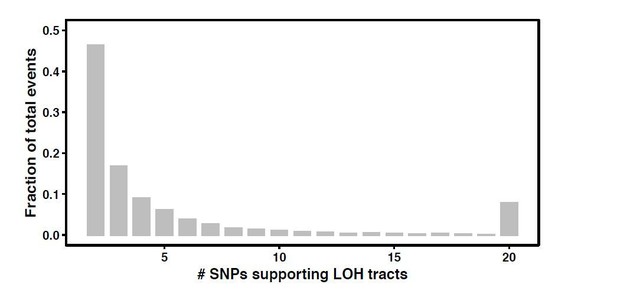
Loss of heterozygosity results in rapid but variable genome homogenization across yeast genetic backgrounds
Figures
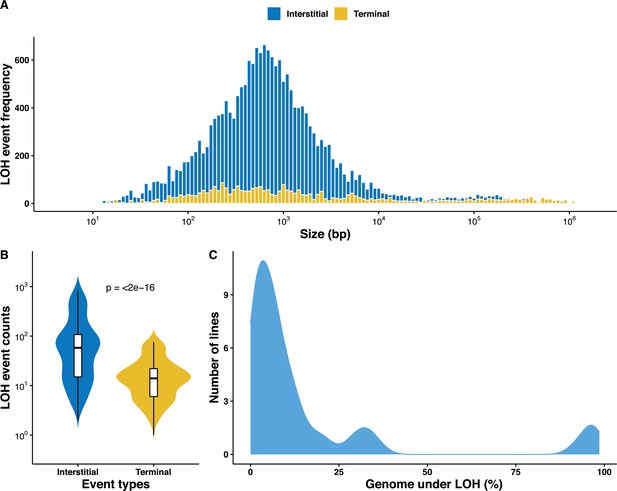
Overall distribution of loss of heterozygosity (LOH) in the 169 mutation accumulation (MA) lines.
(A) LOH event tract size distribution across all 169 MA lines, the average tract sizes of the interstitial LOH (I-LOH) events (7.4 kb) and terminal LOH (T-LOH) events (55.3 kb), respectively. The global average LOH event size was 14.1 kb. (B) Violin plot of the LOH event counts in the MA lines population, I-LOH events were found to be significantly greater than T-LOH events (Wilcoxon test, p<2×10−16). (C) Distribution of MA lines based on the proportion of genome under LOH (%), dashed line indicates average proportion of genome under LOH across the 169 MA lines, 15.9% (±1.86).

Distribution of the heterozygous single nucleotide polymorphism (SNP) densities as a fraction of total heterozygous SNPs in 5 kb windows across the ancestral diploids as described in Table 1.
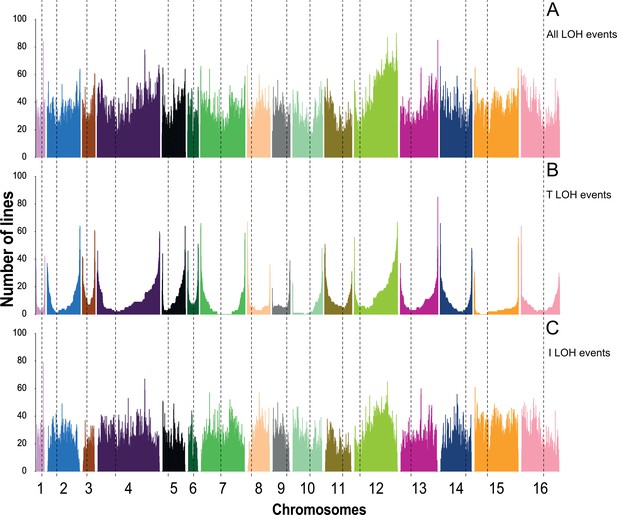
Chromosome-wide distribution of loss of heterozygosity (LOH) events across all mutation accumulation (MA) lines.
