Regulation of human mTOR complexes by DEPTOR
Figures
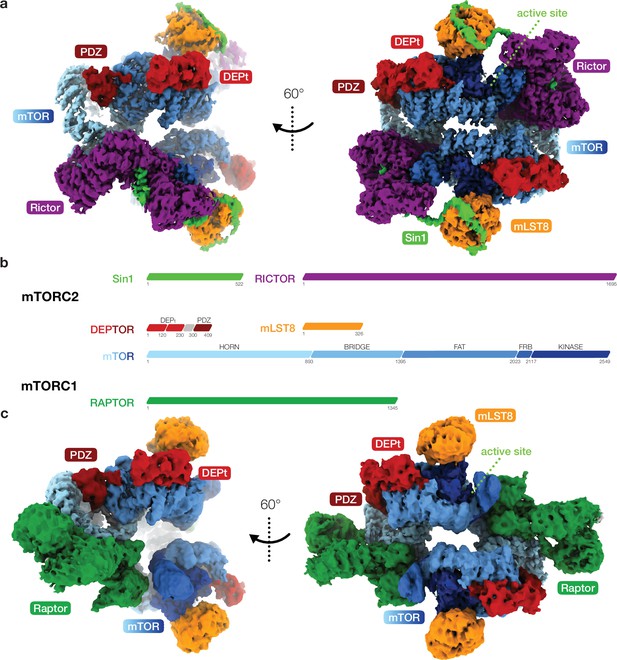
Cryo-EM reconstruction of DEP domain-containing mammalian target of rapamycin (mTOR) interacting protein (DEPTOR)-bound mTOR complexes 1 and 2 (mTORC1 and mTORC2).
(a) Composite map of overall and local focused cryo-EM reconstructions of DEPTOR-bound mTORC2. (b) Schematic representation of the domain architecture of mTORC1, mTORC2, and DEPTOR. (c) Composite map of overall and local focused cryo-EM reconstructions of DEPTOR-mTORC1. In (a) and (c) proteins are colored according to the schemes in (b). DEPTOR binds to mTORC1 and mTORC2 in virtually identical manner via its extended PDZ-linker and DEP domain tandem (DEPt) regions associating with the FAT domain of mTOR.
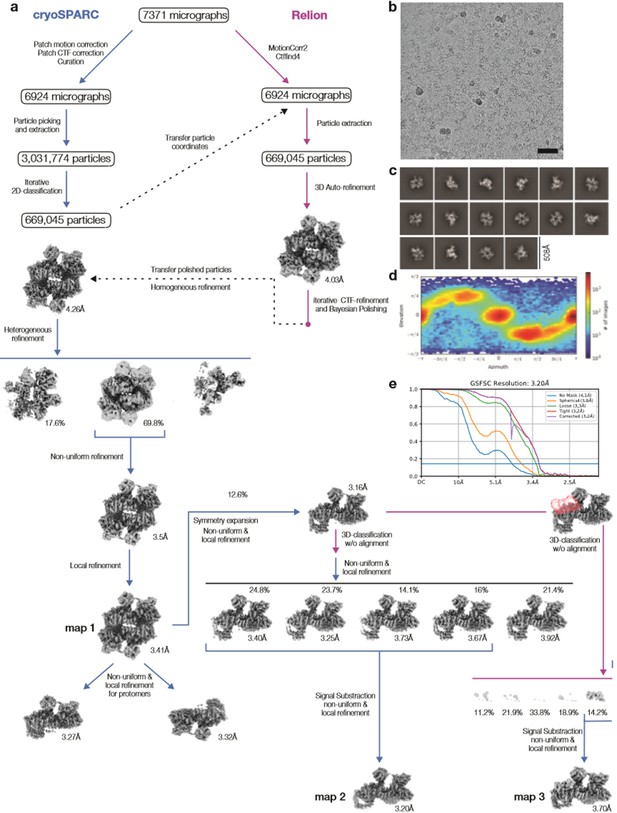
Cryo-EM data processing DEP domain-containing mammalian target of rapamycin (mTOR) interacting protein (DEPTOR)-mTOR complex 2 (mTORC2).
