7 cKO cells at each timepoint. p-values in the legend represent the overall comparison between WT and CD23-cre ABCB7 cKO cultures in unstimulated or stimulated conditions. Statistics were obtained by using a repeated measures two-way ANOVA with Geisser–Greenhouse correction and Holm–Šídák’s multiple comparisons test. (B) Quantification of B220+CD19+ cell viability in class switch-stimulating culture conditions. Cells were analyzed for FVD staining by flow cytometry after 4 days in culture. (C) Quantification of B220+ CD19+ events during flow cytometry after 4 days in class switch-stimulating culture. (A, B) Data collected from five independent experiments (total of five mice/group). Error bars represent SEM, and p-values are indicated above the data. Statistics were obtained by using an unpaired Student’s t-test.
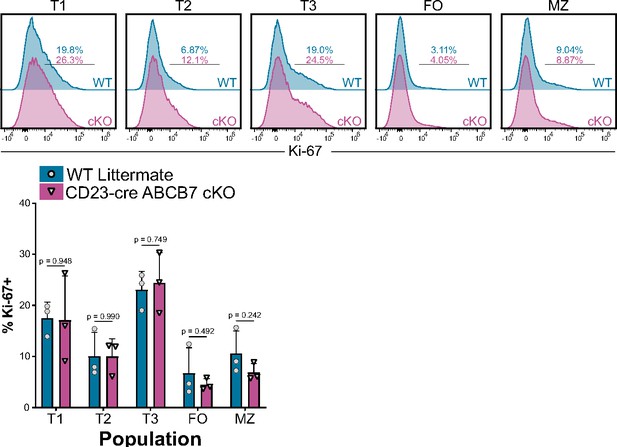
Ki-67 expression in ABCB7-deficient splenic B cells.
Intracellular flow cytometry analysis of Ki-67 expression in peripheral B cell populations from wild-type (WT) and CD23-cre ABCB7 conditional knockout (cKO) mice. Populations were defined as in Figure 1. Offset histograms are representative of three independent experiments (total of three mice/group). Quantification of the percentage of Ki-67+ cells is shown on the graph below. Error bars represent SEM, and p-values are indicated above the data. Statistics were obtained by using an unpaired Student’s t-test.
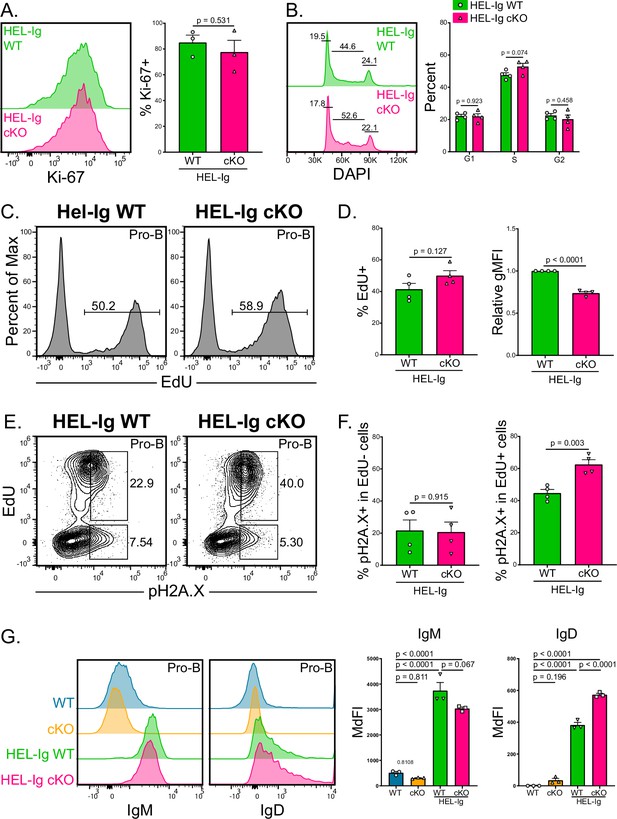
Improved proliferation of B cells from HEL-Ig Mb1-cre ABCB7 conditional knockout (cKO) mice.
