The TRRAP transcription cofactor represses interferon-stimulated genes in colorectal cancer cells
Figures
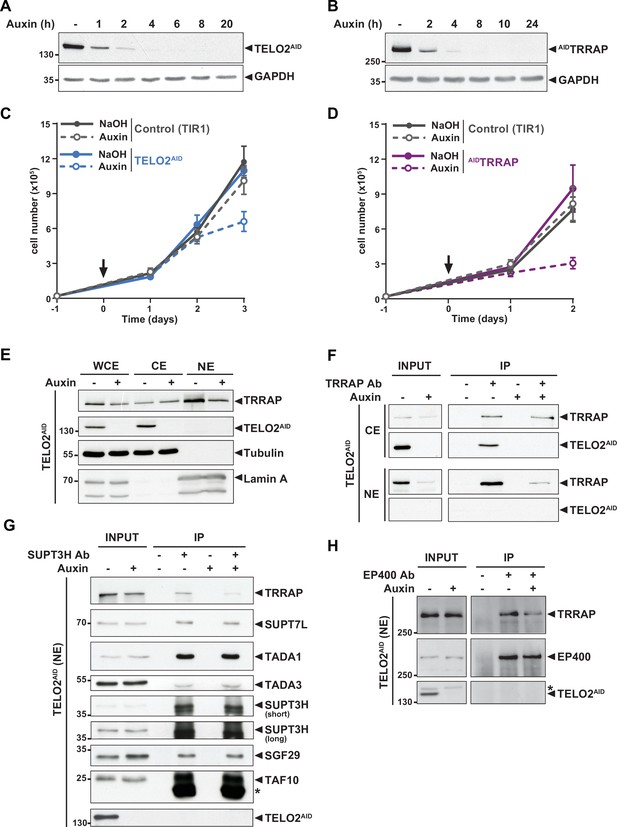
TELO2 promotes TRRAP assembly into the SAGA and TIP60 complexes.
(A, B) Western blot analyses of total extracts from TELO2AID (A) or AIDTRRAP (B) cell lines harvested at different time points after auxin addition, as indicated (hours). Blots were probed with an anti-TELO2 antibody (A), an anti-HA antibody to detect the HA-AID-TRRAP fusion protein (B), and an anti-GAPDH antibody to control for equal loading. (C, D) Proliferation rates of parental TIR1 (control, gray lines), TELO2AID (C, blue lines), and AIDTRRAP cells (D, purple lines). Cells were seeded 1 day before treatment (arrow) with either NaOH (solid lines) or auxin (dashed lines) and counted using trypan blue at the indicated time points. Each value represents average cell counts from four independent experiments, overlaid with the standard deviation (SD). (E) Immunoblotting of TRRAP and TELO2AID in whole cell (WCE), cytoplasmic (CE), and nuclear (NE) extracts of TELO2AID cells treated with auxin (+) or NaOH (-) for 48 hr. Tubulin and Lamin A were used as cytoplasmic and nuclear markers, respectively. (F) TRRAP immunoprecipitation (Ab +) from CE and NE fractions of TELO2AID cells treated with auxin (+) or NaOH (-) for 48 hr. TRRAP and TELO2AID were revealed by immunoblotting a fraction (2%) of each extract (INPUT) and the entire immunopurified eluate (IP). (G, H) SUPT3H and EP400 immunoprecipitation (Ab+) from nuclear extracts of TELO2AID cells treated with auxin (+) or NaOH (-) for 24 hr. SUPT3H (G) and EP400 (H), TRRAP, and each indicated SAGA subunits were revealed by immunoblotting a fraction (2%) of the extract (INPUT) and the entire IP eluate. Short and long indicate various exposure times. * indicates antibody light chain contamination (G) or nonspecific detection (H). (F–H) Control IPs (Ab-) were performed using beads only. Data are representative of three independent experiments. Source data are available in supplementary material (Figure 1A–H, Source data 1).
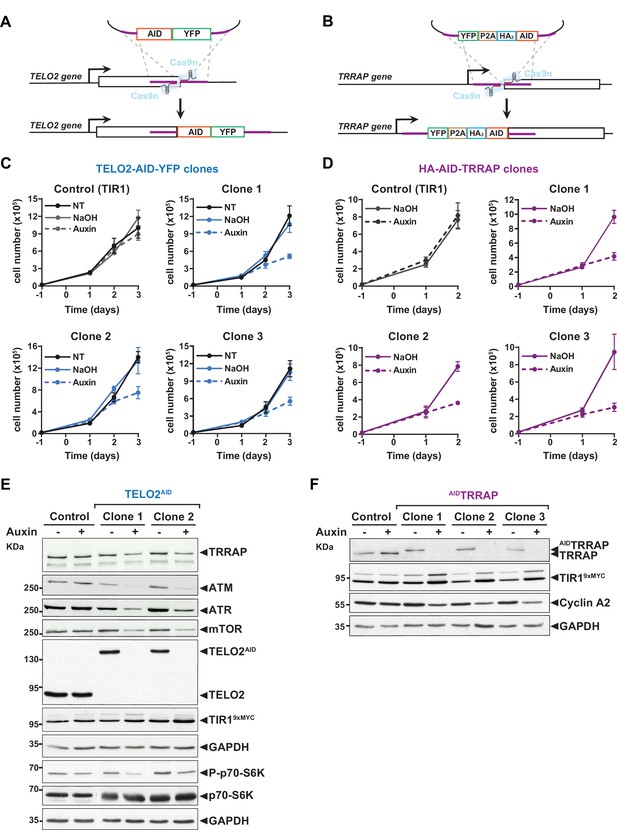
Characterization of TELO2-AID and AID-TRRAP cell lines.
