Minian, an open-source miniscope analysis pipeline
Figures
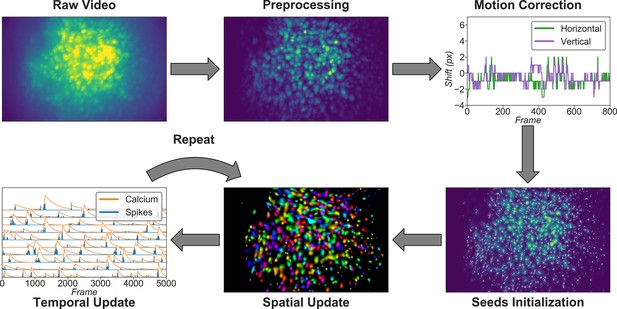
Overview of the analysis pipeline.
The analysis is divided into five stages: preprocessing, where sensor noise and background fluorescence from scattered light are removed; motion correction, where rigid motion of the brain is corrected; seeds initialization, where the initial spatial and temporal matrices for later steps are generated from a seed-based approach; spatial update, where the spatial footprints of cells are further refined; and temporal update, where the temporal signals of cells are further refined. The last two steps of the pipeline are iterative and can be repeated multiple times until a satisfactory result is reached.
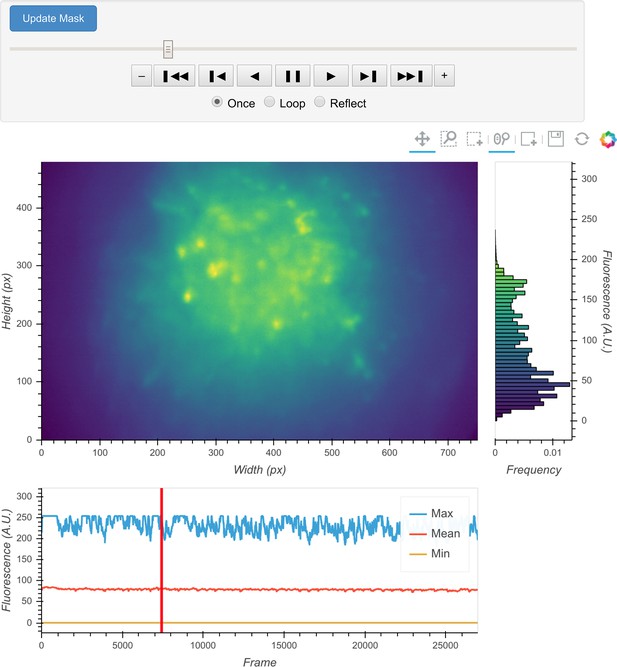
Interactive visualization of raw input video.
One frame is shown in the central panel of the visualization that can be interactively updated with the player toolbar on the top. A histogram of fluorescence intensity of the current frame is shown on the right and will update in response to zooming in on the central frame. A line plot of summary values across time is shown at the bottom. Here, the maximum, mean, and minimum fluorescence values are plotted. These summaries are useful in checking whether there are unexpected artifacts or gaps in the recording. Finally, the user can draw an arbitrary box in the central frame, and the position of this boxed region can be recorded and used as a mask during later steps. For example, during motion correction a subregion of the data containing a stable landmark might provide better information on the motion.
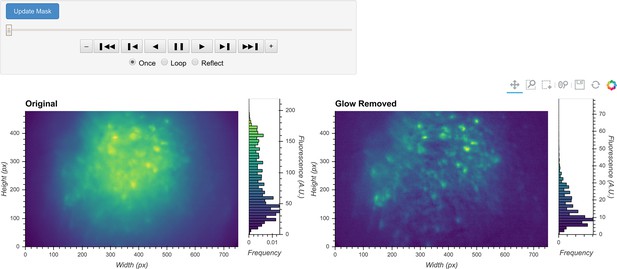
General visualization of preprocessing.
The same visualization of input video can be used to visualize the whole video before and after specific preprocessing steps side-by-side. The effect of vignetting correction is visualized here. The image and accompanying histogram on the left side show the original data; the data after vignetting correction are shown on the right side. Any frame of the data can be selected with the player toolbar and histograms are responsive to all updates in the image.