(A) Intracellular flow cytometry analysis of Ki-67 expression in pro-B cells (B220+ CD19+ CD43+ CD127+) from HEL-Ig wild-type (WT) and HEL-Ig Mb1-cre ABCB7 cKO mice. Quantification of the percent of Ki-67+ cells is shown on the right. Offset histogram is representative of three independent experiments (total of three mice/group). (B) Analysis of cell cycle status using intracellular DAPI staining in CD127+ pro-B cells from HEL-Ig WT and HEL-Ig Mb1-cre ABCB7 cKO mice. Leftmost gate marks cells in G1, middle gate marks cells in S phase, and rightmost gate marks cells in G2/M phases. Values shown above gates were derived from the FlowJo cell cycle analysis modeling tool. Quantification of the proportion of cells in G1, S, and G2/M phases is shown on the right of the plot. Proportions were determined by using the FlowJo cell cycle analysis modeling tool. Offset histograms are representative of four independent experiments (total of four mice/group). (C) Intracellular flow cytometric analysis of the proportion of pro-B cells from HEL-Ig WT and Hel-Ig Mb1-cre ABCB7 cKO mice that incorporated EdU. Cells were pulsed with EdU for 3 hr in culture. Histograms are representative of four independent experiments (total of four mice/group). (D) Quantification of the proportion of cells that incorporated EdU (left plot) and EdU gMFI (right plot) in pro-B cells from (C). (E) Intracellular flow cytometry analysis of pH2A.X expression in pro-B cells from (C). Contour plots are representative of four independent experiments (total of four mice/group). (F) Quantification of the proportion of EdU- cells (left graph) and EdU+ cells (right graph) that were positive for pH2A.X expression. (G) Flow cytometry analysis of IgM (left) and IgD (right) expression in pro-B cells from WT, Mb1-cre ABCB7 cKO, HEL-Ig WT, and HEL-Ig Mb1-cre ABCB7 cKO mice. Quantification of the MdFI is shown on the right. Offset histograms are representative of three independent experiments (total of three mice/group). Error bars represent SEM, and p-values are indicated above the data. Statistics were obtained by using a one-way ANOVA with Tukey’s multiple comparisons test. (A, B, D, F) Error bars represent SEM, and p-values are indicated above the data. Statistics were obtained by using an unpaired Student’s t-test.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Mus musculus) | Abcb7 | GenBank | GeneID:11306 | |
| Strain, strain background (M. musculus) | B6.129S4-Abcb7tm1Mdf/J (ABCB7fl/ABCB7fl/fl) | Ordered from The Jackson Laboratory, described in PMID:16424901 | Cat#:006490;MGI: 3628655; RRID:IMSR_JAX:006490 | ABCB7 floxed mice |
| Strain, strain background (M. musculus) | B6.C(Cg)-Cd79atm1(cre)Reth/EhobJ (Mb1-cre) | Ordered from The Jackson Laboratory, described in PMID:16940357 | Cat#:020505;MGI: 3687451; RRID:IMSR_JAX:020505 | Bone marrow B cell-specific cre |