(A, B) Schematic depiction of the strategy designed to tag endogenous TELO2 (A) and TRRAP (B) with an auxin-inducible degron (AID) in in an HCT116 cell line stably expressing Oryza sativa TIR1. Gene editing was performed using the D10A nickase Cas9n mutant (Cong et al., 2013), which facilitates homology-directed repair and reduces off targets. Repair donor plasmids comprise sequences encoding a full-length IAA17-derived degron (Nishimura et al., 2009), a YFP fluorescent tag, and three HA epitopes. To reduce the risks of affecting TRRAP function with a long fusion sequence at its N-terminus, we inserted a 2A peptide (P2A) between HA-AID and YFP, which is then cleaved off during translation. (C, D) Proliferation rates of parental HCT116-OsTIR1 cells (control) and three distinct TELO2AID (C) or AIDTRRAP (D) clones. The number of viable cells was measured using the trypan blue exclusion assay. 20,000 cells were plated at day –1. At day 0, cells were treated with auxin (dashed line), vehicle (NaOH, full line), or left untreated (NT, black) and monitored at the indicated time points. (E, F) Immunoblotting of the indicated proteins in extracts from HCT116-OsTIR1 cells (control), two distinct TELO2AID clones treated with auxin (+) or NaOH (-) for 48 hr(E), and three distinct AIDTRRAP clones treated with auxin (+) or NaOH (-) for 24 hr. The OsTIR1 F-box protein was detected using an anti-MYC antibody. GAPDH was used as an internal control for equal loading. Source data are available in supplementary material (E, F: Source data 1).
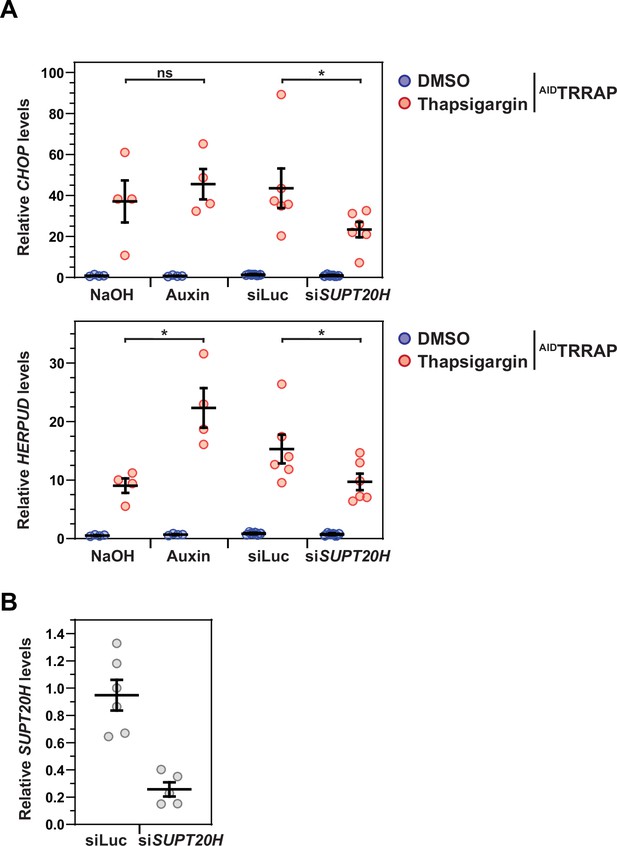
TRRAP is not involved in SAGA-dependent endoplasmic reticulum (ER) stress gene induction.
(A) Expression of CHOP and HERPUD using quantitative RT-PCR of RNA extracted from AIDTRRAP cells treated with either thapsigargin (red dots) or DMSO (blue dots) for 3 hr. Prior to ER stress induction, AIDTRRAP cells were treated with either auxin or NaOH for 48 hr, or transfected with siRNAs targeting luciferase (Luc) or SUPT20H for 48 hr. PPIB served as a control for normalization across samples. Values from one control sample (DMSO, NaOH) were set to 1 to allow comparisons across conditions and replicates. Each line represents the mean value of either four (NaOH, Auxin) or six (siLuc, siSUPT20H) independent experiments, overlaid with individual data points and error bars showing the standard error of the mean (SEM). Statistical significance was determined by two-way ANOVA followed by Tukey’s multiple comparison tests. *p≤0.05; ns: p>0.05. (B) Quantitative RT-PCR analysis of SUPT20H expression normalized to PPIB upon siRNA-mediated knockdown compared to control siRNAs targeting Luc. RNAs were from AIDTRRAP cells 48 hr after transfection. Each line represents the mean value of six independent experiments, overlaid with individual data points and error bars showing the SEM.
