A tRNA processing enzyme is a key regulator of the mitochondrial unfolded protein response
Figures
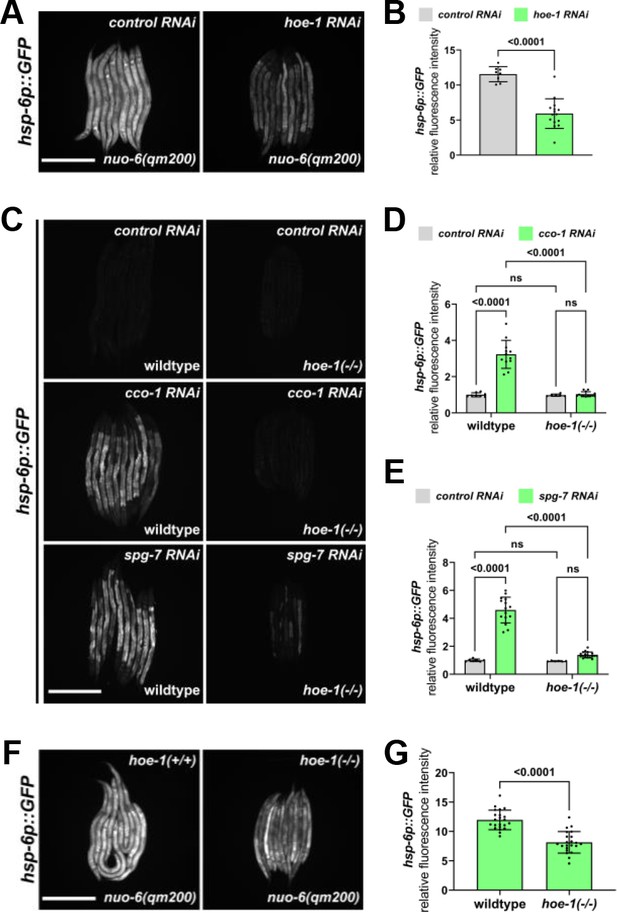
hoe-1 is required for maximal UPRmt activation.
(A) Fluorescence images of UPRmt reporter (hsp-6p::GFP) activation in L4 nuo-6(qm200) animals on control and hoe-1 RNAi. Scale bar 200 μm. (B) Fluorescence intensity quantification of hsp-6p::GFP in individual L4 nuo-6(qm200) animals on control and hoe-1 RNAi normalized to hsp-6p::GFP in a wildtype background on control RNAi (n = 8 and 15 respectively, mean and SD shown, unpaired t-test). (C) Fluorescence images of UPRmt reporter (hsp-6p::GFP) activation in L3/L4 wildtype and hoe-1 null (hoe-1(-/-)) animals on control, cco-1, and spg-7 RNAi. Scale bar 200 μm. (D) Fluorescence intensity quantification of hsp-6p::GFP in individual L3/L4 wildtype and hoe-1(-/-) animals on control and cco-1 RNAi (n = 8,12,6 and 13 respectively, mean and SD shown, ordinary two-way ANOVA with Tukey’s multiple comparisons test). (E) Fluorescence intensity quantification of hsp-6p::GFP in individual L3/L4 wildtype and hoe-1(-/-) animals on control and spg-7 RNAi (n = 7,15,6 and 18 respectively, mean and SD shown, ordinary two-way ANOVA with Tukey’s multiple comparisons test). (F) Fluorescence images of UPRmt reporter (hsp-6p::GFP) activation in L3/L4 nuo-6(qm200) animals with (hoe-1(+/+)) and without (hoe-1(-/-)) hoe-1. Scale bar 200 μm. (G) Fluorescence intensity quantification of hsp-6p::GFP in individual L3/L4 nuo-6(qm200) animals with (hoe-1(+/+)) and without (hoe-1(-/-)) hoe-1 normalized to hsp-6p::GFP in a wildtype background (n = 22 for each condition, mean and SD shown, unpaired t-test).
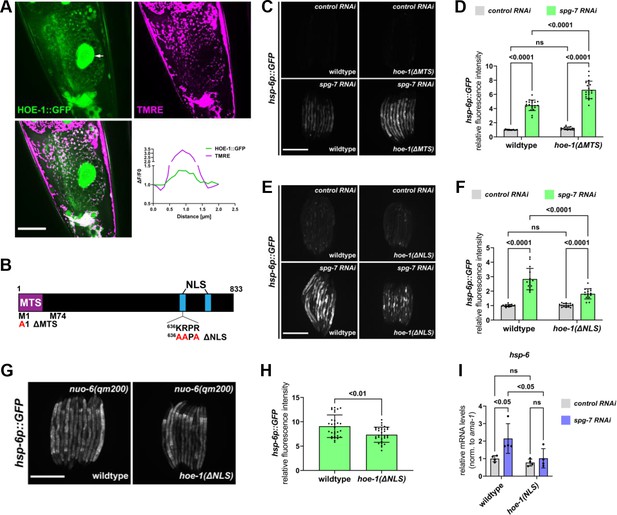
Nuclear HOE-1 is required for maximal UPRmt activation.
(A) Fluorescence images of a terminal intestinal cell in a wildtype animal expressing HOE-1::GFP (green) stained with TMRE (magenta) to visualize mitochondria. GFP and TMRE co-localization shown in white in merged image. Arrow indicates nuclei. Scale bar 20 μm. Representative line segment analysis of individual mitochondrion. (B) Schematic of HOE-1 protein showing the mitochondrial targeting sequence (MTS) and nuclear localization signals (NLS). ΔMTS allele created by replacing START codon with an alanine (M1A). Transcription begins at M74 for nuclear localized HOE-1. ΔNLS allele created by compromising the most N-terminal NLS (636KRPR > AAPA). (C) Fluorescence images of UPRmt reporter (hsp-6p::GFP) in L4 wildtype and hoe-1(ΔMTS) animals on control and spg-7 RNAi. Scale bar 200 μm. (D) Fluorescence intensity quantification of hsp-6p::GFP in individual L4 wildtype and hoe-1(ΔMTS) animals on control and spg-7 RNAi (n = 15,20,17, and 19 respectively, mean and SD shown, ordinary two-way ANOVA with Tukey’s multiple comparisons test). (E) Fluorescence images of UPRmt reporter (hsp-6p::GFP) in L4 wildtype and hoe-1(ΔNLS) animals on control and spg-7 RNAi. Scale bar 200 μm. (F) Fluorescence intensity quantification of hsp-6p::GFP in individual L4 wildtype and hoe-1(ΔNLS) animals on control and spg-7 RNAi (n = 15 for each condition, mean and SD shown, ordinary two-way ANOVA with Tukey’s multiple comparisons test). (G) Fluorescence images of UPRmt reporter in L4 nuo-6(qm200) animals in wildtype and hoe-1(ΔNLS) backgrounds. Scale bar 200 μm. (H) Fluorescence intensity of hsp-6p::GFP in individual L4 nuo-6(qm200) animals in wildtype and hoe-1(ΔNLS) backgrounds (n = 30 for each condition, mean and SD shown, unpaired t-test). (I) mRNA transcript quantification of hsp-6 in L4 wildtype and hoe-1(ΔNLS) animals on control and spg-7 RNAi normalized to ama-1 (n = 4 for each condition, mean and SD shown, ordinary two-way ANOVA with Tukey’s multiple comparisons test).
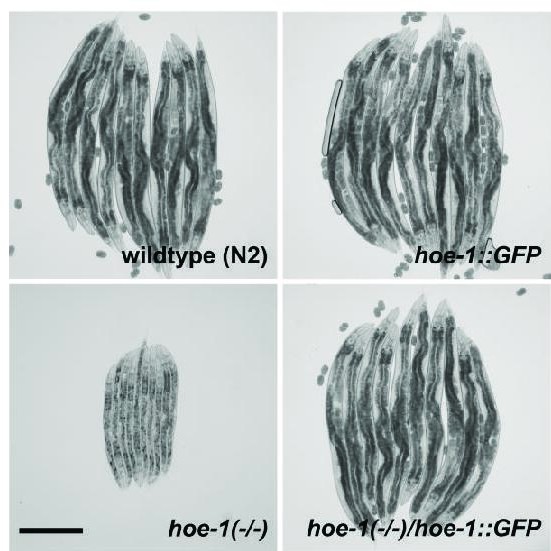
hoe-1::GFP does not compromise growth or development and is sufficient to rescue the developmental arrest of hoe-1(-/-) animals.