| Strain, strain background (M. musculus) | B6.Cg-Tg(Fcer2a-cre)5Mbu/J (CD23-cre) | Ordered from The Jackson Laboratory, described in PMID:18538592 | Cat#:028197;MGI: 3803652; RRID:IMSR_JAX:028197 | Peripheral B cell-specific cre, uses B cell-specific Cd23 promoter |
| Strain, strain background (M. musculus) | C57BL/6-Tg(IghelMD4)4Ccg/J (HEL-Ig) | Ordered from The Jackson Laboratory, described in PMID:7926785 | Cat#:002595;MGI:2384162; RRID:IMSR_JAX:002595 | Transgenic mice with fully rearranged BCR |
| Cell line (M. musculus) | RAG2KO Pro-B cells | Kay Medina Lab | RRID:CVCL_B3QD | Cell line maintained by Medina labSee Materials and methods |
| Antibody | Anti-B220 BV785 (Rat monoclonal, RA3-6B2) | BioLegend | Cat#:103246; RRID:AB_2563256 | FC (1:200) |
| Antibody | Anti-B220 BV510 (Rat monoclonal, RA3-6B2) | BioLegend | Cat#:103248; RRID:AB_2650679 | FACS (1:200) |
| Antibody | Anti-Bcl2 PE (Mouse monoclonal, BCL/10C4) | BioLegend | Cat#:633508; RRID:AB_2290367 | FC (1:100) |
| Antibody | Anti-Bcl-xL AF488 (Rabbit monoclonal, 54H6) | Cell Signaling Technology | Cat#:2767S; RRID:AB_2274763 | FC (1:100) |
| Antibody | Anti-BP-1 biotin (Rat monoclonal, 6C3) | eBioscience | Cat#:12-5891-82; RRID:AB_466015 | FC (1:50) |
| Antibody | Anti-BP-1 BV605 (Mus spretus monoclonal, BP-1) | BD Biosciences | Cat#:745238; RRID:AB_2742824 | FC (1:50) |
| Antibody | Anti-BP-1 PE (Rat monoclonal, BP-1) | eBioscience | Cat#:12-5891-83; RRID:AB_466016 | FACS, FC (1:50) |
| Antibody | Anti-CD1d FITC (Rat monoclonal, 1B1) | BioLegend | Cat#:123508; RRID:AB_1236549 | FACS (1:100) |
| Antibody | Anti-CD2 FITC (Rat monoclonal, RM2-5) | eBioscience | Cat#:11-0021-81; RRID:AB_464872 | FC (1:100) |
| Antibody | Anti-CD4 biotin (Rat monoclonal, RM4-5) | BioLegend | Cat#:100508; RRID:AB_312711 | Negative selection (1:100) |
| Antibody | Anti-human CD5 APC (Mouse monoclonal, L17F12) | Tonbo Biosciences | Cat#:20-0058; RRID:AB_2621548 | FC [Cre reporter] (1:200) |
| Antibody | Anti-CD8α biotin (Rat monoclonal, 53-6.7) | BioLegend | Cat#:100704; RRID:AB_312743 | Negative selection (1:500) |
| Antibody | Anti-CD11b biotin (Rat monoclonal, M1/70) | BioLegend | Cat#:101204; RRID:AB_312787 | Negative selection (1:100) |
| Antibody | Anti-CD11c biotin (Armenian Hamster monoclonal, N418) | BioLegend | Cat#:117304; RRID:AB_313773 | Negative selection (1:100) |
| Antibody | Anti-CD19 BV510 (Rat monoclonal, 1D3) | BD Biosciences | Cat#:562956; RRID:AB_2737915 | FC (1:200) |
| Antibody | Anti-CD19 eFluor 450 (Rat monoclonal, eBio1D3) | eBioscience | Cat#:48-0193-82; RRID:AB_2734905 | FACS, FC (1:500) |
| Antibody | Anti-CD19 PE-Cy7 (Rat monoclonal, 6D5) | BioLegend | Cat#:115520; RRID:AB_313655 | FACS, FC (1:500) |