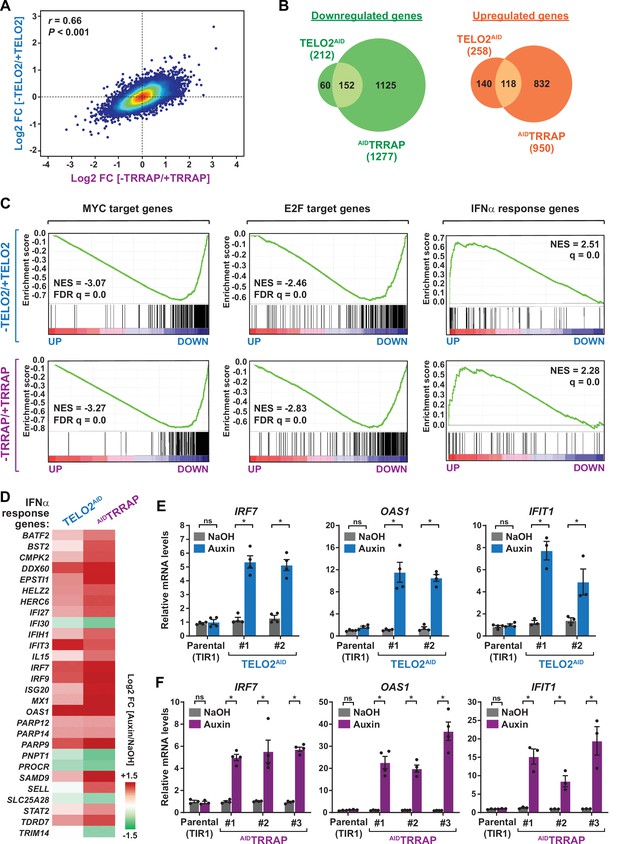
TELO2 and TRRAP regulate an overlapping set of genes and repress type I interferon-stimulated genes.
(A) Density scatter plot comparing gene expression changes between TELO2 and TRRAP-depleted cells. The Pearson correlation coefficient and corresponding p-value are indicated. (B) Venn diagrams showing the overlap between genes differentially expressed upon TELO2 and TRRAP depletion (false discovery rate [FDR] ≤ 1%). The number of transcripts whose levels decrease and increase is shown separately, as indicated. (C) Gene Set Enrichment Analysis (GSEA) showing the most strongly enriched hallmarks in the ranked transcriptome profiles of TELO2- (upper panels) and TRRAP-depleted cells (lower panels). Green lines represent enrichment profiles. NES: normalized enrichment score. Each hit from the hallmark gene set is represented by a vertical black bar, positioned on the ranked transcriptome profile with color-coded fold change values, as indicated. MYC- and E2F-target genes are enriched in the set of genes that are downregulated upon TELO2 and TRRAP depletion, whereas genes from the type I interferon response hallmark are enriched in the set of upregulated genes. (D) Heatmap representation of deregulated IFN α-responsive genes in TELO2 and TRRAP-depleted cells (from the 97 genes of hallmark M5911). The Log2 ratio between auxin- and NaOH-treated cells for each transcript is represented using a sequential color scale. All data are from RNA-seq experiments performed in three distinct TELO2AID and AIDTRRAP clones treated with either auxin or NaOH for 48 and 24 hr, respectively. (E, F) Quantitative RT-PCR analysis of three ISGs, IRF7, OAS1, and IFIT1, following the depletion of TELO2 (E) and TRRAP (F). mRNAs levels were measured in two TELO2AID clones treated with either auxin or NaOH for 48 hr (E), and in three AIDTRRAP clones treated with either auxin or NaOH for 24 hr (F). The corresponding parental TIR1-expressing cell lines were also analyzed and treated identically. Each value represents mean mRNA levels from at least three independent experiments, overlaid with individual data points and error bars showing the SD. PPIB served as a control for normalization across samples. Values from one NaOH-treated parental control replicate were set to 1, allowing comparisons across culture conditions and replicates. Statistical significance was determined by two-way ANOVA followed by Bonferroni’s multiple comparison tests. *p≤0.01.
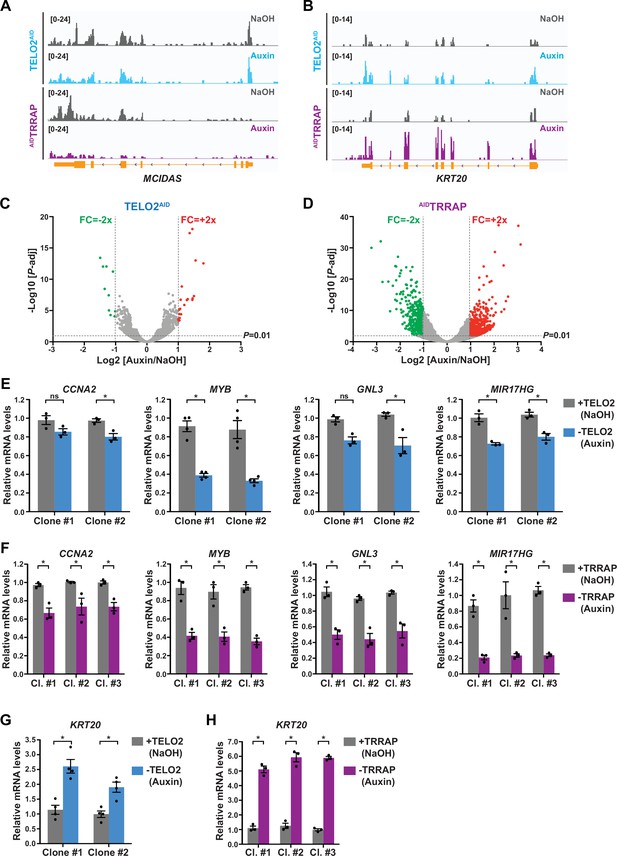
Gene expression changes upon TELO2 and TRRAP depletion.