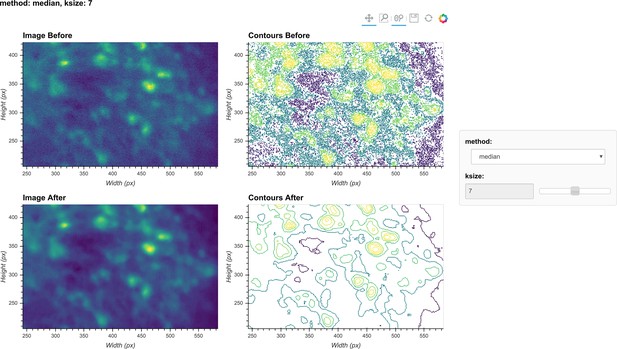
Visualization of denoising.
Here, a single frame from the data is passed through the background removal, and both the image and a contour plot are shown for the frame before and after the process. The contour plots show the iso-contour of five intensity levels spaced linearly across the full intensity range of the corresponding image. The plots are interactive and responsive to the slider of the window size on the right, thus the effect of different window sizes for denoising can be visualized.
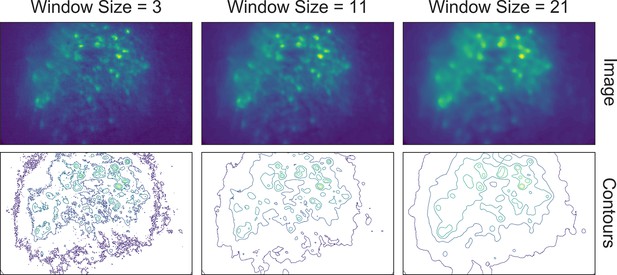
Effect of window size on denoising.
One example frame is chosen from the data, and the resulting images (top row) and contour plots (bottom row) are shown to demonstrate the effect of window size on denoising. Here, a window size of 11 (middle column) is appropriate while both smaller and larger window sizes result in artifacts.
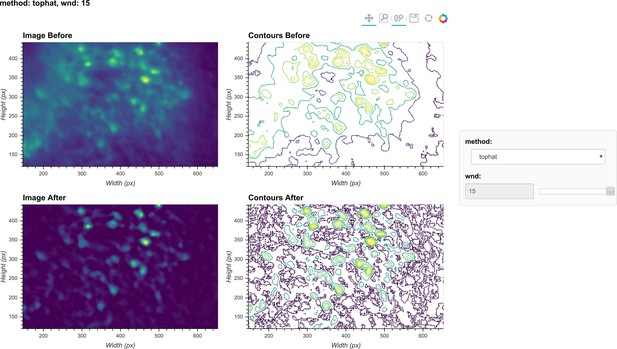
Visualization of background removal.
Here, a single frame from the data is passed through background removal, and both the image and a contour plot are shown for the frame before and after the process. The plots are interactive and responsive to the slider of the window size on the right, thus the effect of different window sizes for background removal can be visualized.
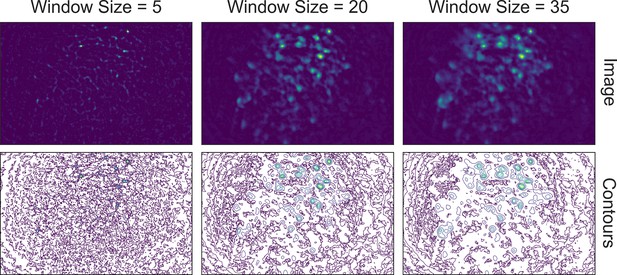
Effect of window size on background removal.
One example frame is chosen from the data, and the resulting images (top row) and contour plots (bottom row) are shown to demonstrate the effect of window size on background removal. The contour plots show the iso-contour of five intensity levels spaced linearly across the full intensity range of the corresponding image. Here, a window size of 20 pixels (middle column) is appropriate while both smaller and larger window sizes produce unsatisfactory results: a window size too small (left column) artificially limits the size of cells, and a window size too large (right column) does not remove the background effectively.
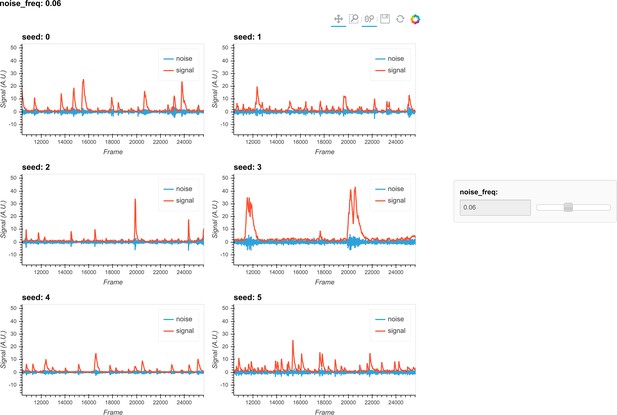
Visualization of noise frequency cutoff.