| Antibody | Anti-CD21/35 PerCP-Cy5.5 (Rat monoclonal, 7E9) | BioLegend | Cat#:123416; RRID:AB_1595490 | FACS (1:100) |
| Antibody | Anti-CD21/35 APC (Rat monoclonal, 7E9) | BioLegend | Cat#:123412; RRID:AB_2085160 | FC (1:200) |
| Antibody | Anti-CD23 APC (Rat monoclonal, B3B4) | BioLegend | Cat#:101614; RRID:AB_2103036 | FC (1:200) |
| Antibody | Anti-CD24 APC (Rat monoclonal, 30-F1) | BioLegend | Cat#:138506; RRID:AB_2565651 | FC (1:1000) |
| Antibody | Anti-CD24 FITC (Rat monoclonal, M1/69) | BioLegend | Cat#:101806; RRID:AB_312839 | FACS, FC (1:1000) |
| Antibody | Anti-CD25 PE (Rat monoclonal, PC61.5) | Tonbo Biosciences | Cat#:50-0251; RRID:AB_2621757 | FC (1:200) |
| Antibody | Anti-CD43 APC (Rat monoclonal, 1B11) | BioLegend | Cat#:121214; RRID:AB_528807 | FC (1:200) |
| Antibody | Anti-CD43 PerCP (Rat monoclonal, 1B11) | BioLegend | Cat#:121222; RRID:AB_893333 | FACS, FC (1:200) |
| Antibody | Anti-CD71 PE (Rat monoclonal, RI7217) | BioLegend | Cat#:113808; RRID:AB_313569 | FC (1:1000) |
| Antibody | Anti-CD93 PE (Rat monoclonal, AA4.1) | eBioscience | Cat#:12-5892-83; RRID:AB_466019 | FC (1:200) |
| Antibody | Anti-CD93 PE-Cy7 (Rat monoclonal, AA4.1) | BioLegend | Cat#:136506; RRID:AB_2044012 | FACS (1:100) |
| Antibody | Anti-CD127 PE-Cy7 (Rat monoclonal, A7R34) | BioLegend | Cat#:135014; RRID:AB_1937265 | FC (1:200) |
| Antibody | Anti-CDK2 PE (Rabbit monoclonal, 78B2) | Cell Signaling Technology | Cat#:14174; RRID:AB_2798413 | FC (1:50) |
| Antibody | Anti-Chk1 (Rabbit monoclonal, E250) | Abcam | Cat#:ab32531; RRID:AB_726821 | FC (1:200) |
| Antibody | Anti-E47/E2A FITC (Mouse monoclonal, G127-32) | BD Biosciences | Cat#:552510; RRID:AB_394408 | FC (1:500) |
| Antibody | Anti-EBF1 (Rabbit polyclonal) | Millipore Sigma | Cat#:ABE1294; RRID:AB_2893472 | FC (1:1000) |
| Antibody | Anti-FOXO1 PE (Rabbit monoclonal, C29H4) | Cell Signaling Technology | Cat#:14262S; RRID:AB_2798437 | FC (1:50) |
| Antibody | Anti-GR-1 biotin (Rat monoclonal, RB6-8C5) | BioLegend | Cat#:108404; RRID:AB_313369 | Negative selection (1:100) |
| Antibody | Anti-HO-1 FITC (Mouse monoclonal, HO-1–2) | Abcam | Cat#:ab69545; RRID:AB_2118659 | FC (1:50) |
| Antibody | Anti-IgA PE (Goat polyclonal IgG) | SouthernBiotech | Cat#:1040-09; RRID:AB_2794375 | FC (1:300) |
| Antibody | Anti-IgD PE (Rat monoclonal, 11–26c.2a) | BioLegend | Cat#:405706; RRID:AB_315028 | FC (1:200) |
| Antibody | Anti-IgG1 PE (Goat polyclonal F(ab')2 IgG) | SouthernBiotech | Cat#:1072-09; RRID:AB_2794434 | FC (1:1000) |
| Antibody | Anti-IgG2a PE (Goat polyclonal F(ab')2 IgG) | SouthernBiotech | Cat#:1082-09; RRID:AB_2794502 | FC (1:300) |
| Antibody | Anti-IgG2b PE (Goat polyclonal F(ab')2 IgG) | SouthernBiotech | Cat#:1092-09; RRID:AB_2794553 | FC (1:300) |