(A) All LOH events, (B) terminal LOH (T-LOH), events, and (C) interstitial LOH (I-LOH) events.
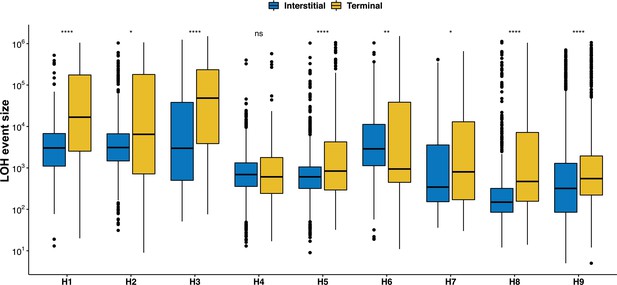
A terminal LOH (T-LOH) events are significantly larger than interstitial LOH (I-LOH) in the 169 mutation accumulation (MA) lines all backgrounds except H4 (Wilcoxon test, *p < 0.05; **p < 0.01; ***p<0.001; ****p < 0.0001; ns – not significant).
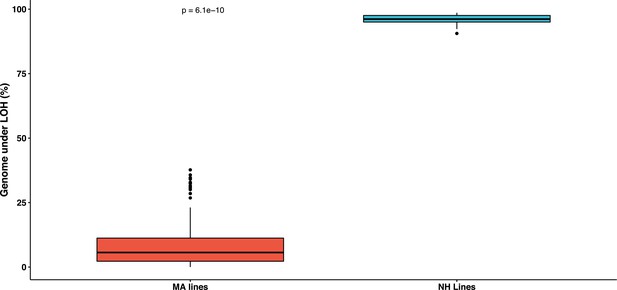
Boxplot depicting the fraction genome under loss of heterozygosity (LOH) in the nearly homozygous lines is significantly greater than the rest of the mutation accumulation (MA) lines (Wilcoxon test, p=6.1×10−10).
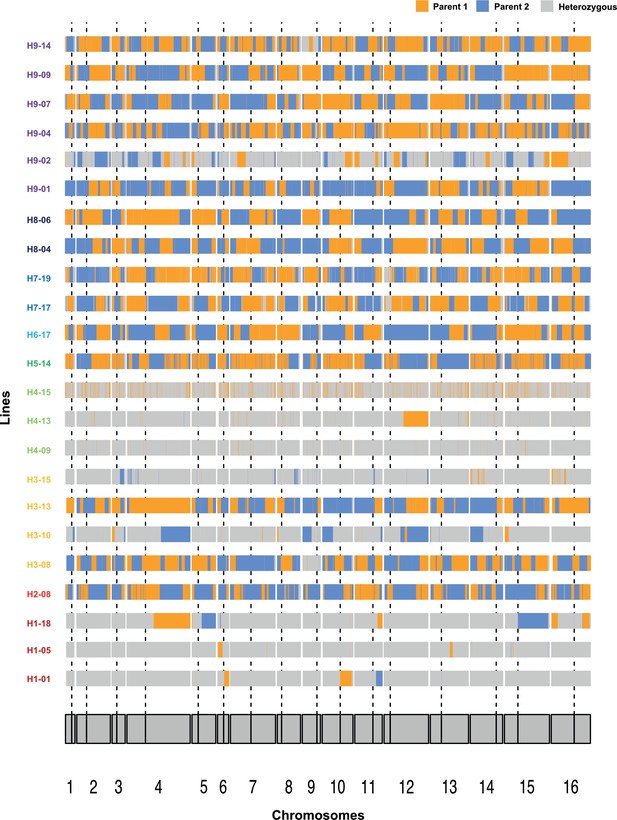
Chromosome-wide loss of heterozygosity (LOH) plots across representative lines from H1, H3, H4, and H9.
Orange and blue colors represent single nucleotide polymorphisms (SNPs) fixed toward either of the parents as described in Table 1 and Supplementary file 1. All the 14 nearly homozygous (NH) lines have been depicted. Dotted vertical lines represent position of the centromere.