The cutoff frequency for noise is one of the critical parameters in the pipeline that affects both the seed initialization process and constrained non-negative matrix factorization’s (CNMF’s) temporal update steps. Here, we help the user determine that parameter by plotting temporal traces from six example seeds. In each plot, the raw signal is passed through a high-pass and low-pass filter at the chosen frequency, and the resulting signals are plotted separately as ‘noise’ and ‘signal.’ The plots are responsive to the chosen frequency controlled by the slider on the right. In this way, the user can visually inspect whether the chosen frequency can effectively filter out high-frequency noise without deforming the calcium signal.
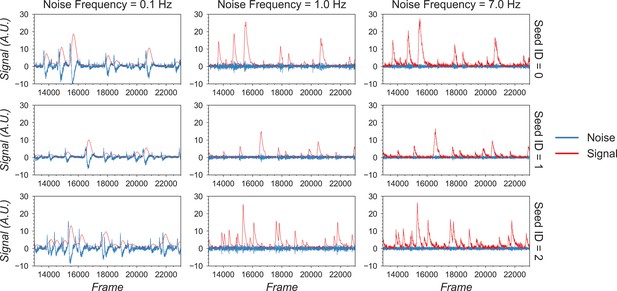
Example of filtered traces with different frequency cutoffs.
Here, the temporal dynamics of three example seeds are chosen, and the low-pass and high-pass filtered traces with different frequency cutoffs are shown. The low-pass filtered trace corresponds to ‘signal,’ while the high-pass filtered trace corresponds to ‘noise.’ Here, a 1 Hz cutoff frequency is considered appropriate since calcium dynamics and random noise are cleanly separated. A cutoff frequency smaller than 1 Hz left the calcium dynamics in the ‘noise’ trace, while a cutoff frequency larger than 1 Hz let random noise bleed into the ‘signal’ trace (i.e., high-frequency fluctuations are presented in periods where the cells seem to be inactive).
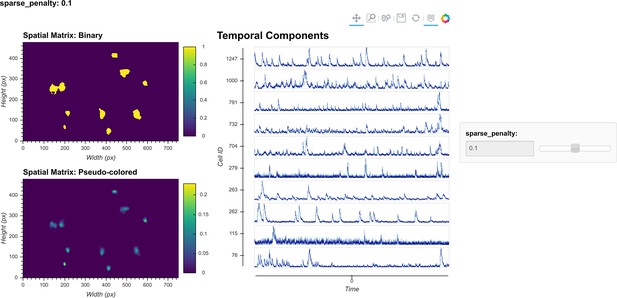
Visualization of spatial updates.
Here, 10 cells are randomly chosen to pass through spatial update with different parameters. The resulting spatial footprints, as well as binarized footprints, are plotted. In addition, the corresponding temporal traces of cells are plotted. The user can visually inspect the size and shape of the spatial footprints and at the same time easily determine whether the results are sparse enough by looking at the binarized footprints.
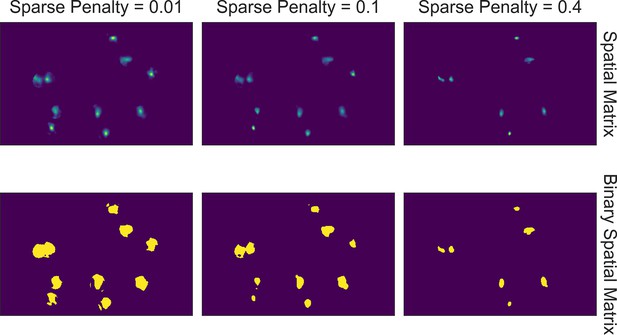
Effect of sparseness penalty in spatial update.
Here, the sum projection of the spatial matrix and binarized spatial matrix is shown for three different sparse penalties. A sparseness penalty of 0.1 is considered appropriate in this case. When the sparseness penalty is set lower, artifacts begin to appear. On the other hand, when the sparseness penalty is set higher, cells are dropped out.
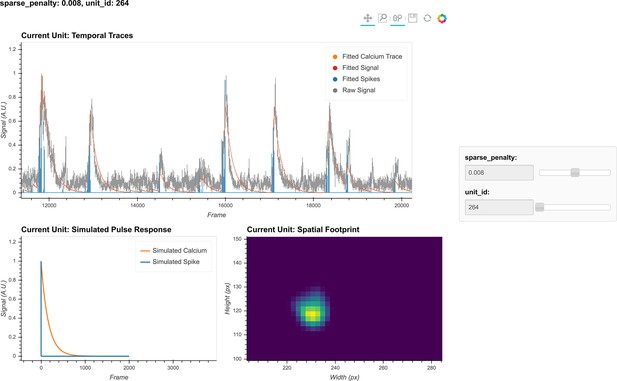
Visualization of temporal update.