Bright-field images of wildtype, hoe-1::GFP, hoe-1(-/-), and hoe-1(-/-)/hoe-1::GFP trans-heterozygous animals 72 hr post-embryo. Scale bar 200 μm.
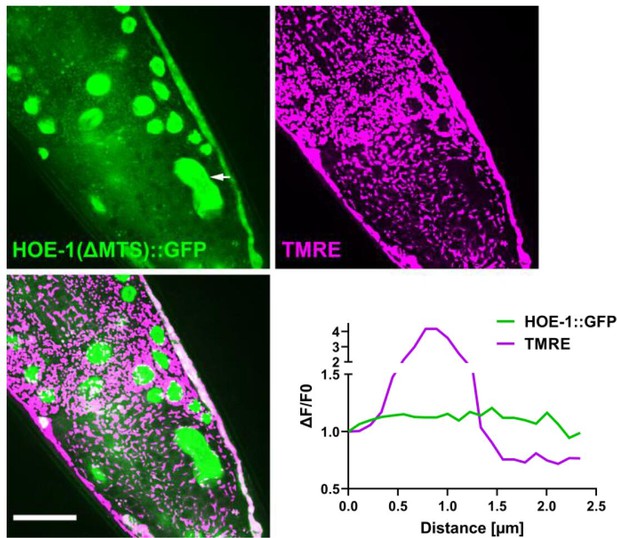
hoe-1(ΔMTS) allele attenuates HOE-1 mitochondrial localization.
Fluorescence images of a terminal intestinal cell in a hoe-1(ΔMTS) day 1 adult animal expressing HOE-1::GFP (green) stained with TMRE (magenta) to visualize mitochondria. GFP and TMRE co-localization shown in white in merged image. Arrow indicates nuclei. Scale bar 20 μm. Representative line segment analysis of individual mitochondrion.
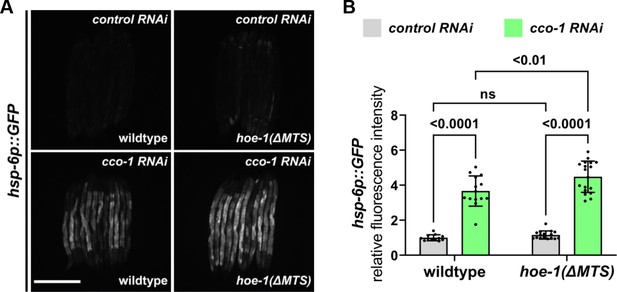
hoe-1(ΔMTS) does not attenuate cco-1 RNAi-induced UPRmt.
(A) Fluorescence images of UPRmt reporter (hsp-6p::GFP) in L4 wildtype and hoe-1(ΔMTS) animals on control and cco-1 RNAi. Scale bar 200 μm. (B) Fluorescence intensity quantification of hsp-6p::GFP in individual L4 wildtype and hoe-1(ΔMTS) animals on control and cco-1 RNAi (n = 12,14,16, and 18 respectively, mean and SD shown, ordinary two-way ANOVA with Tukey’s multiple comparisons test).
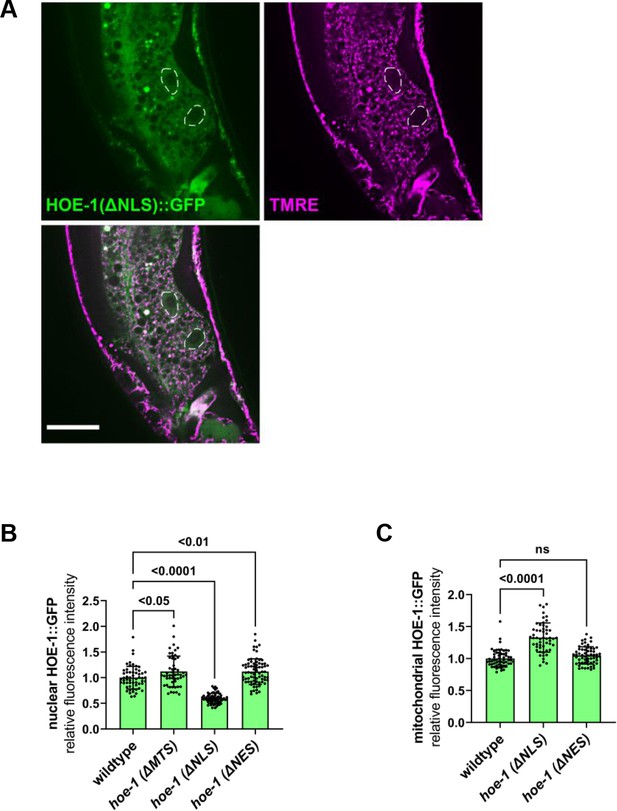
hoe-1(ΔNLS) allele attenuates nuclear HOE-1 localization.
(A) Fluorescence images of a terminal intestinal cell individual a hoe-1(ΔNLS) day 1 adult animal expressing HOE-1::GFP(green) stained with TMRE (magenta) to visualize mitochondria. GFP and TMRE co-localization shown in white in merged image. Nuclei are traced with dashed white line. Scale bar 20 μm (B) Fluorescence intensity quantification of HOE-1::GFP in intestinal nuclei of wildtype, hoe-1(ΔMTS), hoe-1(ΔNLS) and hoe-1(ΔNES) backgrounds (n = 57, 52, 60, and 73 respectively, mean and SD shown, ordinary one-way ANOVA with Dunnett’s multiple comparisons test). (C) Fluorescence intensity quantification of HOE-1::GFP in intestinal mitochondria of wildtype, hoe-1(ΔNLS) and hoe-1(ΔNES) backgrounds (n = 57, 53, and 60 respectively, mean and SD shown, ordinary one-way ANOVA with Dunnett’s multiple comparisons test).
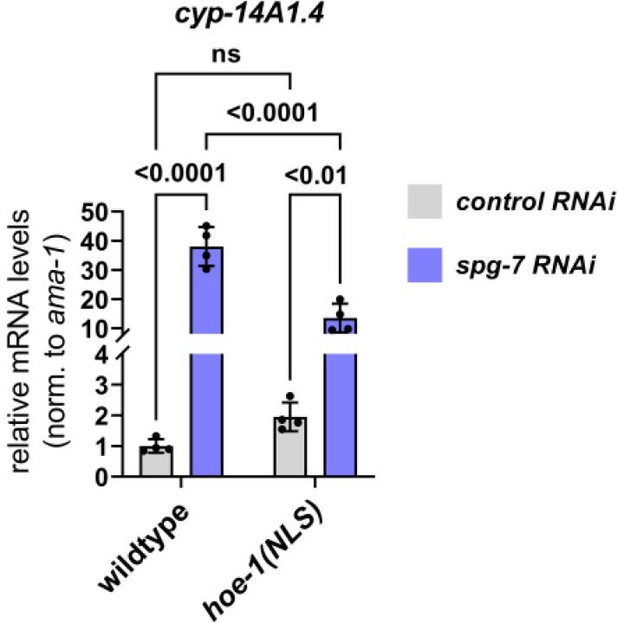
UPRmt-responsive gene cyp-14A1.4 is downregulated under mitochondrial stress conditions in hoe-1(ΔNLS) animals relative to wildtype.
mRNA transcript quantification of cyp-14A1.4 in L4 wildtype and hoe-1(ΔNLS) animals on control and spg-7 RNAi normalized to ama-1 (n = 4 for each condition, mean and SD shown, ordinary two-way ANOVA with Tukey’s multiple comparisons test).
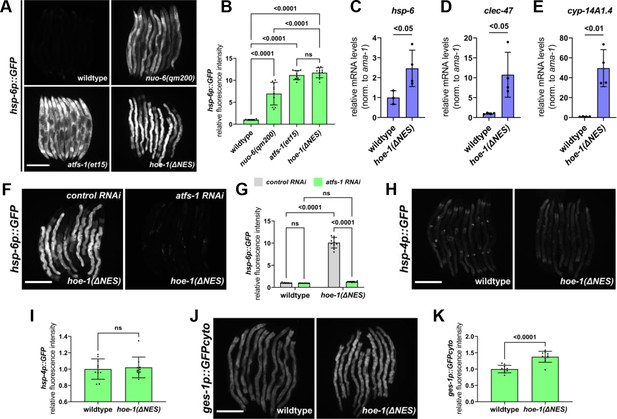
Nuclear export defective HOE-1 is sufficient to specifically activate UPRmt.