(a) Scheme of the cryo-EM data processing workflow. DEPTOR-mTORC2 overall refinement (map 1), focused refinement on symmetry-expanded protomer (map 2), and focused refinement on one protomer classified for the DEP domain tandem (DEPt) region (map 3) were used for modeling and illustration. (b) Representative micrograph of the DEPTOR-mTORC2 dataset is shown; scale bar equals 500 Å. (c) 2D class averages. (d) Viewing direction distribution of the DEPTOR-mTORC2 overall refinement (map 1). (e) Fourier shell correlation (FSC) curves for unmasked, spherical, loose, and tight masks, and corrected FSC curve for the map 2 reconstruction, yielding a gold standard FSC resolution of 3.20 Å.
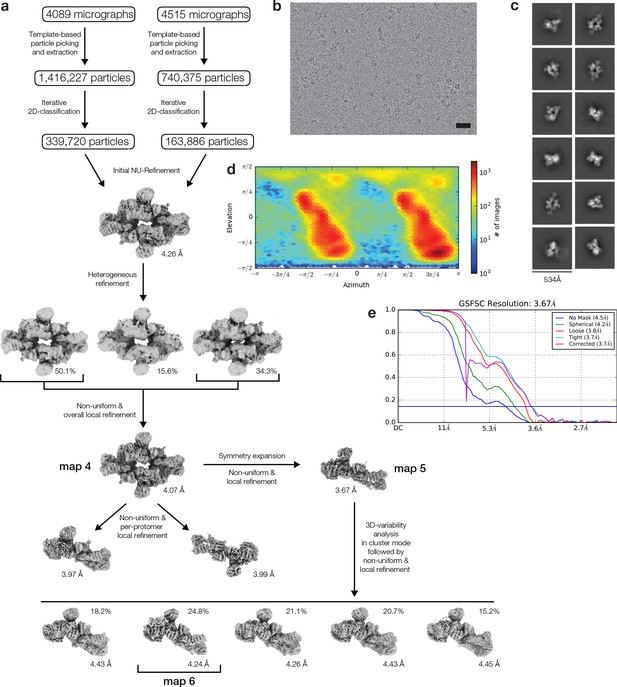
Cryo-EM data processing of DEP domain-containing mammalian target of rapamycin interacting protein (DEPTOR)-mTOR complex 1 (mTORC1).
(a) Scheme of the cryo-EM data processing workflow. DEPTOR-mTORC1 overall refinement (map 4), focused refinement on symmetry-expanded protomer (map 5) and focused refinement on one protomer classified for the DEPt region (map 6) were used for modeling and illustration. (b) Representative micrograph of the DEPTOR-mTORC2 dataset is shown; scale bar equals 500 Å. (c) 2D class averages. (d) Viewing direction distribution of the DEPTOR-mTORC1 overall refinement (map 1). (e) Fourier shell correlation (FSC) curves for unmasked, spherical, loose, and tight masks, and corrected FSC curve for the map 5 reconstruction, yielding a gold standard FSC resolution of 3.67 Å.
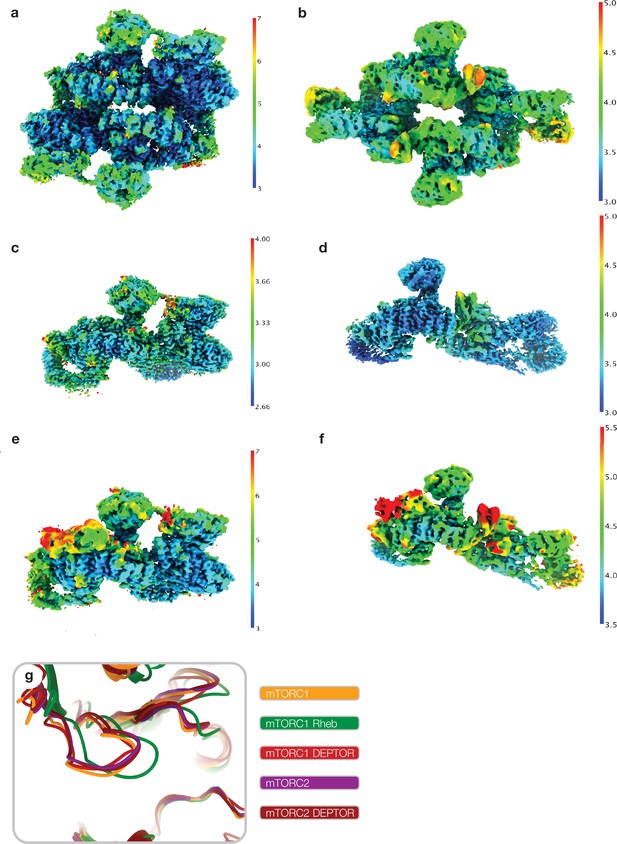
Local resolution and active site state of mammalian target of rapamycin (mTOR) complexes in cryo-EM reconstructions.