Here, a subset of cells is randomly chosen to pass through temporal updates with different parameters. Only one cell is visualized at a given time, and the cell can be selected using the slider on the right. The raw signal, fitted signal, fitted calcium traces, and spike signals are overlaid in the same plot. In addition, a simulated pulse response based on the estimated autoregressive parameters is plotted with the same time scale. Furthermore, the corresponding spatial footprint of the cell is plotted for cross-reference. With a given set of parameters, the user can visually inspect whether the pulse response captures the typical calcium dynamics of the cell, and whether the timing and sparsity of the spike signal fit well with the raw data. The data shown here was acquired with a frame rate of 30 fps.
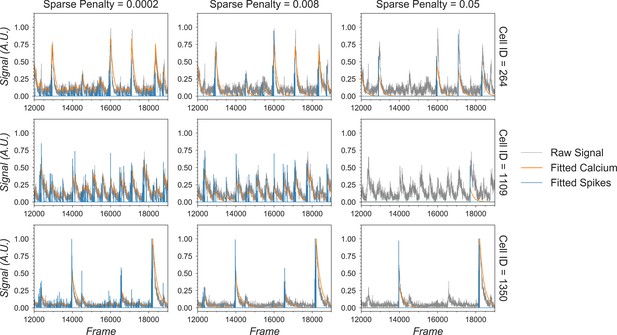
Effect of the sparseness penalty in temporal update.
Here, three example cells are selected and passed to the temporal update with different sparseness penalties. The ‘Raw Signal’ corresponds to the input video projected onto predetermined spatial footprints. The ‘Fitted Calcium’ and ‘Fitted Spikes’ correspond to the resulting model-fitted calcium dynamics and spike signals. A sparseness penalty of 0.008 (middle column) is considered appropriate in this case. The data shown here was acquired with a frame rate of 30 fps.
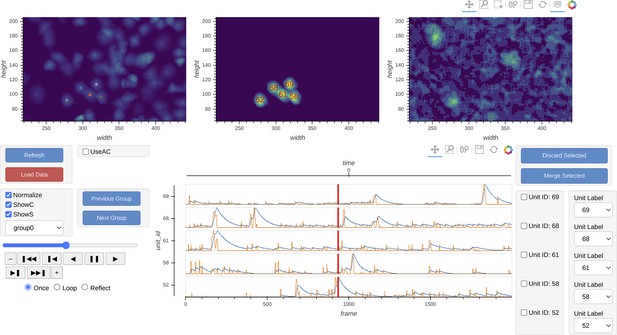
Interactive visualization of Minian output.
The three images on the top show the spatial footprints of all the cells (left), spatial temporal activities of selected subset of cells (middle), and preprocessed data. The bottom row shows the display control panel (left), temporal dynamics of selected subset of cells (middle), and manual curation panel (right). The field of view, current frame, and selection of cells are all synced across different plots to help user focus on a specific region and time. The users can use the control panel to select groups of cells, change display options for temporal dynamics and spatial temporal activities, and change the current frame or play the movie. In addition, the users can directly select cells from the spatial footprints plot on the top left. The users can also directly jump to frames by double-clicking on the temporal dynamic plots. These interactive features help the users quickly focus on region and time of interests. The manual curation menu on bottom right can be used to assign unit labels to each cell, which indicate whether a cell should be dropped or merged.
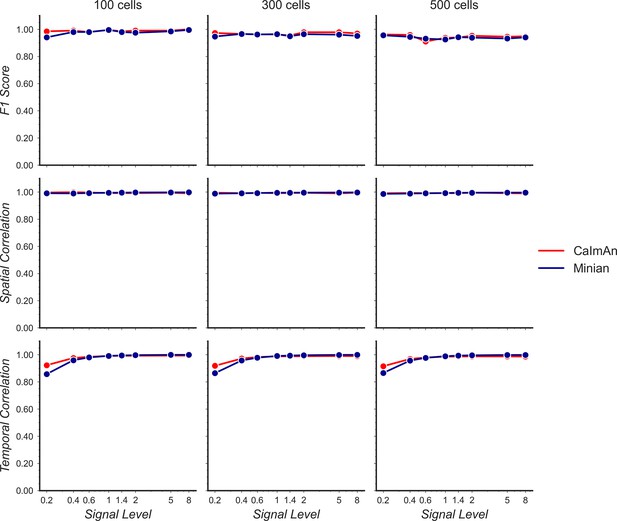
Validation of Minian with simulated datasets.