(A, B) Scaled Integrative Genomics Viewer (IGV) snapshots of two differentially expressed genes, MCIDAS (A) and KRT20 (B), from RNA sequencing (RNA-seq) performed in three distinct TELO2AID and AIDTRRAP clones treated with auxin or NaOH for 48 hr or 24 hr, respectively. (C, D) Volcano plots showing transcript level fold change (FC) measured upon TELO2 (C) and TRRAP (D) depletion plotted against the adjusted p-value (false discovery rate [FDR]). FC was calculated as the Log2 of the ratio of the expression value of each gene between NaOH- (+TELO2 or +TRRAP) and auxin-treated (-TELO2 or -TRRAP) cells. Twofold change thresholds and a 1% FDR cutoff are shown. Blue dots represent mRNAs whose levels decrease at least twofold, and red dots represent mRNAs whose levels increase at least twofold. (E–H) Quantitative RT-PCR analysis of selected downregulated (CCNA2, MYB, GNL3, and MIR17HG) (E, F) and one upregulated gene (KRT20) (G, H). mRNA levels were measured in two TELO2AID clones treated with auxin or NaOH for 48 hr (E, G), and in three AIDTRRAP clones treated with auxin or NaOH for 24 hr (F, H). Each value represents mean mRNA levels from at least three independent experiments, overlaid with individual data points and error bars showing the SD. PPIB served as a control for normalization across samples. Values from one NaOH-treated control sample were set to 1, allowing comparisons across culture conditions and replicates. Statistical significance was determined by two-way ANOVA followed by Bonferroni’s multiple comparison tests. *p≤0.01.
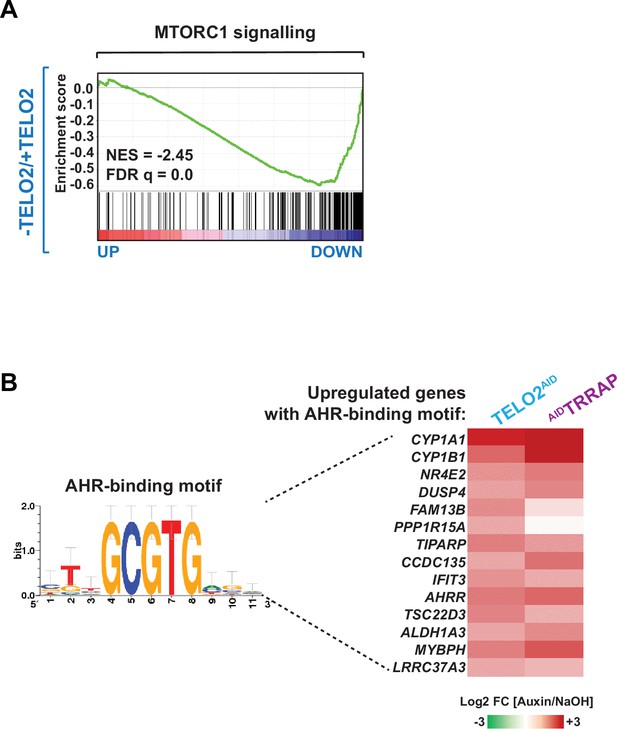
Auxin treatment induces aryl hydrocarbon receptor (AHR)-responsive genes.
(A) Gene Set Enrichment Analysis (GSEA) showing the enrichment of genes encoding MTORC1 signaling components in the ranked transcriptome profiles of TELO2-depleted cells. Green lines represent enrichment profiles. NES: normalized enrichment score. Each hit from the hallmark gene set is represented by a vertical black bar, positioned on the ranked transcriptome profile with color-coded fold change values. (B) Heatmap representation of genes containing a consensus AHR-binding motif and whose expression changes in TELO2 and TRRAP-depleted cells. The Log2 ratio between auxin- and NaOH-treated cells for each transcript is represented using a sequential color scale. All data are from RNA-seq experiments performed in three distinct TELO2AID and AIDTRRAP clones treated with auxin or NaOH for 48 hr or 24 hr, respectively. Shown is the sequence logo of the minimal core AHR-binding motif 5′-GCGTG-3′ (transfac_pro__M00778) (Lo and Matthews, 2012).
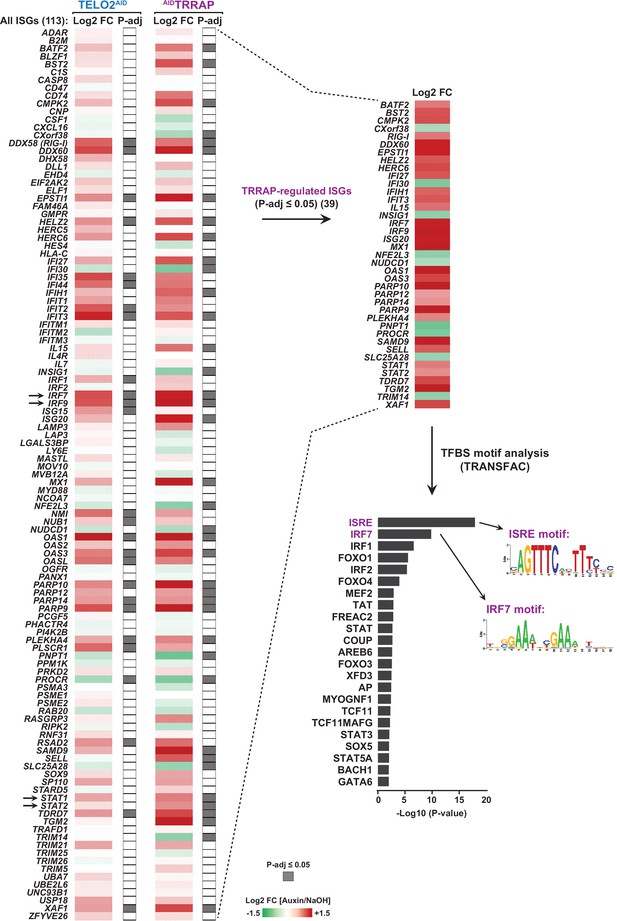
TELO2 and TRRAP repress type I interferon-stimulated genes (ISGs).