(A) Fluorescence images of UPRmt reporter (hsp-6p::GFP) activation in day 2 adult wildtype, nuo-6(qm200), atfs-1(et15), and hoe-1(ΔNES) animals. Scale bar 200 μm. (B) Fluorescence intensity quantification of hsp-6p::GFP in individual day 2 adult wildtype, nuo-6(qm200), atfs-1(et15), and hoe-1(ΔNES) animals (n = 10 for each condition, mean and SD shown, ordinary one-way ANOVA with Tukey’s multiple comparisons test). (C–E) mRNA transcript quantification of hsp-6, clec-47, and cyp-14A1.4, respectively, in day 2 adult wildtype and hoe-1(ΔNES) animals normalized to ama-1 mRNA levels (n = 4 for each condition, mean and SD shown, unpaired t-test). (F) Fluorescence images of UPRmt reporter (hsp-6p::GFP) activation in day 2 adult hoe-1(ΔNES) animals on control and atfs-1 RNAi. Scale bar 200 μm. (G) Fluorescence intensity quantification of hsp-6p::GFP in individual day 2 adult wildtype and hoe-1(ΔNES) animals on control and atfs-1 RNAi (n = 10 for each condition, mean and SD shown, ordinary two-way ANOVA with Tukey’s multiple comparisons test). (H) Fluorescence images of UPRER reporter (hsp-4p::GFP) activation in day 2 adult wildtype and hoe-1(ΔNES) animals. Scale bar 200 μm. (I) Fluorescence intensity quantification of hsp-4p::GFP in individual day 2 adult wildtype and hoe-1(ΔNES) animals (n = 10 for each condition, mean and SD shown, unpaired t-test). (J) Fluorescence images of intestinal-specific basal protein reporter (ges-1p::GFPcyto) activation in day 2 adult wildtype and hoe-1(ΔNES) animals. Scale bar 200 μm. (K) Fluorescence intensity quantification of ges-1p::GFPcyto in individual day 2 adult wildtype and hoe-1(ΔNES) animals (n = 10 for each condition, mean and SD shown, unpaired t-test).
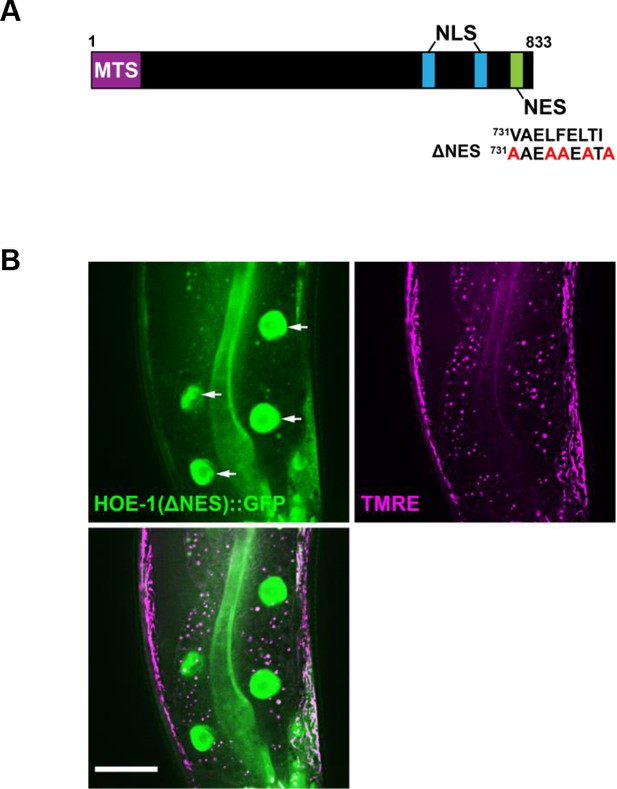
Nuclear export defective HOE-1 has increased nuclear accumulation relative to wildtype.
(A) Schematic of HOE-1 protein showing the mitochondrial targeting sequence (MTS), nuclear localization signals (NLS) and nuclear export signal (NES). hoe-1(ΔNES) mutant generated by changing the strong hydrophobic residues of NES to alanines (731VAELFELTI739>731AAEAAEATA739) (B) Fluorescence images of a terminal intestinal cell in a hoe-1(ΔNES) day 1 adult animal expressing HOE-1::GFP (green) stained with TMRE (magenta) to visualize mitochondria. GFP and TMRE co-localization shown in white in merged image. Arrow indicates nuclei. Scale bar 20 μm. Quantification of nuclear and mitochondrial HOE-1::GFP levels in hoe-1(ΔNES) animals shown in Figure 2—figure supplement 4.
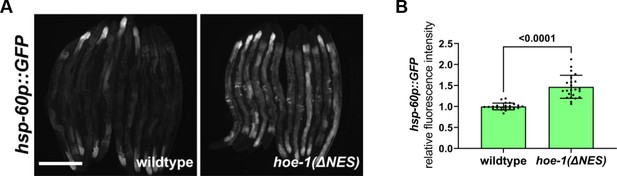
Nuclear export defective HOE-1 activates UPRmt.
(A) Fluorescence images of UPRmt reporter (hsp-60p::GFP) activation in day 2 adult wildtype and hoe-1(ΔNES) animals. Scale bar 200 μm. (B) Fluorescence intensity quantification of hsp-60p::GFP in individual day 2 adult wildtype and hoe-1(ΔNES) animals (n = 24 for each condition, mean and SD shown, unpaired t-test).
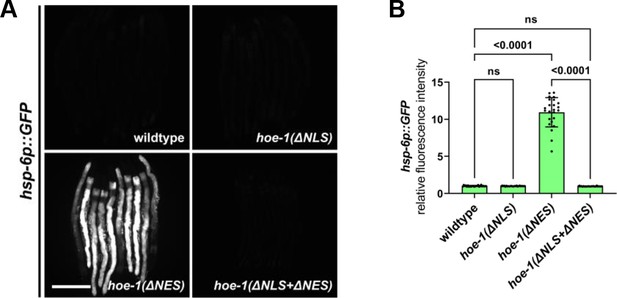
Compromised nuclear import of HOE-1 completely attenuates hoe-1(ΔNES)-induced UPRmt.
(A) Fluorescence images of UPRmt reporter (hsp-6p::GFP) activation in day 2 adult wildtype, hoe-1(ΔNLS), hoe-1(ΔNES), and hoe-1(ΔNLS+ΔNES) animals. Scale bar 200 μm. (B) Fluorescence intensity quantification of hsp-6p::GFP in individual day 2 adult wildtype, hoe-1(ΔNLS), hoe-1(ΔNES), and hoe-1(ΔNLS+ΔNES) animals (n = 24 for each condition, mean and SD shown, ordinary one-way ANOVA with Tukey’s multiple comparisons test).
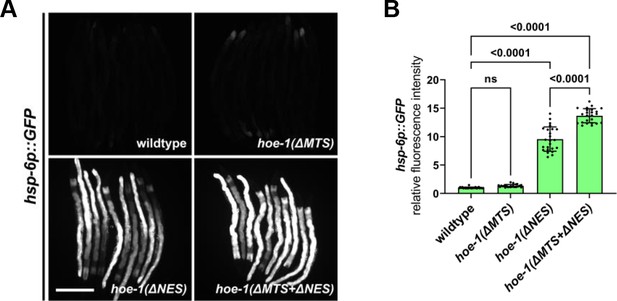
Compromised mitochondrial import of HOE-1 exacerbates hoe-1(ΔNES)-induced UPRmt.
(A) Fluorescence images of UPRmt reporter (hsp-6p::GFP) activation in day 2 adult wildtype, hoe-1(ΔMTS), hoe-1(ΔNES), and hoe-1(ΔMTS+ΔNES) animals. Scale bar 200 μm. (B) Fluorescence intensity quantification of hsp-6p::GFP in individual day 2 adult wildtype, hoe-1(ΔMTS), hoe-1(ΔNES), and hoe-1(ΔMTS+ΔNES) animals (n = 24 for each condition, mean and SD shown, ordinary one-way ANOVA with Tukey’s multiple comparisons test).