Simulated datasets with varying signal level and number of cells are processed through Minian and CaImAn. The F1 score (top), median correlation of spatial footprints (middle), and median correlation of temporal dynamics (bottom) are plotted as a function of signal level. Both pipelines achieve near-perfect (>0.95) F1 scores and spatial footprint correlation across all conditions. The correlation of temporal dynamics is lower when the signal level is 0.2, but remains similar across the two pipelines overall.
-
Figure 15—source data 1
Raw validation performance with simulated data.
- https://cdn.elifesciences.org/articles/70661/elife-70661-fig15-data1-v2.csv
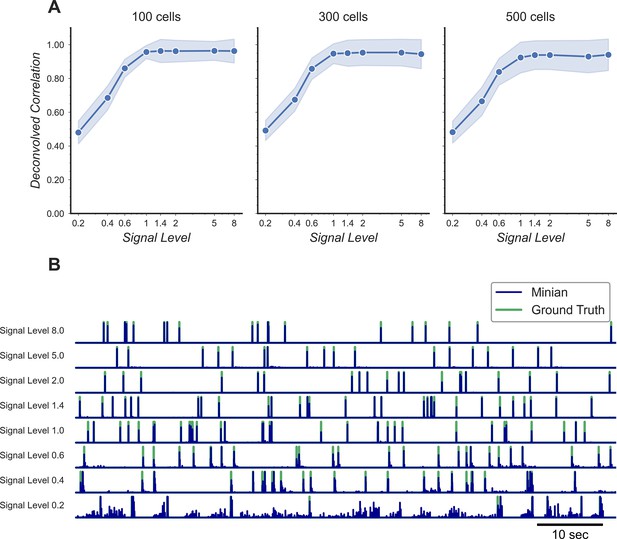
Validation of deconvolved signal from Minian.
(A) Correlation of deconvolved signals from Minian output with simulated ground truth. The mean correlation across all cells (blue line) and standard deviation (light blue shade) are shown separately for different signal levels and number of cells. The correlation asymptote and approach 1 when signal level is higher than 1. (B) Example deconvolved traces from Minian output overlaid with simulated ground truth. One representative cell is drawn from each signal level. The binary-simulated spikes are shown in green, with the real-valued Minian deconvolved output overlaid on top in blue. The deconvolved signals closely match the ground truth, and the main difference between the two signals is in the amplitude of the deconvolved signals, which tend to be influenced by local background.
-
Figure 16—source data 1
Raw correlations between Minian deconvolved traces and simulated ground truth.
- https://cdn.elifesciences.org/articles/70661/elife-70661-fig16-data1-v2.csv
-
Figure 16—source data 2
Raw example traces from Minian and simulated ground truth.
File names indicate signal level and source of trace.
- https://cdn.elifesciences.org/articles/70661/elife-70661-fig16-data2-v2.zip
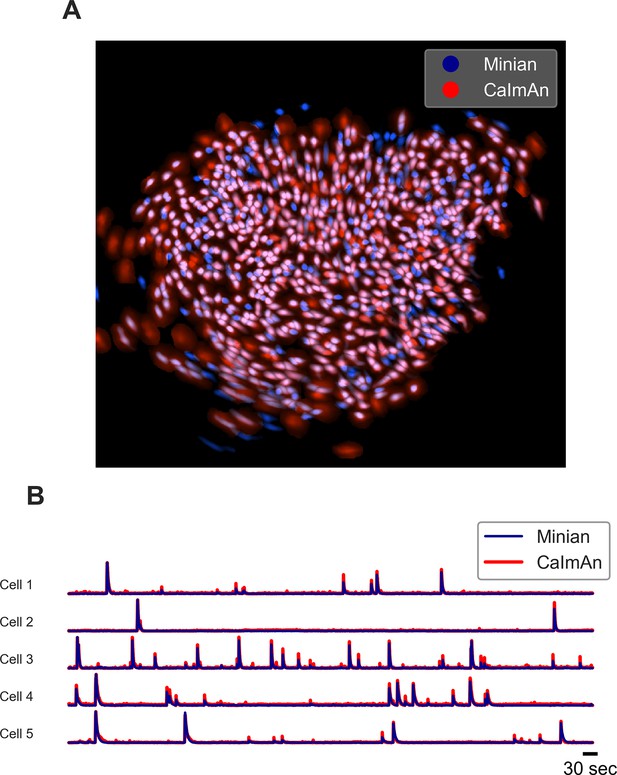
Example output of Minian and CaImAn with experimental datasets.