Heatmap representation of the expression of a list of IFN-I-stimulated genes (113 ISGs) in TELO2 and TRRAP-depleted cells. The list of 113 ISGs results from merging the list of 97 genes from the IFN α-responsive hallmark set with the lists of genes regulated by U-ISGF3 (Wang et al., 2018) and ISGF3 (Brown and Gromeier, 2017), as defined in Cheon et al., 2013. The Log2 ratio between auxin- and NaOH-treated cells for each transcript is represented using a sequential color scale. Gray: adjusted p-values ≤ 0.05 (false discovery rate [FDR] ≤ 5%); white: >0.05. All data are from RNA-seq experiments performed in three distinct TELO2AID and AIDTRRAP clones treated with auxin or NaOH for 48 hr and 24 hr, respectively. Arrows indicate the genes encoding the IRF7 and ISGF3 transcription factors. A heatmap highlighting the 39 ISGs whose expression changes upon TRRAP depletion (FDR ≤ 5%) is shown on the right. Below is a histogram of p-values of transcription factor binding sites (TFBS) enrichment in the sequences of the 39 TRRAP-regulated ISGs. Shown are the sequence logos of the two most enriched motifs, the ISRE motif (transfac_public__M00258) and the IRF7 motif (transfac_public__M00453). The number of genes in each category is indicated in parentheses.
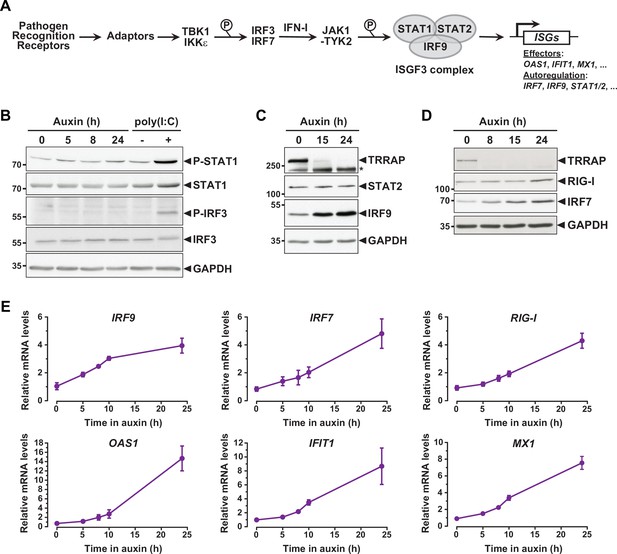
Interferon-stimulated gene (ISG) induction in the absence of an innate immune response.
(A) Schematic representation of the innate immune and IFN-I signaling pathways. (B) Western blot analyses of phosphorylated and total STAT1 and IRF3 levels in extracts from AIDTRRAP cells treated with either NaOH for 24 hr and auxin for various time points, or transfected with polyI:C, as indicated. (C) Western blot analyses of TRRAP, STAT2, and IRF9 levels in extracts from AIDTRRAP cells treated with NaOH for 24 hr or auxin for various time points, as indicated. * indicates nonspecific detection. (D) Western blot analyses of TRRAP, RIG-I, and IRF7 levels in extracts from AIDTRRAP cells treated with NaOH for 24 hr or auxin for various time points, as indicated. (E) RT-qPCR analysis of IRF9, IRF7, RIG-I, OAS1, IFIT1, and MX1 mRNA levels in AIDTRRAP cells over a time course of auxin treatment. RNAs were extracted from cells treated with auxin and harvested at various time points, as indicated. Each value represents mean mRNA levels from at least three independent experiments with distinct AIDTRRAP clones, overlaid with error bars showing the SD for each time point. PPIB served as a control for normalization across samples. Values from one untreated replicate were set to 1, allowing comparisons across culture conditions and replicates. Source data are available in supplementary material (B–D: Source data 1).
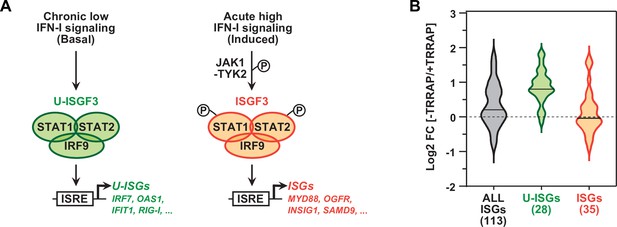
TRRAP specifically represses U-ISGF3-regulated genes.