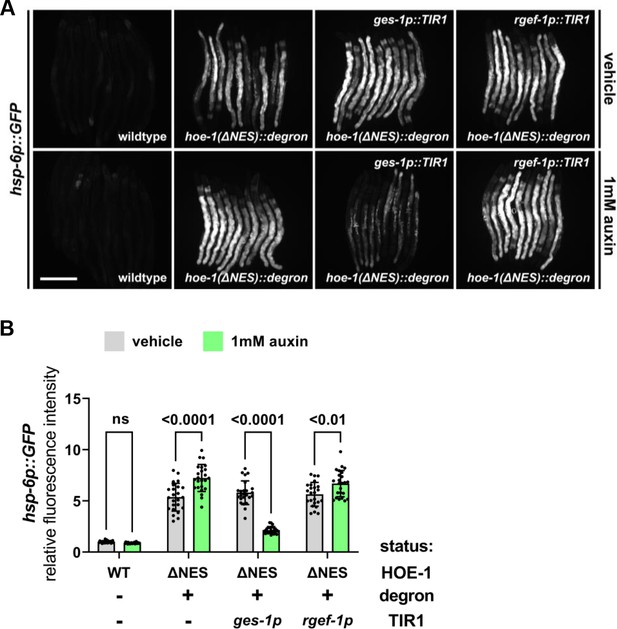
Nuclear export defective HOE-1 activates UPRmt in the intestine cell autonomously.
(A) Fluorescence images of UPRmt reporter (hsp-6p::GFP) activation in day 2 adult wildtype, hoe-1(ΔNES::degron), hoe-1(ΔNES::degron) with intestinal-specific AID (ges-1p::TIR1), and hoe-1(ΔNES::degron) with neuronal-specific AID (rgef-1p::TIR1) animals on vehicle and 1 mM auxin. (B) Fluorescence intensity quantification of hsp-6p::GFP in individual day 2 adult wildtype, hoe-1(ΔNES::degron), hoe-1(ΔNES::degron) with intestinal-specific AID (ges-1p::TIR1), and hoe-1(ΔNES::degron) with neuronal-specific AID (rgef-1p::TIR1) animals on vehicle and 1 mM auxin (n = 24 for each condition, mean and SD shown, ordinary one-way ANOVA with Tukey’s multiple comparisons test). Note that the degron tagged hoe-1(ΔNES) allele has modestly diminished UPRmt activation relative to the untagged hoe-1(ΔNES) allele.
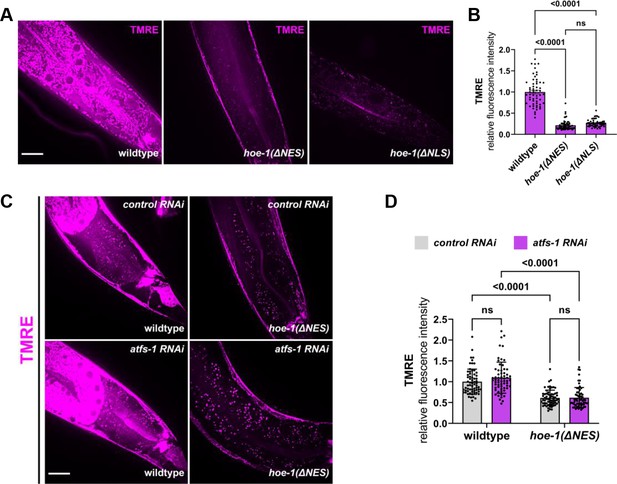
Nuclear export defective HOE-1 activates UPRmt, correlating with reduced mitochondrial membrane potential.
(A) Fluorescence images of TMRE stained day 1 adult wildtype, hoe-1(ΔNES), and hoe-1(ΔNLS) individuals. Scale bar 20 μm. (B) Fluorescence intensity quantification of TMRE staining in individual day 1 adult wildtype, hoe-1(ΔNES), and hoe-1(ΔNLS) animals (n = 57, 60, and 63 respectively, mean and SD shown, ordinary one-way ANOVA with Tukey’s multiple comparisons test). (C) Fluorescence images of TMRE stained day 1 adult wildtype and hoe-1(ΔNES) animals on control and atfs-1 RNAi. Scale bar 20 μm. (D) Fluorescence intensity quantification of TMRE staining in individual day 1 adult wildtype and hoe-1(ΔNES) animals on control and atfs-1 RNAi (n = 65, 62, 65, and 61 respectively, mean and SD shown, ordinary two-way ANOVA with Tukey’s multiple comparisons test).
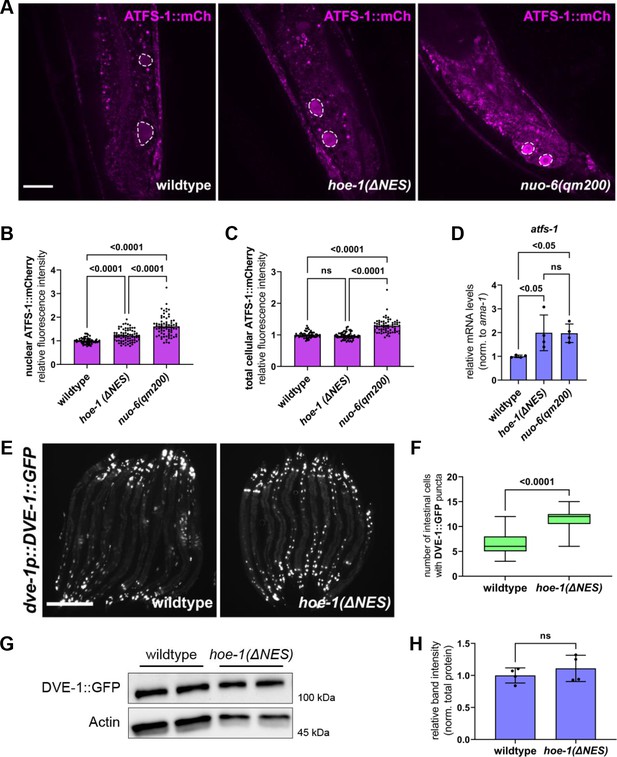
Nuclear export defective HOE-1 animals have increased nuclear accumulation of UPRmt transcription factors ATFS-1 and DVE-1.
(A), Fluorescence images of ATFS-1::mCherry in the terminal intestine of day 2 adult wildtype hoe-1(ΔNES), and nuo-6(qm200) individuals (tip of the tail is in the bottom of each panel). Intestinal nuclei outlined with dashed white line. Scale bar 20 μm. (B) Fluorescence intensity quantification of nuclear ATFS-1::mCherry in wildtype, hoe-1(ΔNES), and nuo-6(qm200) individuals (n = 65, 74, and 72 respectively, mean and SD shown, ordinary one-way ANOVA with Tukey’s multiple comparisons test). (C) Fluorescence intensity quantification of total cellular ATFS-1::mCherry in wildtype, hoe-1(ΔNES), and nuo-6(qm200) individuals (n = 61, 62, and 67 respectively, mean and SD shown, ordinary one-way ANOVA with Tukey’s multiple comparisons test). (D) mRNA transcript quantification of atfs-1 in day 2 adult wildtype, nuo-6(qm200), and hoe-1(ΔNES) animals normalized to ama-1 (n = 4 for each condition, mean and SD shown, ordinary one-way ANOVA with Tukey’s multiple comparisons test). (E) Fluorescence images of dve-1p::DVE-1::GFP in day 2 adult wildtype and hoe-1(ΔNES) animals. Scale bar 200 μm. (F) Number of intestinal cell nuclei with DVE-1::GFP puncta above brightness threshold of 25 in day 2 adult wildtype and hoe-1(ΔNES) animals (n = 33 and 41 respectively, unpaired t-test). (G) Western blot for DVE-1::GFP and actin from day 1 adult wildtype and hoe-1(ΔNES) animals. (H) Quantification of DVE-1::GFP western blot band intensity from day 1 adult wildtype and hoe-1(ΔNES) animals normalized to total protein (n = 4 for each condition, mean and SD shown, unpaired t-test).