Cryo-EM reconstructions used for modeling colored by local resolution calculated using cryoSPARC at 0.143 FSC cutoff (a) map 1, (b) map 4, (c) map 2, (d) map 5, (e) map 3, (f) map 6, (Figure 1—figure supplement 1a; Figure 1—figure supplement 2a). (g) Superimposition of free mTOR complex 1 (mTORC1) (6BCX; Yang et al., 2017), Rheb-activated mTORC1(6BCU; Yang et al., 2017), free mTORC2 (6ZWM; Scaiola et al., 2020), and DEP domain-containing mTOR interacting protein (DEPTOR)-bound mTORC1 and mTORC2 (this study). DEPTOR-bound mTOR complexes resemble the non-Rheb activated state of the mTOR kinase active site.
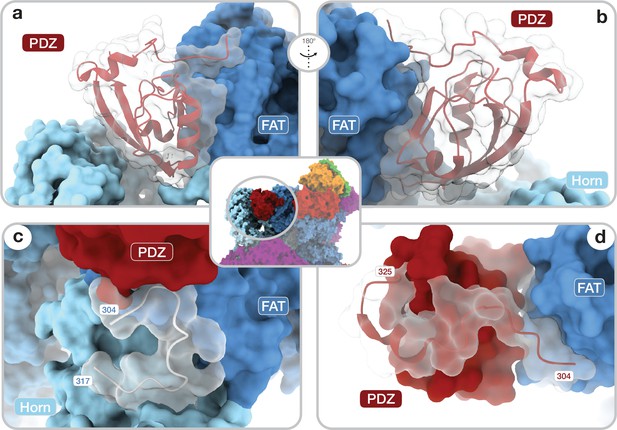
Architecture of the DEP domain-containing mammalian target of rapamycin (mTOR) interacting protein (DEPTOR) PDZ domain and its interaction with mTOR.
(a, b) Front (a) and back (b) view of DEPTOR PDZ bound to the mTOR FAT domain. The PDZ domain (shown as transparent surface with red cartoon) binds to a hinge in the FAT domain of mTOR. (c) The PDZ domain N-terminal extension stretches toward the FAT domain. The adjacent N-terminal linker inserts into a groove on the FAT domain and substantially contributes the PDZ-mTOR interface. (d) Loop region (aamTOR290–350) in the mTOR Horn-region (transparent with cartoon) is disordered in free mTOR complexes and contributes to the mTOR-PDZ interface and thereby creates a link between the Horn-region and the FAT domain of mTOR and the DEPTOR PDZ domain.
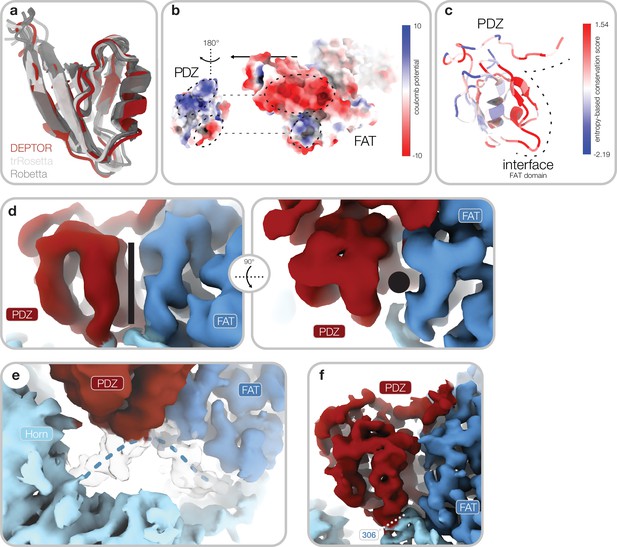
PDZ domain interaction with mammalian target of rapamycin (mTOR) complexes.