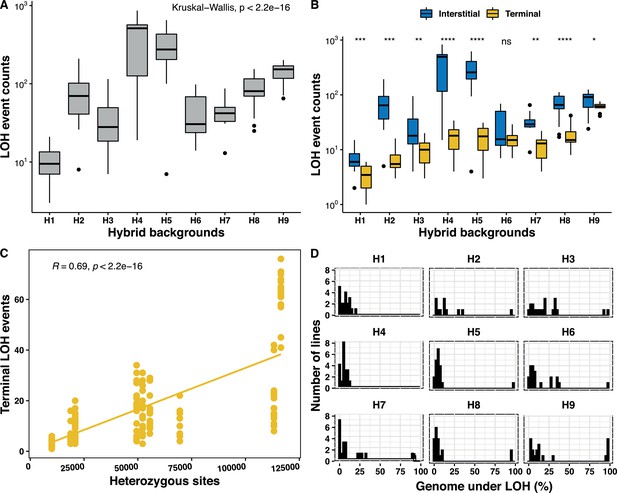
Genetic background-dependent variation in the loss of heterozygosity (LOH) repertoire.
(A) The frequency of total LOH events across the nine genetic backgrounds are highly variable (Kruskal-Wallis test, p<2×10−16). (B) Variability in interstitial and terminal LOH events counts across all backgrounds H1–H9 (Kruskal-Wallis test, p<10−16), interstitial events were always in excess, except for in H6 (Wilcoxon test, *p < 0.05; **p < 0.01; ***p<0.001; ****p < 0.0001; ns – not significant). (C) Frequency of terminal LOH events increases with increasing heterozygosity in the nine genetic backgrounds (Pearson’s correlation; r = 0.69, p<2×10−16), interstitial and total events do not bear any correlation with the background heterozygosity (Pearson’s correlation; p>0.05). (D) Proportion of genome under LOH is significantly variable across the nine backgrounds (H1–H9) (Kruskal-Wallis test, p<2×10−16).
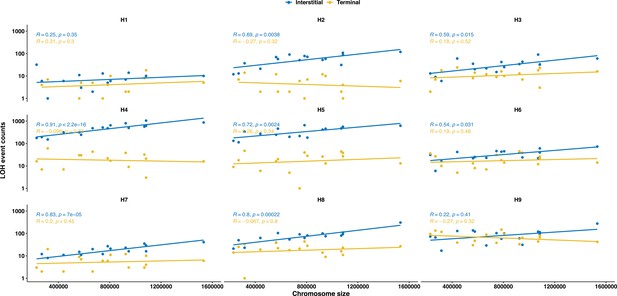
The frequency of interstitial LOH (I-LOH) events increases with increasing chromosome size for all backgrounds except for H1 and H9; no such correlation was observed for terminal LOH (T-LOH) events.
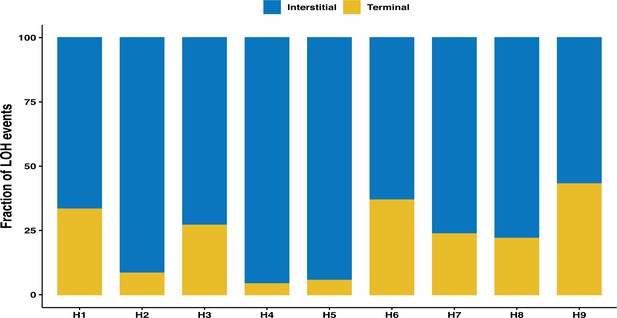
Overall fraction of interstitial and terminal loss of heterozygosity (LOH) tracts across the nine genetic backgrounds.