-
Figure 5—source data 1
Blots for wildtype and hoe-1(ΔNES) animals with DVE-1::GFP (Figure 4G and H).
All panels are the same membrane. (A) Image of stain-free blot for total protein from day 1 adult wildtype and hoe-1(ΔNES) animals. Four biological replicates of each condition: Lane #1 BR Spectra Protein Ladder – ladder bands in kDa denoted, Lane #2–5 wildtype and #6–9 hoe-1(ΔNES). (B) Chemiluminescence image of blot for DVE-1::GFP using GFP primary antibody. (C) Composite image of chemiluminescence and colorimetric images of blot for DVE-1::GFP to show bands relative to ladder. (D) Chemiluminescence image of blot for actin using β-actin primary antibody. (E) Composite image of chemiluminescence and colorimetric images of blot for actin to show bands relative to ladder.
- https://cdn.elifesciences.org/articles/71634/elife-71634-fig5-data1-v3.zip
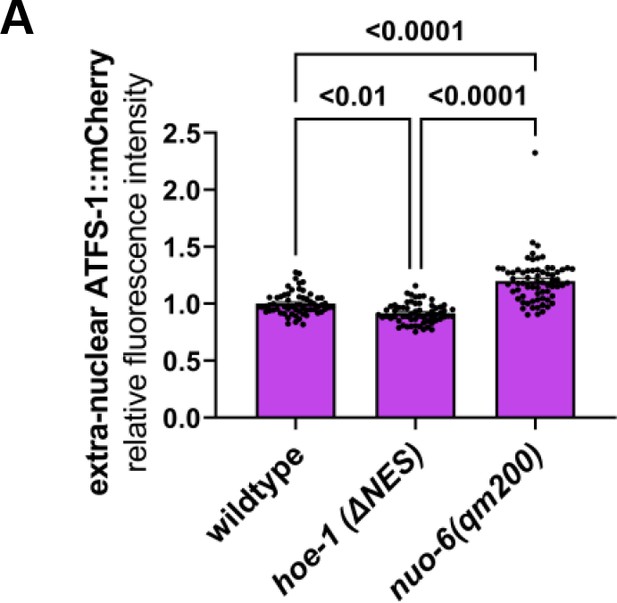
Nuclear export defective HOE-1 does not elevate extra-nuclear ATFS-1::mCherry levels.
Fluorescence intensity quantification of extra-nuclear ATFS-1::mCherry in wildtype, hoe-1(ΔNES), and nuo-6(qm200) individuals (n = 61, 62, and 67 respectively, mean and SD shown, ordinary one-way ANOVA with Tukey’s multiple comparisons test).
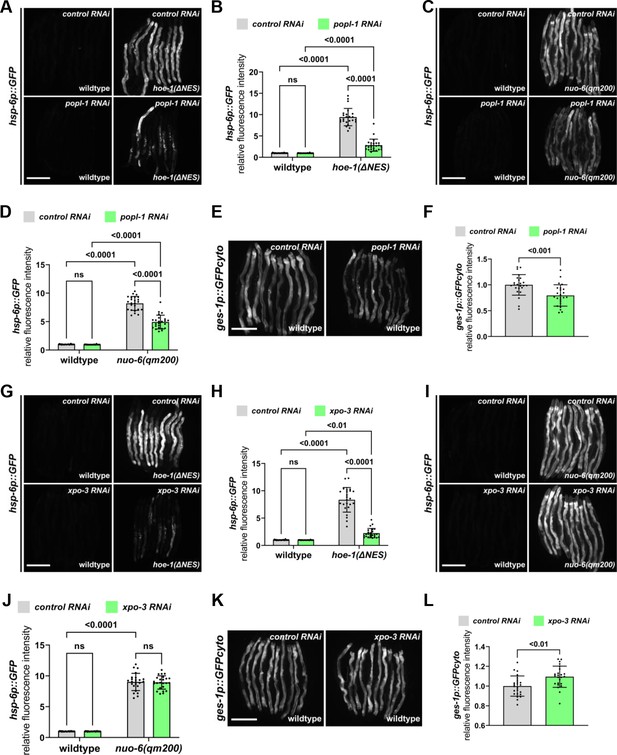
Nuclear export defective HOE-1 activates UPRmt via altered tRNA processing.
(A) Fluorescence images of UPRmt reporter (hsp-6p::GFP) activation in day 2 adult wildtype and hoe-1(ΔNES) animals on control and popl-1 RNAi. Scale bar 200 μm. (B) Fluorescence intensity quantification of hsp-6p::GFP in individual day 2 adult wildtype and hoe-1(ΔNES) animals on control and popl-1 RNAi (n = 24 for each condition, mean and SD shown, ordinary two-way ANOVA with Tukey’s multiple comparisons test). (C) Fluorescence images of UPRmt reporter (hsp-6p::GFP) activation in day 2 adult wildtype and nuo-6(qm200) animals on control and popl-1 RNAi. Scale bar 200 μm. (D) Fluorescence intensity quantification of hsp-6p::GFP in individual day 2 adult wildtype and nuo-6(qm200) animals on control and popl-1 RNAi (n = 24 for each condition, mean and SD shown, ordinary two-way ANOVA with Tukey’s multiple comparisons test). (E) Fluorescence images of intestinal-specific basal protein reporter (ges-1p::GFPcyto) activation in day 2 adult wildtype animals on control and popl-1 RNAi. Scale bar 200 μm. (F) Fluorescence intensity quantification of ges-1p::GFPcyto in individual day 2 adult wildtype animals on control and popl-1 RNAi (n = 24 for each condition, mean and SD shown, unpaired t-test). (G) Fluorescence images of UPRmt reporter (hsp-6p::GFP) activation in day 2 adult wildtype and hoe-1(ΔNES) animals on control and xpo-3 RNAi. Scale bar 200 μm. (H) Fluorescence intensity quantification of hsp-6p::GFP in individual day 2 adult wildtype and hoe-1(ΔNES) animals on control and xpo-3 RNAi (n = 24 for each condition, mean and SD shown, ordinary two-way ANOVA with Tukey’s multiple comparisons test). (I) Fluorescence images of UPRmt reporter (hsp-6p::GFP) activation in day 2 adult wildtype and nuo-6(qm200) animals on control and xpo-3 RNAi. Scale bar 200 μm. (J) Fluorescence intensity quantification of hsp-6p::GFP in individual day 2 adult wildtype and nuo-6(qm200) animals on control and xpo-3 RNAi (n = 24 for each condition, mean and SD shown, ordinary two-way ANOVA with Tukey’s multiple comparisons test). (K) Fluorescence images of intestinal-specific basal protein reporter (ges-1p::GFPcyto) activation in day 2 adult wildtype animals on control and xpo-3 RNAi. Scale bar 200 μm. (L) Fluorescence intensity quantification of ges-1p::GFPcyto in individual day 2 adult wildtype animals on control and xpo-3 RNAi (n = 24 for each condition, mean and SD shown, unpaired t-test).
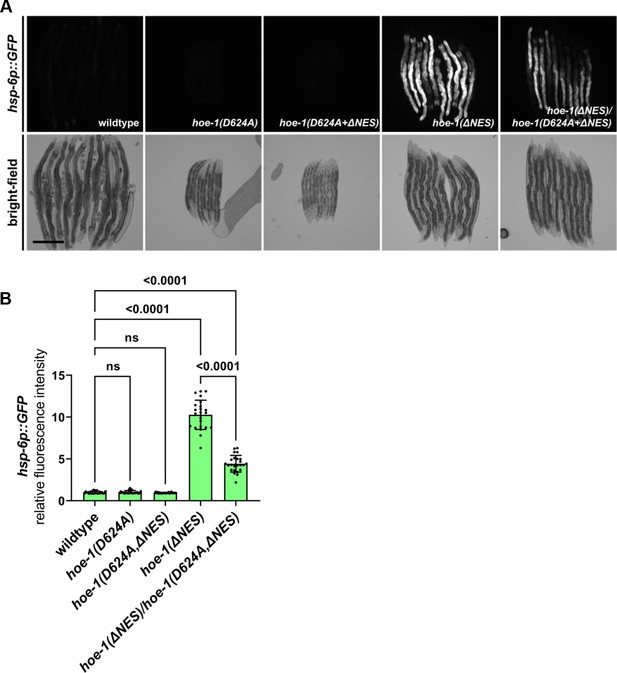
Nuclear export defective HOE-1 induced UPRmt is dependent upon the catalytic activity of HOE-1.