(a) Superimposition of models for the PDZ core obtained from trRosetta (light gray) and Robetta (dark gray) and the final model based on the cryo-EM reconstruction (red) (map 2, Figure 1—figure supplement 1a). (b) Complementary surface electrostatic potential is observed for the two binding interfaces between DEP domain-containing mTOR interacting protein (DEPTOR) and the mTOR FAT domain. (c) Sequence conservation of the PDZ domain mapped onto the structure. The interface to mTOR is schematically indicated by a dashed line. mTOR interacting residues are highly conserved. (d) Map 2 (Figure 1—figure supplement 1a) lowpass-filtered to 5 Å. The canonical binding groove of the PDZ domain (indicated by a black rod/dot) is empty and not peptide-bound. This mode of interaction allows regulation of the PDZ mTOR association by binding of additional interaction partners to the canonical binding groove. (e) Map 2 (Figure 1—figure supplement 1a) lowpass-filtered to 3.5 Å resolution. A linker of the Horn-region (indicated by blue dotted line) adopts a structured conformation upon PDZ binding and provides a structural link between the Horn-region, the FAT domain, and the PDZ domain. (f) Quality of the cryo-EM reconstruction for the PDZ. The binding interface is well defined with a local resolution of around 3 Å. Local resolution for the PDZ domain decreases due to flexibility with increasing distance from the interface to around 4 Å (Figure 1—figure supplement 3c). Continuous density is observed for the PDZ N-terminal extension and the transition into the FAT-bound linker. FmTOR306 (labeled) is an integral part of the PDZ-mTOR interface.
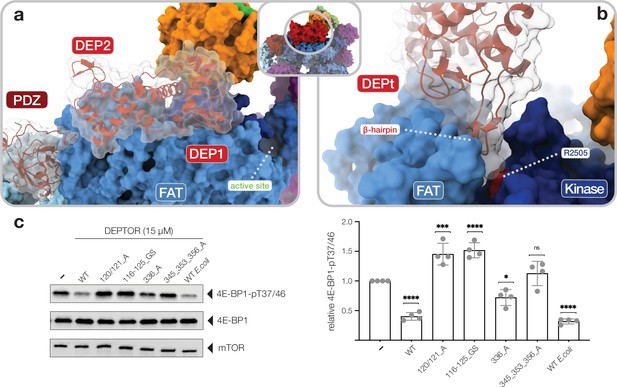
Interactions of the DEP domain-containing mammalian target of rapamycin (mTOR) interacting protein (DEPTOR) DEP domain tandem (DEPt) region with mTOR.
(a) Surface representation of DEPTOR (transparent with cartoon in red) bound to mTOR complex 2 (mTORC2). The DEPt region binds centrally on top of the helical repeats of the FAT domain. (b) The protruding hairpin of the first DEP domain of DEPt inserts into a crevice between the kinase and FAT domain of mTOR. The DEPTOR-displacing mutant R2505P (Grabiner et al., 2014) is located in close proximity. (c) Analysis of the impact of wild-type and mutant forms of DEPTOR on Rheb-stimulated mTORC1 activity. Mutants are described in Figure 3—figure supplement 2. mTORC1 was incubated with 4E-BP1 and Rheb for stimulation, in the presence of DEPTOR wild-type and mutants. Reactions were separated by SDS-PAGE and analyzed by western blot. 4E-BP1 phosphorylation was detected with an antibody specific to phosphorylation of residues T37/46. Quantification (mean ± SD) of western blots in 4E-BP1-pT37/46 signals were normalized to total 4E-BP1 signals and the statistical significance of changes between control (0 µM DEPTOR) and DEPTOR variants determined by one-way ANOVA. ****p < 0.0001, ***p < 0.001, *p < 0.05, nsp >0.05, n = 4.
-
Figure 3—source data 1
Source data of kinase assay.
Uncropped blots of all four replicates (bands shown in Figure 3c indicated) and statistical analysis of western blot quantification.