(A) Schematic representation of the transcription regulation of distinct subsets of interferon-stimulated genes (ISGs), depending on whether ISGF3 unphosphorylated, in basal conditions (U-ISGF3, left), or phosphorylated in response to IFN-I signaling (ISGF3, right). (B) Violin plots showing the distribution of gene expression changes induced by TRRAP depletion for all ISGs (gray), for ISGs controlled by U-ISGF3 (green), and for ISGs controlled by ISGF3 (orange). Log2 fold changes (Log2[FC]) were calculated as the Log2 of the ratio of the expression value of each gene between NaOH- (+TRRAP) and auxin-treated (-TRRAP) AIDTRRAP cells. The number of genes in each category is indicated.
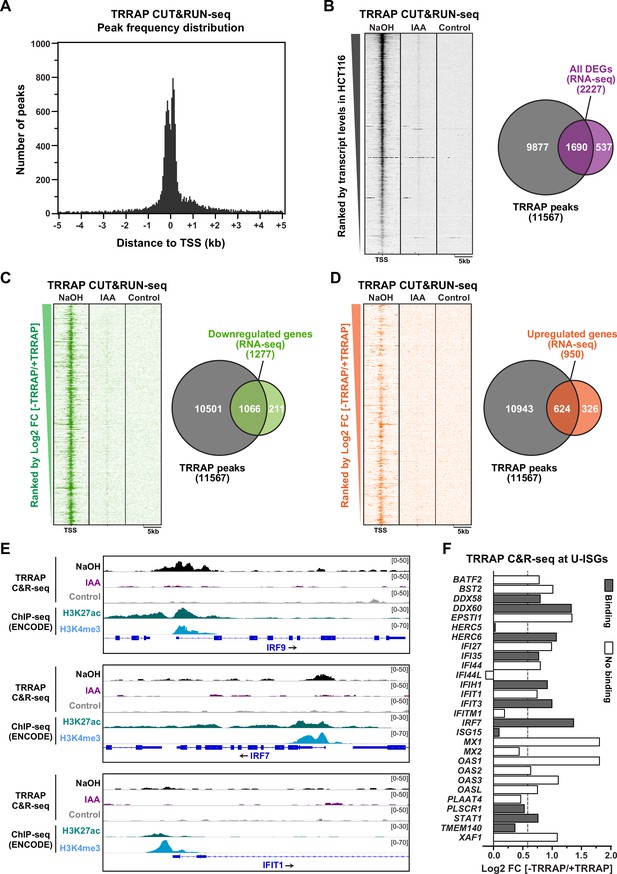
Genome-wide profiling of TRRAP occupancy.
(A) Histogram showing the frequency distribution of TRRAP-bound loci across the genome, relative to annotated transcription start sites (TSS). Shown are the number of peaks computed by MACS2 in bins of 50 bp within ±5 kb of each TSS. (B–D) Heatmaps showing anti-HA CUT&RUN-seq profiles of TRRAP, along with control IgG profiles, obtained from AIDTRRAP HCT116 cells treated with either NaOH or auxin (IAA) for 12 hr, as indicated. For each gene, 5 kb regions upstream and downstream of the TSS are shown and ranked either by transcript abundance in control conditions (B) or the Log2 fold change (FC) upon auxin-mediated TRRAP depletion (C, D). Right panels show Venn diagrams of the overlap between the number of TRRAP-bound promoters and all differentially expressed genes in TRRAP-depleted HCT116 cells (B), downregulated genes (C), and upregulated genes (D) (false discovery rate [FDR] ≤ 1). All transcript-level measurements were obtained from our RNA-seq analyses (Figure 2). All anti-HA CUT&RUN-seq experiments were performed in duplicates. (E) Scaled snapshots of TRRAP CUT&RUN-seq profiles from AIDTRRAP HCT116 cells treated as in (B) with NaOH (black) and IAA (purple), along with control IgG profiles (gray). Shown are the IRF9, IRF7, and IFIT1 loci, from top to bottom. ChIP-seq profiles of the H3K27ac and H3K4me3 marks in HCT116 cells were obtained from the ENCODE database and serve to locate the proximal-promoter regions of each gene. (F) Overview of TRRAP binding at promoters of the 28 annotated U-ISGs (Figure 4). The bar graph shows the FC of each transcript in TRRAP-depleted HCT116 cells, computed from our RNA-seq analyses (Figure 2), color-coded according to either the presence (filled bar) or absence (empty bar) of a TRRAP CUT&RUN peak in their promoters. The dashed line corresponds to a 1.5-fold change threshold.
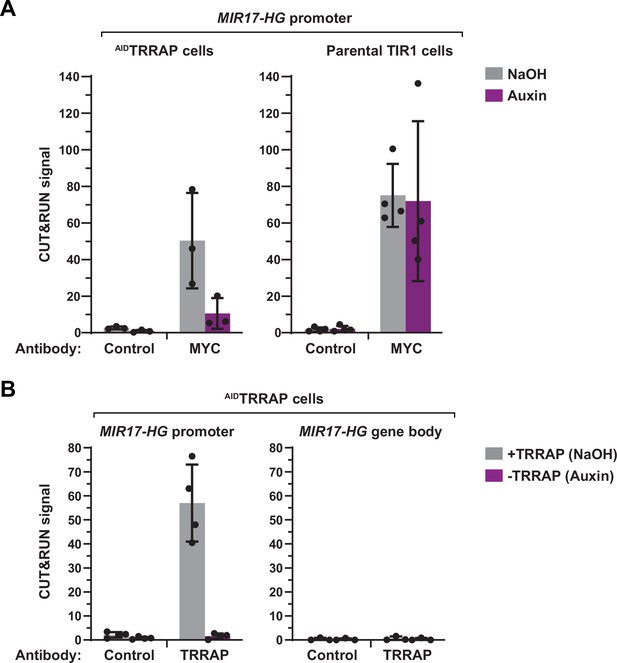
Implementation of the CUT&RUN procedure.