(A) Fluorescence images of UPRmt reporter (hsp-6p::GFP) activation and corresponding bright-field images of wildtype, catalytically-dead hoe-1 (hoe-1(D624A)) mutant, catalytically-dead nuclear export defective hoe-1 (hoe-1(D624A+ΔNES)) mutant, hoe-1(ΔNES), and hoe-1(ΔNES)/hoe-1(D624A+ΔNES) trans-heterozygous mutant animals 96 hr post-embryo. Scale bar 200 μm. (B) Fluorescence intensity quantification of hsp-6p::GFP in individual wildtype, hoe-1(D624A), hoe-1(D624A+ΔNES), hoe-1(ΔNES), and hoe-1(ΔNES)/hoe-1(D624A+ΔNES) trans-heterozygous animals 96 hr post-embryo (n = 24 for each condition, mean and SD shown, ordinary one-way ANOVA with Tukey’s multiple comparisons test).
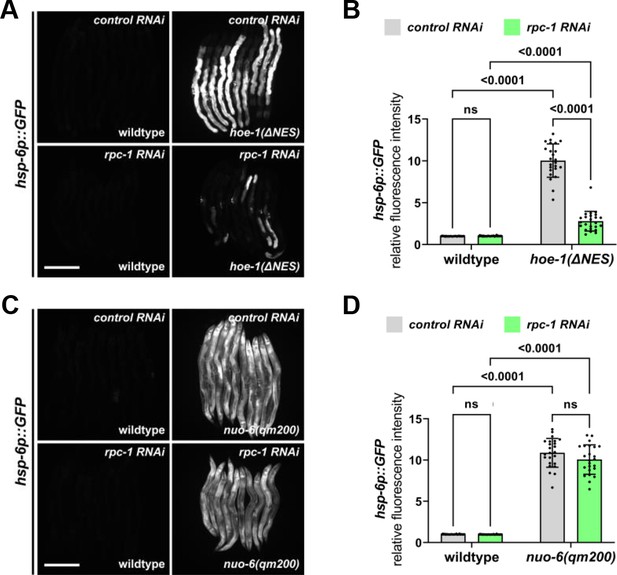
RNAi against RNA polymerase III subunit, rpc-1, preferentially attenuates hoe-1(ΔNES)-induced UPRmt.
(A) Fluorescence images of UPRmt reporter (hsp-6p::GFP) activation in wildtype and hoe-1(ΔNES) day 2 adult animals on control and rpc-1 RNAi. Scale bar 200 μm. (B) Fluorescence intensity quantification of hsp-6p::GFP in individual wildtype and hoe-1(ΔNES) day 2 adult animals on control and rpc-1 RNAi (n = 24 for each condition, mean and SD shown, ordinary two-way ANOVA with Tukey’s multiple comparisons test). (C) Fluorescence images of UPRmt reporter (hsp-6p::GFP) activation in wildtype and nuo-6(qm200) day 2 adult animals on control and rpc-1 RNAi. Scale bar 200 μm. (D) Fluorescence intensity quantification of hsp-6p::GFP in individual wildtype and nuo-6(qm200) day 2 adult animals on control and rpc-1 RNAi (n = 24 for each condition, mean and SD shown, ordinary two-way ANOVA with Tukey’s multiple comparisons test).
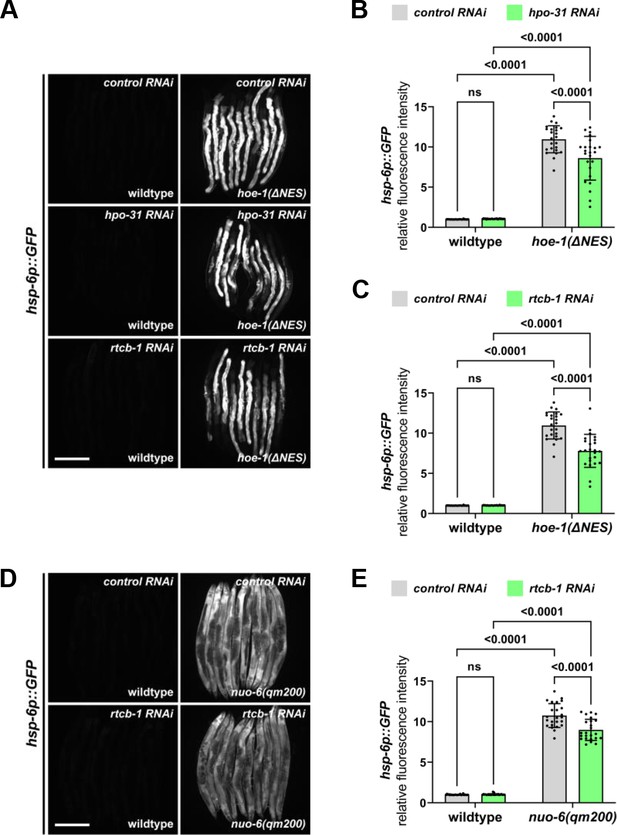
RNAi against tRNA nucleotidyl transferase, hpo-31, and tRNA ligase, rtcb-1, mildly attenuate both hoe-1(ΔNES)- and nuo-6(qm200)-induced UPRmt.
(A) Fluorescence images of UPRmt reporter (hsp-6p::GFP) activation in wildtype and hoe-1(ΔNES) day 2 adult animals on control, hpo-31, and rtcb-1 RNAi. Scale bar 200 μm. (B) Fluorescence intensity quantification of hsp-6p::GFP in individual wildtype and hoe-1(ΔNES) day 2 adult animals on control and hpo-31 RNAi (n = 24 for each condition, mean and SD shown, ordinary two-way ANOVA with Tukey’s multiple comparisons test). (C) Fluorescence intensity quantification of hsp-6p::GFP in individual wildtype and hoe-1(ΔNES) day 2 adult animals on control and rtcb-1 RNAi (n = 24 for each condition, mean and SD shown, ordinary two-way ANOVA with Tukey’s multiple comparisons test). Note that the same control RNAi animals were used for analysis in both panels B and C as experiments were conducted simultaneously. (D) Fluorescence images of UPRmt reporter (hsp-6p::GFP) activation in wildtype and nuo-6(qm200) day 2 adult animals on control, and rtcb-1 RNAi. Scale bar 200 μm. (E) Fluorescence intensity quantification of hsp-6p::GFP in individual wildtype and nuo-6(qm200) day 2 adult animals on control and rtcb-1 RNAi (n = 24 for each condition, mean and SD shown, ordinary two-way ANOVA with Tukey’s multiple comparisons test).
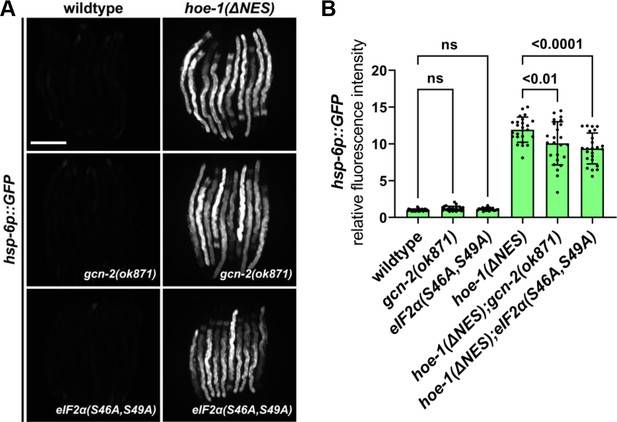
Nuclear export defective HOE-1 induced UPRmt is not gcn-2 or eIF2α dependent.
(A) Fluorescence images of UPRmt reporter (hsp-6p::GFP) activation in day 2 adult wildtype, gcn-2(ok871), eIF2α(S46A,S49A), hoe-1(ΔNES), hoe-1(ΔNES);gcn-2(ok871), and hoe-1(ΔNES);eIF2α(S46A,S49A) animals. Scale bar 200 μm. (B) Fluorescence intensity quantification of hsp-6p::GFP in individual day 2 adult wildtype, gcn-2(ok871), eIF2α(S46A,S49A), hoe-1(ΔNES), hoe-1(ΔNES);gcn-2(ok871), and hoe-1(ΔNES);eIF2α(S46A,S49A) animals (n = 24 for each condition, mean and SD shown, ordinary two-way ANOVA with Tukey’s multiple comparisons test).