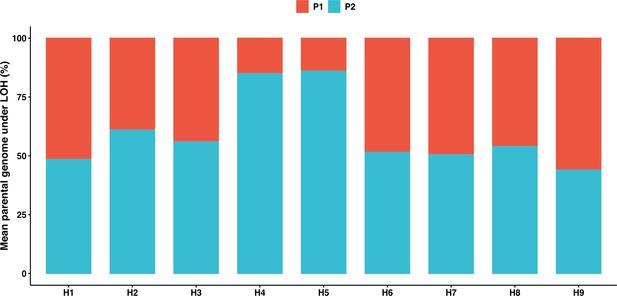
Fraction of the genome fixed toward either of the parental genomes across the nine genetic backgrounds.
Biased fixation was only observed in H4 and H5 backgrounds (binomial test, p<0.05).
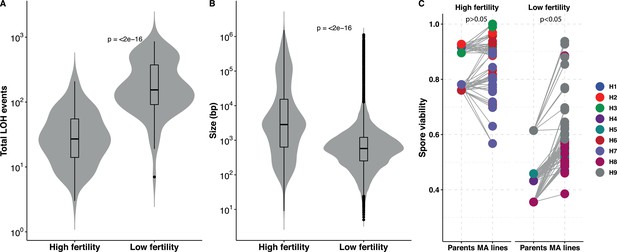
Loss of heterozygosity (LOH) accumulation is associated with spore fertility.
(A) Total LOH events accumulated in mutation accumulation (MA) lines derived from ancestral diploids with high spore fertility, that is, meiotic spore viability greater than or equal to 75% and the fraction of four-spore viable tetrads greater than 50% (H1, H2, H3, H6, H7) is significantly lower than in MA lines derived from ancestral diploids with low spore fertility, that is, meiotic spore viability lesser than 75% and the fraction of four-spore viable tetrads less than 50% (H4, H5, H8, H9) (Wilcoxon test, p<2×10−16). (B) The size (in bp) of the LOH events accumulated in MA lines derived from ancestral diploids with high spore fertility is significantly larger than in MA lines derived from ancestral diploids with low spore fertility (Wilcoxon test, p<2×10−16). The average LOH event size in the high spore fertility and low spore fertility MA lines are 45.8 and 7.4 kb, respectively. (C) Spore viabilities in both the high and low spore fertility groups compared respective to their ancestral diploids. There is a significant improvement in the spore viabilities of the MA lines derived from the low spore fertility ancestors (Mann-Whitney U test, p=0.04), whereas the viabilities do not change in the MA lines derived from the high spore fertility ancestral diploids (Mann-Whitney U test, p=0.34). The spore viabilities of the individual MA lines and the ancestral diploids detailed in Supplementary file 4.
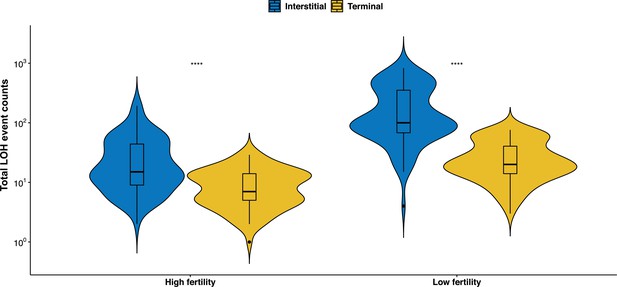
The total number of interstitial LOH (I-LOH) and terminal LOH (T-LOH) events are significantly greater in the mutation accumulation (MA) lines derived from ancestral diploids with low fertility (Wilcoxon test, *p < 0.05; **p < 0.01; ***p<0.001; ****p < 0.0001; ns – not significant).
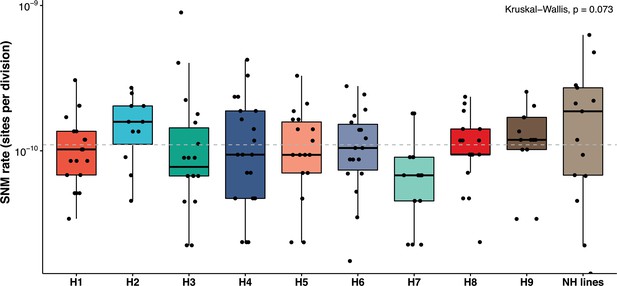
Mutation rates are constant across the backgrounds and within the nearly homozygous (NH) lines (range 0.67–1.9 × 10−10 mutations/site/division; Kruskal-Wallis test, p>0.05).