(A) An example field of view from one of the experimental datasets. The spatial footprints from Minian and CaImAn are colored as blue and red, respectively, and overlaid on top of each other. Most of the spatial footprints from both pipelines overlap with each other. (B) Five example matched temporal activity from Minian and CaImAn overlaid on top of each other. The extracted temporal activity is highly similar across the two pipelines.
-
Figure 17—source data 1
Raw spatial footprint values shown in the overlay plot.
- https://cdn.elifesciences.org/articles/70661/elife-70661-fig17-data1-v2.zip
-
Figure 17—source data 2
Raw example traces from Minian and Caiman.
File names indicate cell id and source of trace.
- https://cdn.elifesciences.org/articles/70661/elife-70661-fig17-data2-v2.zip
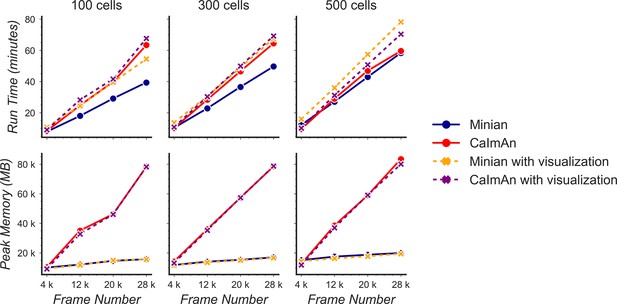
Benchmarking of computational performance.
Data with varying number of cells and frames were processed through Minian and CaImAn. The run time (top) and peak memory usage (bottom) were recorded and plotted as a function of frame number. For both pipelines, the run time scales linearly as a function of the number of frames and remains similar across the pipelines. However, the peak memory usage for CaImAn also scales linearly as the number of frames increases, while Minian maintains a relatively constant peak memory usage across different frame numbers and cell numbers.
-
Figure 18—source data 1
Raw memory usage and running time with different datasets for both pipelines.
- https://cdn.elifesciences.org/articles/70661/elife-70661-fig18-data1-v2.csv
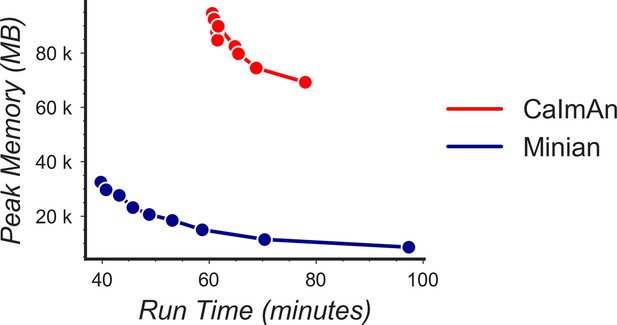
Tradeoff between run time and memory usage.
Simulated data with 500 cells and 28,000 frames were processed through Minian and CaImAn with different numbers of parallel processes. We varied the number of parallel processes from 2 to 10, and the resulting memory usage is plotted as a function of run time. For both pipelines, the curve takes a hyperbola shape, showing the tradeoff between run time and memory usage.
-
Figure 19—source data 1
Raw memory usage and running time with different parallel processes for both pipelines.
- https://cdn.elifesciences.org/articles/70661/elife-70661-fig19-data1-v2.csv
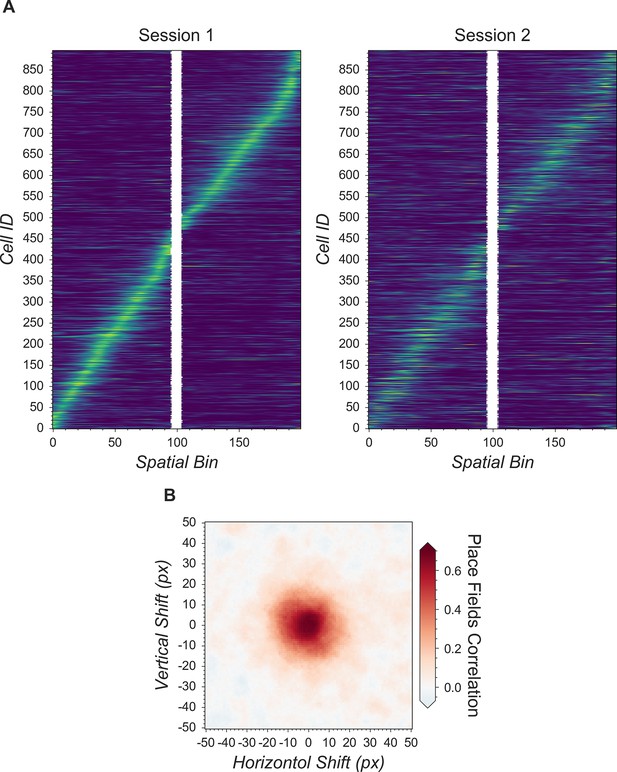
Validation of Minian with hippocampal CA1 place cells.