| Antibody | Anti-IgG3 PE (Goat polyclonal F(ab')2 IgG) | SouthernBiotech | Cat#:1102-09; RRID:AB_2784525 | FC (1:300) |
| Antibody | Anti-IgM APC-Cy7 (Rat monoclonal, RMM-1) | BioLegend | Cat#:406516; RRID:AB_10660305 | FC (1:100) |
| Antibody | Anti-IgM biotin (Rat monoclonal, RMM-1) | BioLegend | Cat#:406504; RRID:AB_315054 | Negative selection (1:100) |
| Antibody | Anti-IgM BV510 (Rat monoclonal, RMM-1) | BioLegend | Cat#:406531; RRID:AB_2650758 | FC (1:100) |
| Antibody | Anti-IgM FITC (Rat monoclonal, RMM-1) | BioLegend | Cat#:406506; RRID:AB_315056 | FC (1:100) |
| Antibody | Anti-IgM PE (Rat monoclonal, RMM-1) | BioLegend | Cat#:406508; RRID:AB_315058 | FACS (1:100) |
| Antibody | Anti-IgM PE-CF594 (Rat monoclonal, R6-60.2) | BD Biosciences | Cat#:562565; RRID:AB_2737658 | FACS (1:100) |
| Antibody | Anti-IKAROS AF488 (Rabbit monoclonal, D6N9Y) | Cell Signaling Technology | Cat#:89389S; RRID:AB_2800139 | FC (1:100) |
| Antibody | Anti-IRF4 PE (Rat monoclonal, 3E4) | eBioscience | Cat#:12-9858-82; RRID:AB_10852721 | FC (1:100) |
| Antibody | Anti-Ki-67 BV421 (Rat monoclonal, 16A8) | BioLegend | Cat#:652411; RRID:AB_2562663 | FC (1:200) |
| Antibody | Anti-Ki-67 PE (Rat monoclonal, 16A8) | BioLegend | Cat#:652404; RRID:AB_2561525 | FC (1:200) |
| Antibody | Anti-Mcl-1 PE (Rabbit monoclonal, D2W9E) | Cell Signaling Technology | Cat#:65617S; RRID:AB_2799688 | FC (1:50) |
| Antibody | Mouse IgG1 kappa Isotype FITC (Mouse monoclonal, P3.6.2.8.1) | eBioscience | Cat#:11-4714-81; RRID:AB_470021 | FC (concentration matched antibodies of interest) |
| Antibody | Mouse IgG1 kappa Isotype PE (Mouse monoclonal, MOPC-21) | BioLegend | Cat#:400111; RRID:AB_2847829 | FC (concentration matched antibodies of interest) |
| Antibody | Mouse IgG2b kappa Isotype FITC (Mouse monoclonal, 27-35) | BD Biosciences | Cat#:555742; RRID:AB_396085 | FC (concentration matched antibodies of interest) |
| Antibody | Anti-p53 AF488 (Mouse monoclonal, 1C12) | Cell Signaling Technology | Cat#:2015S; RRID:AB_2206297 | FC (1:50) |
| Antibody | Anti-PARP (Rabbit monoclonal, 46D11) | Cell Signaling Technology | Cat#:9532S; RRID:AB_659884 | FC (1:100) |
| Antibody | Anti-pATM (Ser1981) PE (Mouse monoclonal, 10H11.E12) | BioLegend | Cat#:651204; RRID:AB_2562655 | FC (1:500) |
| Antibody | Anti-PAX5 APC (Rat monoclonal, 1H9) | eBioscience | Cat#:17-9918-80; RRID:AB_10734230 | FC (1:200) |
| Antibody | Anti-pChk1 (Ser317) PE (Rabbit monoclonal, D12H3) | Cell Signaling Technology | Cat#:13959; RRID:AB_2893473 | FC (1:50) |
| Antibody | Anti-pH2A.X (Ser139) AF647 (Rabbit monoclonal, 20E3) | Cell Signaling Technology | Cat#:9720S; RRID:AB_10692910 | FC (1:100) |
| Antibody | Rabbit IgG Isotype Control AF488 (Rabbit monoclonal) | Cell Signaling Technology | Cat#:4340; RRID:AB_561545 | FC (concentration matched antibodies of interest) |