The overall, mean single nucleotide mutation (SNM) rate in the 169 MA lines 1.1 × 10−10 per site per division is not different from previous estimates in various diploid Saccharomyces cerevisiae strains. NH lines represent the SNM rates in the NH lines. SNMs have been detailed in Supplementary files 5 and 6.
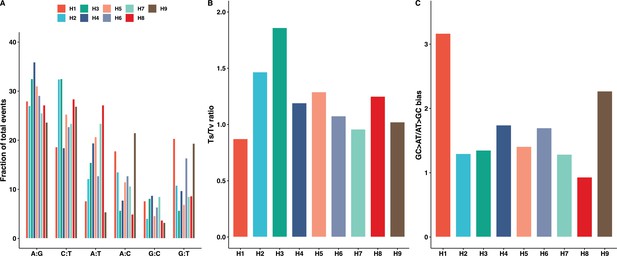
SNM spectrum across the nine hybrid backgrounds.
(A) The spectrum of single nucleotide mutations (SNMs) in the mutation accumulation (MA) lines is variable across all genetic backgrounds (Chi-square test, p<0.05; Supplementary file 5–6). (B) The transition to transversion ratio is similar across all genetic backgrounds, significantly increased only in H3 (Chi-square test, p<0.05; Supplementary file 5–6). (C) The GC > AT/AT > GC mutational bias was similar in all genetic backgrounds, significantly increased only in H1 (Chi-square test, p<0.05; Supplementary file 5–6).
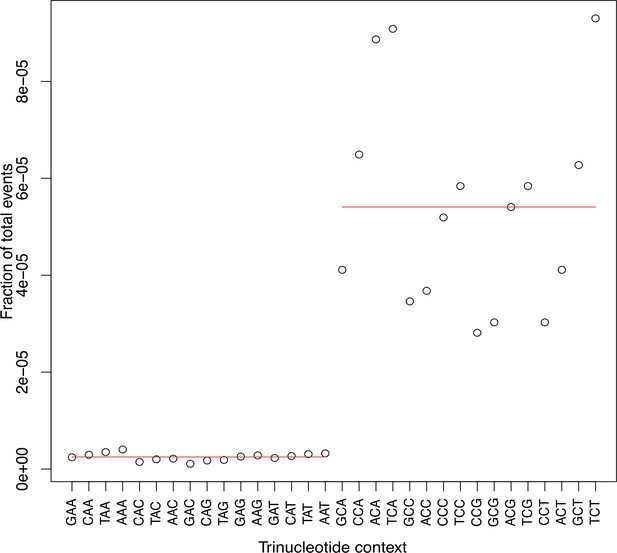
The frequency of mutations was significantly impacted at C/G sites than A/T sites by the neighboring nucleotides (Chi-square test, p<0.05; Supplementary file 5–6).
Horizontal red lines indicate mean events.
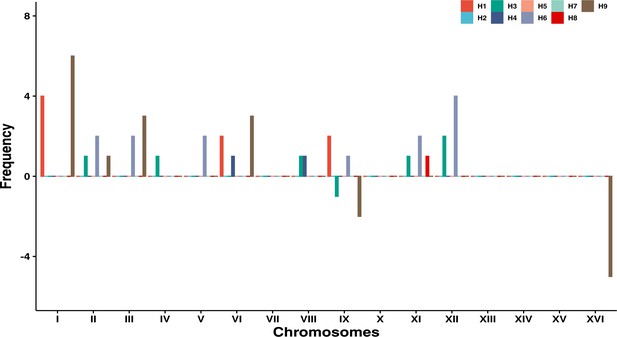
The frequency of aneuploid chromosomes across the nine genetic backgrounds (H1–H9).