(A) Matching place cells from two recording sessions. The cells are matched from one session to the other using the cross-session registration algorithm and sorted based on place field in the first session. In both sessions, animals run on a 2-m-long linear track with water reward at both ends. The track is divided into 200 spatial bins. The mean ‘firing’ rate calculated from the spike signal for each cell is shown. Cell IDs are assigned by Minian when each session is analyzed independently. (B) Averaged correlations of spatial firing rates with different artificial shifts. We artificially shifted the spatial footprints of the second linear track session, then carried out registration and calculated a mean correlation of spatial firing rates for all place cells. The artificial shifts were relative to the aligned spatial footprints and range from –50 to 50 pixels.
-
Figure 20—source data 1
Raw correlation of spatial firing pattern with different shifts in field of view.
- https://cdn.elifesciences.org/articles/70661/elife-70661-fig20-data1-v2.csv
-
Figure 20—source data 2
Raw spatial firing activity for the two sessions shown.
- https://cdn.elifesciences.org/articles/70661/elife-70661-fig20-data2-v2.zip
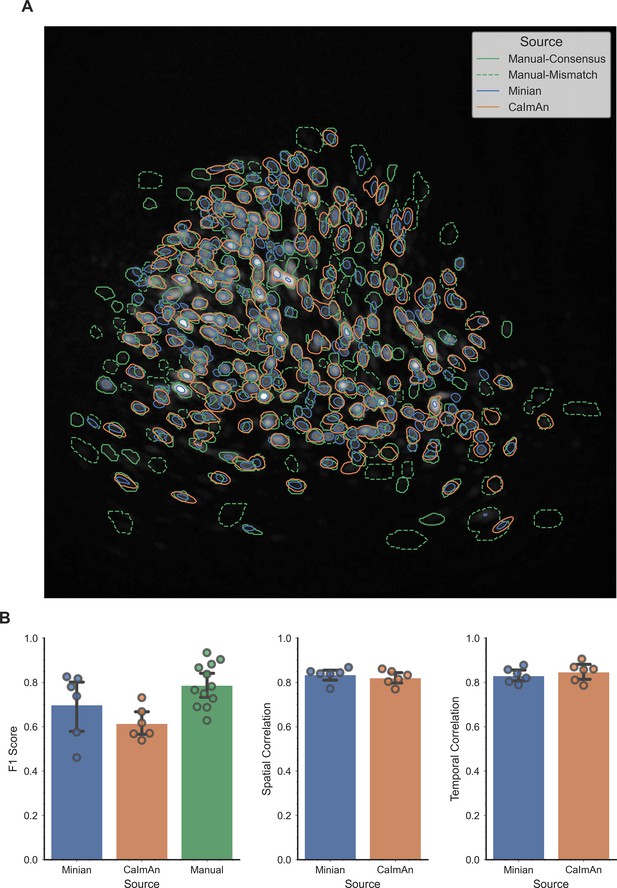
Validation of Minian with experimental datasets.
An example field of view from one of the experimental datasets is shown in panel A, with contours of detected ROIs overlaid on top. ROIs labeled by human labelers are shown in green, where solid lines indicate ROIs labeled by both labelers (labeled as “Manual-Consensus”) and dashed lines indicate ROIs labeled by one of the labelers (labeled as “Manual-Mismatch”). The F1 scores, spatial footprints correlation and temporal dynamics correlation are plotted for the two pipelines in panel B. The F1 scores of the two human labelers are also included in the plot for comparison (labeled as “Manual”). The F1 scores, spatial footprints correlation and temporal dynamics correlation were all not significantly different across the two pipelines (One-way ANOVA, p > 0.05).