| Antibody | Rabbit IgG Isotype Control AF647 (Rabbit monoclonal) | Cell Signaling Technology | Cat#:3452; RRID:AB_10695811 | FC (concentration matched antibodies of interest) |
| Antibody | Rabbit IgG Isotype Control PE (Rabbit monoclonal, DA1E) | Cell Signaling Technology | Cat#:5742; RRID:AB_10694219 | FC (concentration matched antibodies of interest) |
| Antibody | Rabbit IgG Monoclonal Isotype Control (Rabbit monoclonal, EPR25A) | Abcam | Cat#:ab172730; RRID:AB_2687931 | FC (concentration matched antibodies of interest) |
| Antibody | Rabbit IgG Polyclonal Isotype Control (Rabbit monoclonal) | Abcam | Cat#:ab171870; RRID:AB_2687657 | FC (concentration matched antibodies of interest) |
| Antibody | Rat IgG2a kappa Isotype APC (Rat monoclonal, RTK2758) | BioLegend | Cat#:400512; RRID:AB_2814702 | FC (concentration matched antibodies of interest) |
| Antibody | Rat IgG2a kappa Isotype PE (Rat monoclonal, RTK2758) | BioLegend | Cat#:400508; RRID:AB_326530 | FC (concentration matched antibodies of interest) |
| Antibody | Rat IgG2b kappa Isotype PE (Rat monoclonal, eB149/10H5) | eBioscience | Cat#:12-4031-82; RRID:AB_470042 | FC (concentration matched antibodies of interest) |
| Antibody | Anti-NK1.1 biotin (Mouse monoclonal, PK136) | BioLegend | Cat#:108704; RRID:AB_313391 | Negative selection (1:100) |
| Antibody | Anti-Ter119 biotin (Rat monoclonal, TER-119) | BioLegend | Cat#:116204; RRID:AB_313705 | Negative selection (1:100) |
| Antibody | Anti-TCRβ biotin (Armenian Hamster monoclonal, H57-597) | BioLegend | Cat#:109204; RRID:AB_313427 | Negative selection (1:100) |
| Antibody | Anti-TCRγδ biotin (Armenian Hamster monoclonal, eBioGL3) | eBioscience | Cat#:13-5711-82; RRID:AB_466668 | Negative selection (1:100) |
| Antibody | Anti-TdT PE (Mouse monoclonal, 19–3) | eBioscience | Cat#:12-5846-82; RRID:AB_1963620 | FC (1:200) |
| Antibody | Anti-VDAC1 (Rabbit polyclonal) | ProteinTech | Cat#:55259-1-AP; RRID:AB_10837225 | FC (1:100) |
| Sequence-based reagent | qPCR: ABCB7 probe | Thermo Fisher | AssayId:Mm01235269_m1 | FAM-MGB |
| Sequence-based reagent | qPCR: Eukaryotic 18S rRNA Endogenous Control | Thermo Fisher | Cat#:4352930 | FAM-MGB |
| Sequence-based reagent | qPCR: Rag1 probe | Thermo Fisher | AssayId:Mm01270936_m1 | FAM-MGB |
| Sequence-based reagent | qPCR: Rag2 probe | Thermo Fisher | AssayId:Mm00501300_m1 | FAM-MGB |
| Peptide, recombinant protein | Annexin V-FITC conjugate | BD Biosciences | Cat#:556420; AB_2665412 | FC (1:500) |
| Peptide, recombinant protein | Streptavidin BV510 | BioLegend | Cat#:405234 | FC (1:200) |
| Peptide, recombinant protein | Recombinant human BAFF | PeproTech | Cat#:310-13 | Class switch cultures |
| Peptide, recombinant protein | Recombinant murine IFN-γ | PeproTech | Cat#:315-05 | Class switch cultures |