Two backgrounds carried ancestral trisomies, H3 (+1 chrIX) and H9 (+1 chrIX; +1 chrXVI).
Tables
Hybrid mutation accumulation (MA) lines.
| Hybrid | Cross* | Het positions | No. of sequenced lines | No. of bottlenecks | Total no. of divisions |
|---|---|---|---|---|---|
| H1 | ABS × BKL | 9972 | 20 | 100 | 2446 |
| H2 | ABP × BFQ | 18789 | 12 | 75 | 1842 |
| H3 | BAP × BAN | 20875 | 20 | 75 | 1863 |
| H4 | BTI × ABA | 49412 | 20 | 75 | 1772 |
| H5 | ACD × AKQ | 52223 | 20 | 75 | 1777 |
| H6 | ACK × CMQ | 55570 | 20 | 100 | 2392 |
| H7 | ACG × BAK | 69456 | 19 | 75 | 1844 |
| H8 | CGD × AKE | 113241 | 19 | 75 | 1769 |
| H9 | BAM × CPG | 116475 | 19 | 100 | 2452 |
-
* Standardized names from Peter et al., 2018.
Mean event counts for 5 kb at chromosome ends vs rest of the genome.
| Hybrid Background | Mean density at chr_ends | Mean density rest_genome |
|---|---|---|
| H1 | 3.66 | 3.98 |
| H2 | 7.06 | 6.81 |
| H3 | 7.57 | 9.26 |
| H4 | 15.00 | 22.19 |
| H5 | 20.05 | 23.67 |
| H6 | 20.27 | 22.80 |
| H7 | 31.54 | 31.78 |
| H8 | 36.20 | 46.73 |
| H9 | 46.58 | 47.70 |
Additional files
-
Supplementary file 1
Haploid strains used to generate the nine hybrid diploids, standardized names as per Peter et al., 2018.
- https://cdn.elifesciences.org/articles/70339/elife-70339-supp1-v2.xlsx
-
Supplementary file 2
Mean growth rate estimates at bottleneck 0 and at the end of the experiment for the nine genetic backgrounds.
These estimates were used to determine the number of divisions at every bottleneck (see Materials and methods).
- https://cdn.elifesciences.org/articles/70339/elife-70339-supp2-v2.xlsx
-
Supplementary file 3
All loss of heterozygosity (LOH) tracts across the 169 mutation accumulation (MA) lines supported by at least two single nucleotide polymorphisms (SNPs).
Tracts were merged if consecutive tracts were disrupted by less than two SNPs. LOH tracts in nearly homozygous lines highlighted in yellow.
- https://cdn.elifesciences.org/articles/70339/elife-70339-supp3-v2.xlsx
-
Supplementary file 4
Meiotic spore viability in the mutation accumulation (MA) lines, H9-8 and H9-11 excluded as they did not sporulate.
SV-percentage spore viability in the MA lines; statistical significance of differences in spore viability between ancestral diploid and the derived MA lines were determined using the p-values from Fisher’s exact test (GraphPad prism). N – number of tetrads analyzed for spore viability, ns – not significant; p>0.05.
- https://cdn.elifesciences.org/articles/70339/elife-70339-supp4-v2.xlsx
-
Supplementary file 5
Mutations detected in the 169 mutation accumulation (MA) lines.
Multi-nucleotide mutations (MNMs) highlighted in purple.
- https://cdn.elifesciences.org/articles/70339/elife-70339-supp5-v2.xlsx
-
Supplementary file 6
Chi-square test p-values for the single nucleotide mutation (SNM) spectrum variation.
Chi-square test was performed using the Microsoft excel function Chisq.test().
- https://cdn.elifesciences.org/articles/70339/elife-70339-supp6-v2.xlsx
-
Supplementary file 7
List of strains used in this study.
- https://cdn.elifesciences.org/articles/70339/elife-70339-supp7-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/70339/elife-70339-transrepform-v2.docx