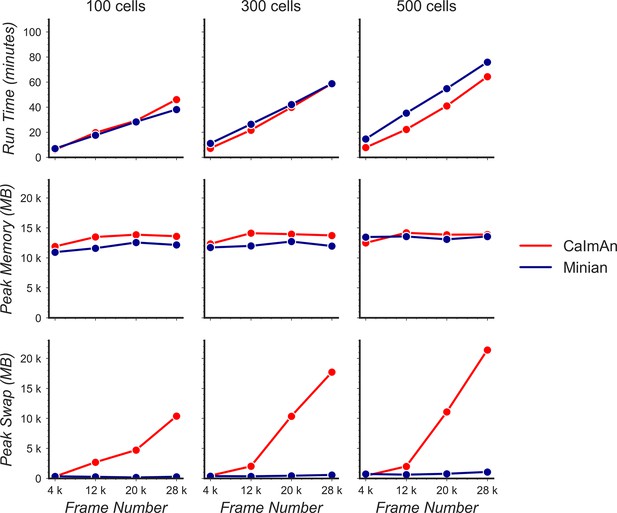
Benchmarking of computational performance in OS X.
Data with varying number of cells and frames were processed through Minian and CaImAn without visualization. The run time (top) and peak physical memory usage (middle) and peak swap memory usage (bottom) were recorded and plotted as a function of frame number.
Tables
A list of computers tested with Minian with specifications (listed roughly by increasing computation power).
| Manufacture | Model | CPU | RAM | Storage | Operating system |
|---|---|---|---|---|---|
| Custom-built | Carbon | AMD Ryzen Threadripper 2950 × 4.4 GHz × 16 | 128 GB | 2TB SSD | Ubuntu 18.04 |
| Microsoft | Surface Pro 6 | Intel Core i5-8250U 1.6 GHz × 4 | 8 GB | 256GB SSD | Windows 10 |
| Dell | Precision 5,530 | Intel Core i5-8400H 2.5 GHz × 4 | 16 GB | 256GB SSD | Ubuntu 18.04 |
| Apple | MacBook Pro 152 | Intel Core i7-8559U 2.7 GHz × 4 | 16 GB | 1TB SSD | macOS 10.14 Mojave |
| Custom-built | Amethyst | Intel Xeon E5-1650 3.6 GHz × 6 | 128 GB | 6TB HDD | Ubuntu 17.1 |
A list of open-source packages and the specific versions on which Minian depends.
| Package | Version |
|---|---|
| av | 7.0 |
| Bokeh | 1.4 |
| Bottleneck | 1.3 |
| cairo | 1.16 |
| CVXPY | 1.0 |
| Dask | 2.11 |
| Datashader | 0.1 |
| distributed | 2.11 |
| ecos | 2.0 |
| FFmpeg | 4.1 |
| FFTW | 3.3 |
| HoloViews | 1.12 |
| IPython | 7.12 |
| ipywidgets | 7.5 |
| Jupyter | 1.0 |
| Matplotlib | 3.1 |
| natsort | 7.0 |
| netCDF4 | 1.5 |
| NetworkX | 2.4 |
| Node.js | 13.9 |
| Numba | 0.48 |
| NumPy | 1.18 |
| openCV | 4.2 |
| pandas | 1.0 |
| Panel | 0.8 |
| Papermill | 2.0 |
| param | 1.9 |
| pip | 20.0 |
| pyFFTW | 0.12 |
| Python | 3.8 |
| SciPy | 1.4 |
| scs | 2.1 |
| statsmodels | 0.11 |
| tifffile | 2020.2 |
| tqdm | 4.43 |
| xarray | 0.15 |
| Zarr | 2.4 |
| MedPy | 0.4 |
| SimpleITK | 1.2 |
List of algorithm implementations in different pipelines.
For a lot of steps, different algorithm implementation can be chosen by the user based on features of the data. In such cases, we only list the default and most commonly used algorithms here.
| Step | Minian implementation | CaImAn implementation | MIN1PIPE implementation | Critical parameters |
|---|---|---|---|---|
| Denoising | Median filter | None | Anisotropic filter | Spatial window size of the filter |
| Background removal | Morphological top-hat transform | None | Morphological top-hat transform | Spatial window size of the top-hat transform |
| Motion correction | FFT-based translational motion correction | Nonrigid patch-wise translational motion correction (NoRMCorre) | Mix of translational motion correction and Demons diffeomorphic motion correction | Different |
| Initialization | Seed-based with peak-noise ratio and KS-test refinement | Pixel-wise correlation and peak-noise ratio thresholding | Seed-based with GMM, peak-noise ratio and KS-test refinement | Threshold for correlation and peak-noise ratio |
| Spatial and temporal updates | CNMF with CVXPY as deconvolution backend | CNMF-E with Oasis as deconvolution backend | CNMF with CVX MATLAB package as deconvolution backend | Noise cutoff frequency Expected size of neurons Sparse penalty |
-
GMM: Gaussian mixture model; KS: Kolmogorov–Smirnov; CNMF: constrained non-negative matrix factorization.