| Peptide, recombinant protein | Recombinant murine IL-4 | PeproTech | Cat#:214-14 | Class switch cultures |
| Peptide, recombinant protein | Recombinant murine IL-5 | PeproTech | Cat#:215-15 | Class switch cultures |
| Peptide, recombinant protein | Anti-δ dextran (mouse) | Fina Biosolutions | Cat#:FINABIO0001 | Class switch cultures |
| Peptide, recombinant protein | Recombinant human TGF-β1 | PeproTech | Cat#:100-21 | Class switch cultures |
| Commercial assay or kit | Click-iT Plus EdU AF488 Flow Cytometry Assay Kit | Thermo Fisher | Cat#:C10632 | EdU Assay |
| Commercial assay or kit | DNeasy Blood and Tissue Kit | Qiagen | Cat#:69504 | DNA purification |
| Commercial assay or kit | EasySep Mouse Streptavidin RapidSpheres Isolation Kit | STEMCELL Technologies | Cat#:19860A | Negative selection |
| Commercial assay or kit | MethoCult M3630 pre-B CFU kit | STEMCELL Technologies | Cat#:03630 | Pre-B CFU assay |
| Commercial assay or kit | OneTaq DNA polymerase | New England Biolabs | Cat#:M0480L | Semiquantitative PCR |
| Commercial assay or kit | RNeasy Mini Kit | Qiagen | Cat#:74104 | RNA isolation |
| Commercial assay or kit | SuperScript IV First-Strand Synthesis System | Thermo Fisher | Cat#:18091050 | cDNA synthesis |
| Chemical compound, drug | CFSE | Sigma | Cat#:21888 | Class switch cultures (2.5 μM) |
| Chemical compound, drug | FBS (qualified) | Thermo Fisher | Cat#:10437-028 | Class switch cultures |
| Chemical compound, drug | DAPI | BioLegend | Cat#:422801 | FC, cell cycle analysis (1:4000) |
| Chemical compound, drug | LPS | Sigma | Cat#:L-2630 | Class switch cultures |
| Software, algorithm | Illustrator CC 2019 | Adobe | RRID:SCR_010279 | Figure and image panel preparation |
| Software, algorithm | FlowJo v10.8 | BD Biosciences | RRID:SCR_008520 | FC analysis |
| Software, algorithm | GraphPad Prism v9 | GraphPad | RRID:SCR_002798 | Graphs and statistics |
| Other | BODIPY 581/591C11C11 | Thermo Fisher | Cat#:D3861 | FC (2 μM) |
| Other | CellROX Green | Thermo Fisher | Cat#:C10444 | FC (5 μM) |
| Other | Ghost Dye Violet 450 (viability dye) | Tonbo Biosciences | Cat#:13-0863 | FACS, FC (1:1000) |
| Other | Ghost Dye Violet 510 (viability dye) | Tonbo Biosciences | Cat#:13-0870 | FACS, FC (1:1000) |
| Other | Ghost Dye Red 780 (viability dye) | Tonbo Biosciences | Cat#:13-0865 | FC (1:1000) |
| Other | MitoSOX Red | Thermo Fisher | Cat#:M36008 | FC (5 μM) |
| Other | MitoTracker Green FM | Thermo Fisher | Cat#:M7514 | FC (100 nM) |
| Other | Phen Green SK, diacetate | Thermo Fisher | Cat#:P14313 | FC (5 μM) |
| Other | Tetramethylrhodamine methyl ester perchlorate (TMRM) | Sigma | Cat#:T5428 | FC (100 nM) |
| Other | ThiolTracker Violet | Thermo Fisher | Cat#:T10095 | FC (4 μM) |