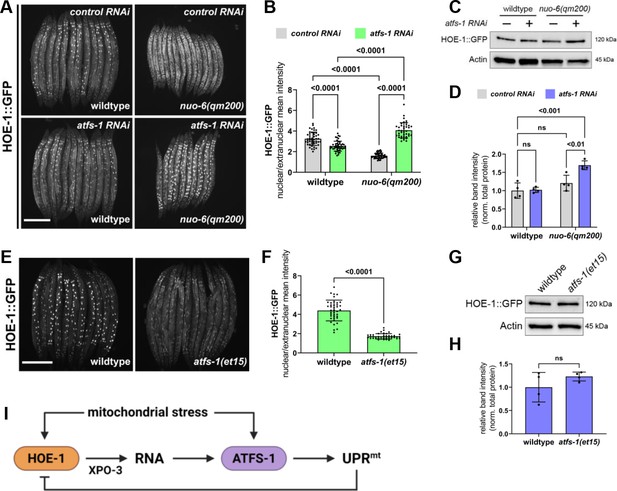
Nuclear HOE-1 levels are elevated during mitochondrial stress in the absence of ATFS-1 but decreased in the presence of ATFS-1.
(A) Fluorescence images of HOE-1::GFP in day 1 adult wildtype and nuo-6(qm200) animals on control and atfs-1 RNAi. Scale bar 200 μm. (B) Fluorescence intensity quantification of intestinal nuclei relative to extranuclear signal in day 1 adult wildtype and nuo-6(qm200) animals on control and atfs-1 RNAi (n = 40 for each condition, mean and SD shown, ordinary two-way ANOVA with Tukey’s multiple comparisons test). (C) Western blot for HOE-1::GFP and actin from day 1 adult wildtype and nuo-6(qm200) animals on control and atfs-1 RNAi. (D) Quantification of HOE-1::GFP western blot band intensity from day 1 adult wildtype and nuo-6(qm200) animals on control and atfs-1 RNAi normalized to total protein (n = 4 for each condition, mean and SD shown, ordinary two-way ANOVA with Tukey’s multiple comparisons test). (E) Fluorescence images of HOE-1::GFP in day 1 adult wildtype and atfs-1(et15) animals. Scale bar 200 μm. (F) Fluorescence intensity quantification of intestinal nuclei relative to extranuclear signal in day 1 adult wildtype and atfs-1(et15) animals (n = 40 for each condition, mean and SD shown, unpaired t-test). (G) Western blot for HOE-1::GFP and actin from day 1 adult wildtype and atfs-1(et15) animals. (H) Quantification of HOE-1::GFP western blot band intensity from day 1 adult wildtype and atfs-1(et15) animals normalized to total protein (n = 4 for each condition, mean and SD shown, unpaired t-test). (I) Mitochondrial stress triggers activation of HOE-1 resulting in altered RNA processing that facilitates UPRmt via ATFS-1. Activation of UPRmt negatively regulates HOE-1.
-
Figure 8—source data 1
Blots for wildtype and nuo-6(qm200) animals on control and atfs-1 RNAi (Figure 8C).
All panels are the same membrane. (A) Image of stain-free blot for total protein from day 1 adult wildtype and nuo-6(qm200) animals on control and atfs-1 RNAi. Two biological replicates of each condition: Lane # 1&11 BR Spectra Protein Ladder – ladder bands in kDa denoted. Lane # 2&7 wildtype on control RNAi, 3&8 wildtype on atfs-1 RNAi, 4&9 nuo-6(qm200) on control RNAi, and 5&10 nuo-6(qm200) on atfs-1 RNAi. Lane # 6 empty. (B) Chemiluminescence image of blot for HOE-1::GFP using GFP primary antibody. (C) Composite image of chemiluminescence and colorimetric images of blot for HOE-1::GFP to show bands relative to ladder. (D) Chemiluminescence image of blot for actin using β-actin primary antibody. (E) Composite image of chemiluminescence and colorimetric images of blot for actin to show bands relative to ladder.
- https://cdn.elifesciences.org/articles/71634/elife-71634-fig8-data1-v3.zip
-
Figure 8—source data 2
Blots for wildtype and nuo-6(qm200) animals on control and atfs-1 RNAi (Figure 8D).
Samples were loaded and ran on two separate membranes simultaneously (Membrane A and Membrane B). All panels in each column are the same membrane. (A) Image of stain-free blots for total protein from day 1 adult wildtype and nuo-6(qm200) animals on control and atfs-1 RNAi. Two biological replicates on each blot of each condition: Lane # 1&10 BR Spectra Protein Ladder – ladder bands in kDa denoted. Lane # 2&6 wildtype on control RNAi, 3&7 wildtype on atfs-1 RNAi, 4&8 nuo-6(qm200) on control RNAi, and 5&9 nuo-6(qm200) on atfs-1 RNAi. (B) Chemiluminescence image of blots for HOE-1::GFP using GFP primary antibody. (C) Composite images of chemiluminescence and colorimetric images of blots for HOE-1::GFP to show bands relative to ladder. (D) Chemiluminescence images of blots for actin using β-actin primary antibody. (E) Composite images of chemiluminescence and colorimetric images of blots for actin to show bands relative to ladder.
- https://cdn.elifesciences.org/articles/71634/elife-71634-fig8-data2-v3.zip
-
Figure 8—source data 3
Blots for wildtype and atfs-1(et15) animals (Figure 8G).
All panels are the same membrane. (A) Image of stain-free blot for total protein from day 1 adult wildtype and atfs-1(et15) animals. Two biological replicates of each condition: Lane # 1&6 BR Spectra Protein Ladder – ladder bands in kDa denoted. Lane #2&4 wildtype and #3&5 atfs-1(et15). (B) Chemiluminescence image of blot for HOE-1::GFP using GFP primary antibody. (C) Composite image of chemiluminescence and colorimetric images of blot for HOE-1::GFP to show bands relative to ladder. (D) Chemiluminescence image of blot for actin using β-actin primary antibody. (E) Composite image of chemiluminescence and colorimetric images of blot for actin to show bands relative to ladder.
- https://cdn.elifesciences.org/articles/71634/elife-71634-fig8-data3-v3.zip
-
Figure 8—source data 4
Blots for wildtype and atfs-1(et15) animals (Figure 8H).
All panels are the same membrane. (A), Image of stain-free blot for total protein from day 1 adult wildtype and atfs-1(et15) animals. Four biological replicates of each condition: Lane # 1 BR Spectra Protein Ladder – ladder bands in kDa denoted. Lanes #2,4,6,8 wildtype and #3,5,7,9 atfs-1(et15). (B) Chemiluminescence image of blot for HOE-1::GFP using GFP primary antibody. (C) Composite image of chemiluminescence and colorimetric images of blot for HOE-1::GFP to show bands relative to ladder. (D) Chemiluminescence image of blot for actin using β-actin primary antibody. (E) Composite image of chemiluminescence and colorimetric images of blot for actin to show bands relative to ladder.
- https://cdn.elifesciences.org/articles/71634/elife-71634-fig8-data4-v3.zip
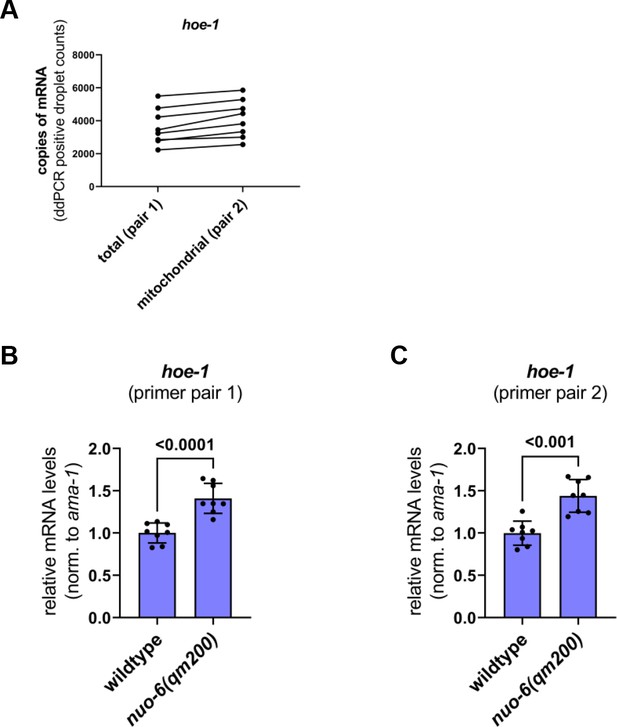
hoe-1 mRNA levels are upregulated under conditions of mitochondrial stress.