(A, B) CUT&RUN-qPCR analysis of MYC (A) and TRRAP (B) occupancy at the MIR17-HG locus in HCT116 cells. Parental TIR-1-expressing and -derived AIDTRRAP cell lines were treated with either NaOH (gray) or auxin (purple) for 12 hr. CUT&RUN were performed using either rabbit IgGs (control), an anti-MYC antibody, or an anti-HA antibody to target endogenous TRRAP. Cleaved DNA fragments were released, extracted, and analyzed by qPCR using oligonucleotides that amplify either the promoter or the gene body of MIR17-HG, as indicated. Each column represents the mean footprint signal measured from at least three independent experiments with distinct AIDTRRAP clones, overlaid with individual data points and error bars showing the SD.
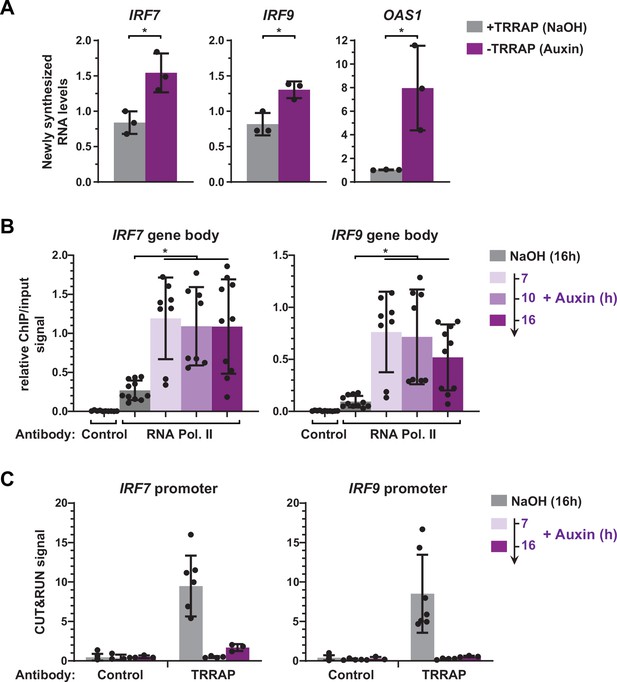
TRRAP directly represses the transcription of IRF7 and IRF9.
(A) RT-qPCR analysis of newly synthesized IRF7, IRF9, and OAS1. AIDTRRAP cells were treated with either NaOH (gray) or auxin (purple) for 10 hr, prior to a 20 min incubation with 4-thiouridine (4sU), extraction, and enrichment of labeled nascent RNAs. Each value represents mean pre-mRNA levels from three independent experiments with distinct AIDTRRAP clones, overlaid with individual data points and error bars showing the SD. PPIB served as a control for normalization across samples. For all genes, primers amplifying intronic regions were used. Values from one untreated replicate were set to 1, allowing comparisons across culture conditions and replicates. Statistical significance was determined by unpaired, two-tailed Student’s t-tests. *p≤0.05. (B) Kinetic ChIP-qPCR analysis of RNA polymerase II occupancy at the IRF7 and IRF9 gene bodies. AIDTRRAP cells were treated with NaOH for 16 hr and auxin for 7, 10, and 16 hr, as indicated, prior to chromatin extraction, sonication, and immunoprecipitation with either control rabbit IgGs or an antibody recognizing total RNA polymerase II (F-12). Each value represents mean IP/input ratios from at least eight independent experiments, overlaid with individual data points and error bars showing the SD. Statistical significance was determined by one-way ANOVA followed by Tukey’s multiple comparison tests. *p≤0.05. (C) CUT&RUN-qPCR analysis of TRRAP occupancy at IRF7 and IRF9 promoters. AIDTRRAP cells were treated with either NaOH for 16 hr or auxin for 7 and 16 hr, as indicated, prior to CUT&RUN-qPCR experiments performed using either control rabbit IgGs or an anti-HA antibody, to target endogenous TRRAP. Each column represents the mean footprint signal measured from at least three independent experiments, overlaid with individual data points and error bars showing the SD.