- https://cdn.elifesciences.org/articles/70871/elife-70871-fig3-data1-v1.pdf
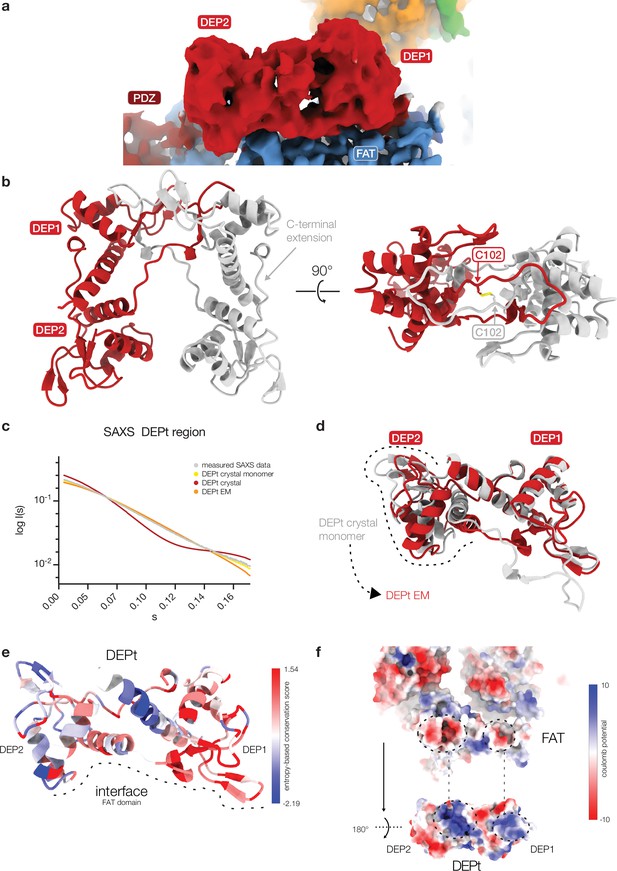
DEP domain tandem (DEPt) in crystals and associated with mTOR complex 2 (mTORC2).
(a) Cryo-EM reconstruction of the DEPt (map 3, Figure 1—figure supplement 1a) based on local refinement. The local resolution of 4–6 Å allows to identify secondary structure elements and fold, but not individual amino acid side chains. (b) The DEPt of DEP domain-containing mTOR interacting protein (DEPTOR) crystallized as a domain-swapped dimer. The domain-swapped dimer is stabilized by a non-native disulfide bridge between C102 of the protomers. (c) Small-angle X-ray scattering (SAXS) data and fitted curves for three different DEPt models. DEPt crystal monomer χ2: 0.59; DEPt crystal dimer χ2: 49.27; DEPt EM χ2: 5.88. The DEPt is monomeric in solution in a conformation corresponding to the conformation found in the crystal structure, which is related to the mTOR-bound state by a simple domain rotation with minor translation component. (d) Superimposition based on the DEP1 of DEPt from the crystal structure and bound to mTOR. The FAT-bound confirmation of DEPt differs from the free form by a 3.6 Å translation and 39° rotation of DEP2 relative to DEP1. (e) Sequence conservation of DEPt mapped onto the structure of DEPt. The interface to mTOR is schematically indicated by a dashed line. mTOR interacting residues are highly conserved in DEPt. DEP1 of DEPt, which mainly mediates interaction of DEPt and mTOR, is more conserved than DEP2. (f) Complementary surface electrostatic potential is observed for the binding interface between DEPt and the mTOR FAT domain. The two positively charged patches in DEPt were recently described to bind to phosphatidic acid (PA) (Weng et al., 2021).
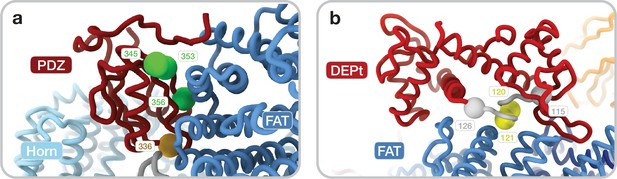
Mutations in PDZ and DEP domain tandem (DEPt) interface.
(a) Mutants targeting the PDZ-mammalian target of rapamycin (mTOR) interface: single mutant D336A (golden) and triple mutant R345A/Q353A/D356A (green). (b) Mutants targeting the DEPt-mTOR interface: double mutant D120A/D121A (yellow) and a linker substitution mutant where 116–125DEPTOR are substituted with a glycine serine linker (GS) (Loewith and Hall, 2011) (gray).