(A) Copies of total hoe-1 mRNA (primer pair 1) versus hoe-1 mRNA that include the mitochondrial targeting sequence (primer pair 2) in day 1 adult wildtype animals. ddPCR droplet counts shown. Paired samples connected with solid black line. (B–C) mRNA transcript quantification of hoe-1 in day 1 adult wildtype and nuo-6(qm200) animals normalized to ama-1 (n = 4 for each condition, mean and SD shown, unpaired t-test) measured with two separate primer pairs.
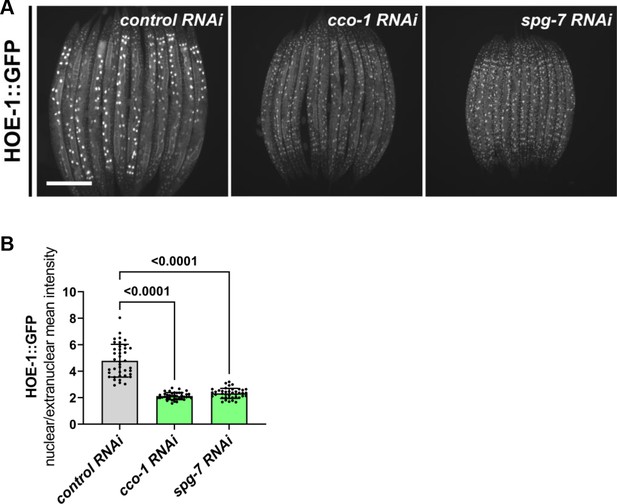
UPRmt-inducing cco-1 and spg-7 RNAi both attenuate HOE-1 nuclear levels.
(A) Fluorescence images of HOE-1::GFP expressing animals on control, cco-1, and spg-7 RNAi. Scale bar 200 μm. (B) Fluorescence intensity quantification of intestinal nuclei relative to extranuclear signal of HOE-1::GFP on control, cco-1, and spg-7 RNAi (n = 40 for each condition, mean and SD shown, ordinary one-way ANOVA with Dunnett’s multiple comparisons test).
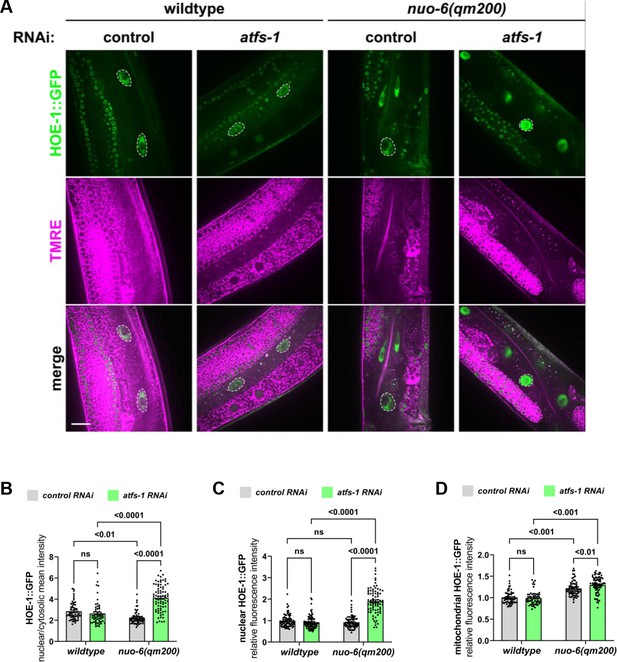
Nuclear HOE-1 levels are elevated during mitochondrial stress in the absence of ATFS-1 but decreased in the presence of ATFS-1.
(A) Fluorescence images of the intestine of individual day 1 adult wildtype and nuo-6(qm200) animals expressing HOE-1::GFP (green) stained with TMRE (magenta) to visualize mitochondria on control and atfs-1 RNAi. GFP and TMRE co-localization shown in white in merged image. Nuclei are traced with dashed white line. Scale bar 20 μm. (B) Fluorescence intensity quantification of the nuclear to cytosolic ratio of HOE-1::GFP in intestine of wildtype and nuo-6(qm200) animals on control and atfs-1 RNAi (n = 59, 64, 57, and 75 respectively, mean and SD shown, ordinary two-way ANOVA with Tukey’s multiple comparisons test). (C) Fluorescence intensity quantification of nuclear HOE-1::GFP in intestine of wildtype and nuo-6(qm200) animals on control and atfs-1 RNAi (n = 76, 76, 80, and 77 respectively, mean and SD shown, ordinary two-way ANOVA with Tukey’s multiple comparisons test). (D) Fluorescence intensity quantification of mitochondrial HOE-1::GFP in intestine of wildtype and nuo-6(qm200) animals on control and atfs-1 RNAi (n = 59, 64, 57, and 75 respectively, mean and SD shown, ordinary two-way ANOVA with Tukey’s multiple comparisons test).
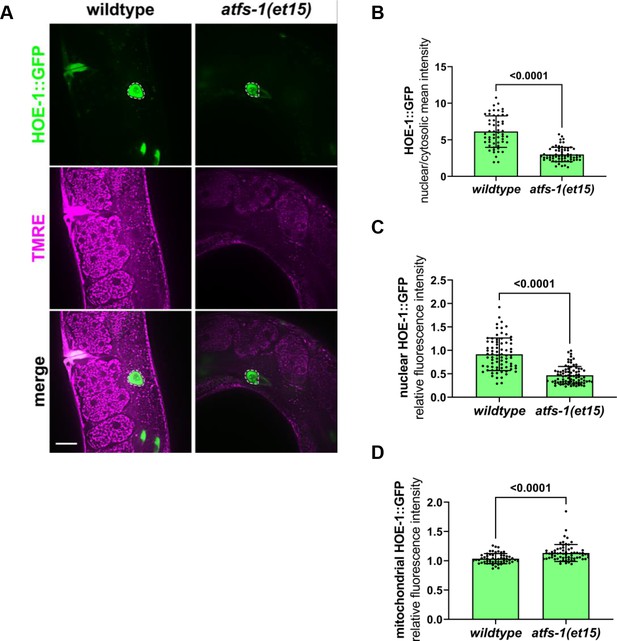
Constitutive activation of UPRmt by atfs-1 gain-of-function (atfs-1(et15)) depletes nuclear HOE-1 levels.
(A) Fluorescence images of the intestine of individual day 1 adult wildtype and atfs-1(et15) animals expressing HOE-1::GFP (green) stained with TMRE (magenta) to visualize mitochondria. GFP and TMRE co-localization shown in white in merged image. Nuclei are traced with dashed white line. Scale bar 20 μm. (B) Fluorescence intensity quantification of the nuclear to cytosolic ratio of HOE-1::GFP in intestine of wildtype and atfs-1(et15) animals (n = 56 and 66 respectively, mean and SD shown, unpaired t-test). (C) Fluorescence intensity quantification of nuclear HOE-1::GFP in intestine of wildtype and atfs-1(et15) animals (n = 77 and 81 respectively, mean and SD shown, unpaired t-test). (D) Fluorescence intensity quantification of mitochondrial HOE-1::GFP in intestine of wildtype and atfs-1(et15) animals (n = 56 and 66 respectively, mean and SD shown, unpaired t-test).
Additional files
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/71634/elife-71634-transrepform1-v3.pdf
-
Supplementary file 1
C. elegans strains used in this study.
- https://cdn.elifesciences.org/articles/71634/elife-71634-supp1-v3.xlsx
-
Supplementary file 2
Oligonucleotides used in this study.
- https://cdn.elifesciences.org/articles/71634/elife-71634-supp2-v3.xlsx