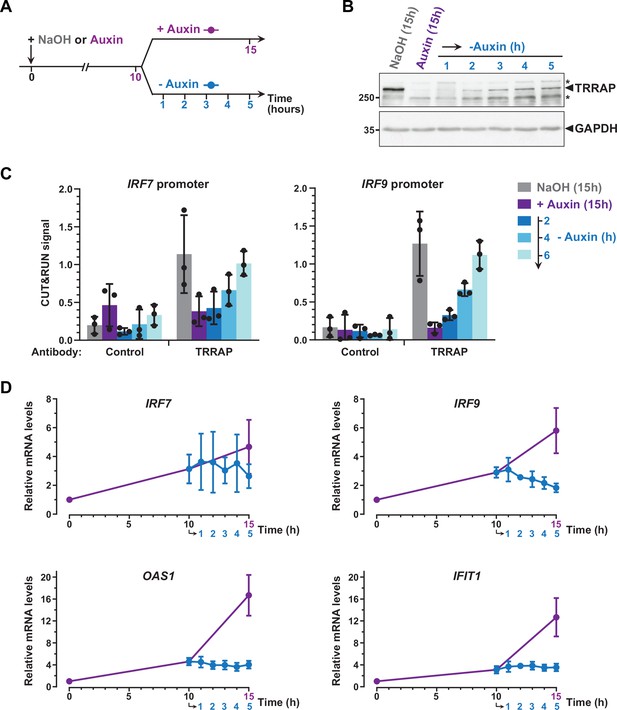
Dynamics of interferon-stimulated gene (ISG) regulation by TRRAP.
(A) Experimental strategy: AIDTRRAP cells were harvested before auxin addition, after incubation with auxin for 10 hr (purple), and at various time points up to 5 hr after auxin removal (blue), as indicated. As a control, AIDTRRAP cells were also harvested following 15 hr of either NaOH (gray) or auxin (purple) treatment. (B) Western blot analysis of TRRAP protein levels after auxin addition and subsequent removal, demonstrating the reversibility of TRRAP auxin-mediated degradation. Blots were probed with anti-TRRAP and anti-GAPDH antibodies. The latter was used to control for equal loading. * marks nonspecific bands detected by the anti-TRRAP antibody. Source data are available in supplementary material (Figure 6B, Source data 1). (C) CUT&RUN-qPCR analysis of TRRAP-bound DNA extracted following micrococcal nuclease (MNase) cleavage directed either by an anti-HA antibody (TRRAP) or control IgG (control). CUT&RUN was performed in AIDTRRAP cells treated with either NaOH (gray) or auxin (purple) for 15 hr as controls. In parallel, AIDTRRAP cells were also treated with auxin for 10 hr and then washed out of auxin for 2, 4, and 6 hr (blue). qPCR was performed using oligonucleotides that amplify proximal regions of the IRF7 and IRF9 promoters, as in Figure 6C. Each column represents the mean footprint signal measured from three distinct AIDTRRAP clones, overlaid with individual data points and error bars showing the SD. (D) RT-qPCR analysis of IRF7, IRF9, OAS1, and IFIT1 mRNA levels after auxin addition and subsequent removal. RNAs were extracted from cells treated as schematized in (A). Each point represents the mean value of three distinct AIDTRRAP clones, overlaid with error bars showing the SD. PPIB served as a control for normalization across samples. Values from untreated samples were set to 1, allowing comparisons across culture conditions and replicates.
Additional files
-
Supplementary file 1
Gene expression changes upon TELO2 depletion.
Shown are DESeq2 results from RNA-seq experiments comparing HCT116 TELO2-AID cells treated with either NaOH or auxin for 48 hr. log2FoldChange indicates the Log2 fold change of the ratio of RNA-seq counts in auxin-treated cells over NaOH-treated cells, averaged from three independent clones. padj indicates the p-values after Benjamin–Hochberg correction for multiple testing.
- https://cdn.elifesciences.org/articles/69705/elife-69705-supp1-v2.csv
-
Supplementary file 2
Gene expression changes upon TRRAP depletion.
Shown are DESeq2 results from RNA-seq experiments comparing HCT116 AID-TRRAP cells treated with either NaOH or auxin for 24 hr. log2FoldChange indicates the Log2 fold change of the ratio of RNA-seq counts in auxin-treated cells over NaOH-treated cells, averaged from three independent clones. padj indicates the p-values after Benjamin–Hochberg correction for multiple testing.
- https://cdn.elifesciences.org/articles/69705/elife-69705-supp2-v2.csv
-
Supplementary file 3
Interferon-stimulated gene (ISG) expression changes upon TELO2 and TRRAP depletion.
Shown are DESeq2 results as in Tables S1 and S2, filtered for IFN-I stimulated genes (113 ISGs). The list of 113 ISGs results from merging the 97 genes from the HALLMARK_INTERFERON_ALPHA_RESPONSE (MSigDB) with all non-redundant U-ISGF3- and ISGF3-regulated genes. Source data for Figure 4 and Figure 2—figure supplement 3.
- https://cdn.elifesciences.org/articles/69705/elife-69705-supp3-v2.csv
-
Supplementary file 4
Supplementary file 4.Genome-wide occupancy profile of TRRAP in HCT116 cells.
'NarrowPeaks' output files from MACS2 peak calling analyses of duplicate anti-HA CUT&RUN-seq experiments performed in AID-TRRAP cells, treated with either NaOH or auxin (IAA) for 12 hr, compared to a control IgG CUT&RUN-seq sample.
- https://cdn.elifesciences.org/articles/69705/elife-69705-supp4-v2.xlsx
-
Supplementary file 5
List of oligonucleotides used in this study.
- https://cdn.elifesciences.org/articles/69705/elife-69705-supp5-v2.xlsx
-
Supplementary file 6
List of antibodies used in this study.
- https://cdn.elifesciences.org/articles/69705/elife-69705-supp6-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/69705/elife-69705-transrepform1-v2.pdf
-
Source data 1
This contains all original uncropped scans of all Western blots.
- https://cdn.elifesciences.org/articles/69705/elife-69705-data1-v2.zip