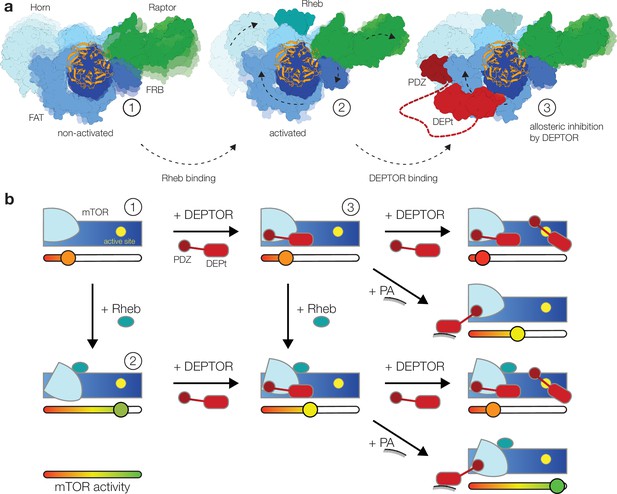
Model for the DEP domain-containing mammalian target of rapamycin (mTOR) interacting protein (DEPTOR)-mediated regulation of mTOR activity.
(a) Structure-based representation of (1) the basal state of non-activated mTOR complex 1 (mTORC1) (based on PDB: 6BCX), (2) the allosteric activation of mTORC1 by Rheb binding (based on PDB: 6BCU), and (3) the impact of DEPTOR association via the PDZ domain and DEP domain tandem (DEPt) on the conformational state and activity of mTORC1. Possible transitions in subpopulations of conformational states are indicated by shadowing. (b) Schematic diagram of the suggested regulatory interactions between DEPTOR and mTOR complexes. Structurally characterized states shown in (a) are indicated by numbers. DEPTOR binding via the PDZ domain and DEPt prevents allosteric activation. At high concentrations, DEPTOR binds to mTORC1 in a secondary binding mode as a substrate and sterically influences access of other substrates to the active site. Phosphatidic acid (PA) may interfere with the DEPt-mTOR association, relieving the allosteric inhibition of mTORCs. The remaining bound PDZ domain mildly stimulates kinase activity in activated and non-activated mTOR complexes.
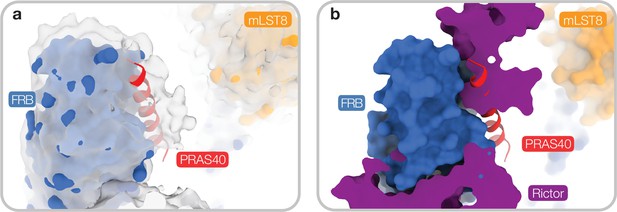
Residual density in a substrate recruitment site at the FRB domain for DEP domain-containing mammalian target of rapamycin (mTOR) interacting protein (DEPTOR)-bound mTOR complex 1 (mTORC1).
(a) In cryo-EM reconstructions of DEPTOR-mTORC1, additional density is observed at a site of the FRB, where substrates and PRAS40 (red, 5WBU; Yang et al., 2017) bind. Unsharpened map at low contour level is shown to illustrate additional density. (b) This substrate recruitment site on the FRB is occupied in mTORC2 by Rictor. Same view shown as in panel (a).
Videos
The DEP domain-containing mammalian target of rapamycin (mTOR) interacting protein (DEPTOR)-mTOR complex 2 (mTORC2).
Overview of cryo-EM reconstruction and model, highlighting functionally relevant sites and interactions discussed in the associated manuscript.
The DEP domain-containing mammalian target of rapamycin (mTOR) interacting protein (DEPTOR)-mTOR complex 1 (mTORC1).
Overview of cryo-EM reconstruction and model, highlighting functionally relevant sites and interactions discussed in the associated manuscript.
Additional files
-
Supplementary file 1
Cryo-EM data collection and refinement statistics of DEP domain-containing mammalian target of rapamycin (mTOR) interacting protein (DEPTOR)-mTOR complex 2 (mTORC2) complex.
- https://cdn.elifesciences.org/articles/70871/elife-70871-supp1-v1.docx
-
Supplementary file 2
Cryo-EM data collection and refinement statistics of DEP domain-containing mammalian target of rapamycin (mTOR) interacting protein (DEPTOR)-mTOR complex 1 (mTORC1) complex.
- https://cdn.elifesciences.org/articles/70871/elife-70871-supp2-v1.docx
-
Supplementary file 3
X-ray data collection and refinement for DEP domain-containing mammalian target of rapamycin (mTOR) interacting protein (DEPTOR) DEP domain tandem (DEPt).
- https://cdn.elifesciences.org/articles/70871/elife-70871-supp3-v1.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/70871/elife-70871-transrepform1-v1.docx





