A conserved neuropeptide system links head and body motor circuits to enable adaptive behavior
Figures
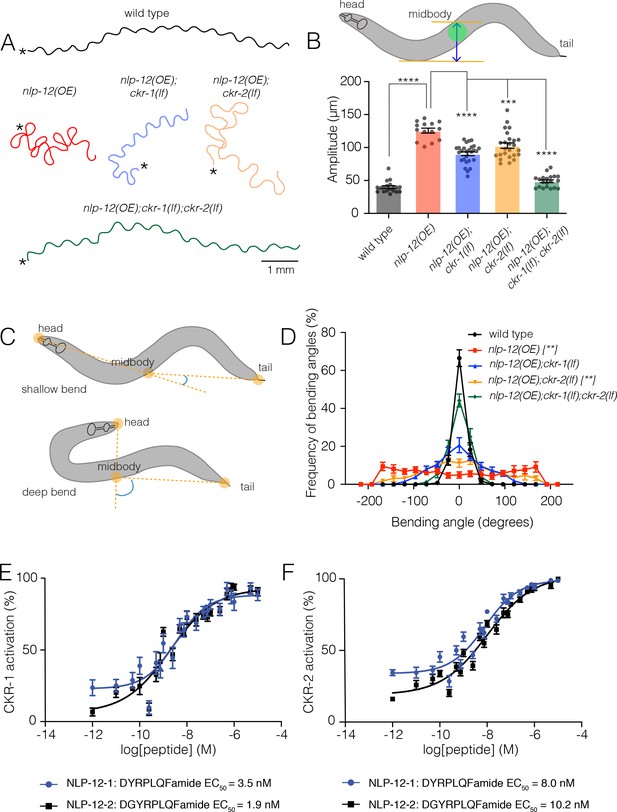
NLP-12/CCK induced locomotor responses require functional ckr-1 signaling.
(A) Representative movement trajectories of wild-type (black), nlp-12(OE) (red), nlp-12(OE);ckr-1(lf) (blue), nlp-12(OE);ckr-2(lf) (orange), and nlp-12(OE);ckr-1(lf);ckr-2(lf) (green) animals during forward runs (30 s) on NGM agar plates seeded with OP50 bacteria. nlp-12(OE) refers to the transgenic strain (ufIs104) stably expressing high levels of wild-type nlp-12 genomic sequence. Note the convoluted nlp-12(OE) movement tracks are restored to wild type by combined ckr-1 and ckr-2 deletion. Scale bar, 1 mm. Asterisks (*) indicate position of worm at start of recording. (B) Average body bend amplitude (indicated in schematic by blue arrow between orange lines, midbody centroid [green] of worm) for the genotypes as indicated. Bars represent mean ± SEM. In this and subsequent figures. ****p<0.0001, ***p<0.001, ANOVA with Holms-Sidak post hoc test. wild-type n=19, nlp-12(OE): n=14, nlp-12(OE);ckr-1(lf): n=27, nlp-12(OE);ckr-2(lf): n=25, nlp-12(OE);ckr-1(lf);ckr-2(lf): n=20. (C) Schematic representation of measured body bending angle, for shallow (top) and deep (bottom) body bends. Solid orange circles indicate the vertices (head, midbody, and tail) of the body bending angle (blue) measured. (D) Frequency distribution of body bending angle (indicated in blue in (C)) for the genotypes indicated. Kolmogorov-Smirnov test: wild-type versus nlp-12(OE)**, wild-type versus nlp-12(OE);ckr-2(lf)**, nlp-12(OE) versus nlp-12(OE);ckr-1(lf);ckr-2(lf)**, **p<0.01. wild-type: n=12, nlp-12(OE): n=10, nlp-12(OE);ckr-1(lf): n=10, nlp-12(OE);ckr-2(lf): n=12, nlp-12(OE);ckr-1(lf);ckr-2(lf): n=12. (E, F) Concentration-response curves of the mean calcium responses (% activation ± SEM) in CHO cells expressing either CKR-1 (E) or CKR-2 (F) for different concentrations of synthetic peptides NLP-12–1 (solid blue circles) or NLP-12–2 (solid black squares). Solid lines indicate curve fits to the data (n=6). 95% confidence intervals (nM), CKR-1: NLP-12–1, 1.79–7.07; NLP-12–2, 0.93–3.77 and CKR-2: NLP-12–1, 5.16–12.51; NLP-12–2, 6.43–16.73. NGM, nematode growth media.
-
Figure 1—source data 1
Source data for body bending amplitude (Figure 1B).
- https://cdn.elifesciences.org/articles/71747/elife-71747-fig1-data1-v2.xlsx
-
Figure 1—source data 2
Source data for frequency of bending angles (Figure 1D).
- https://cdn.elifesciences.org/articles/71747/elife-71747-fig1-data2-v2.xlsx
-
Figure 1—source data 3
Source data for in vitro analysis of CKR-1 activation (Figure 1E).
- https://cdn.elifesciences.org/articles/71747/elife-71747-fig1-data3-v2.xlsx
-
Figure 1—source data 4
Source data for in vitro analysis of CKR-2 activation (Figure 1F).
- https://cdn.elifesciences.org/articles/71747/elife-71747-fig1-data4-v2.xlsx
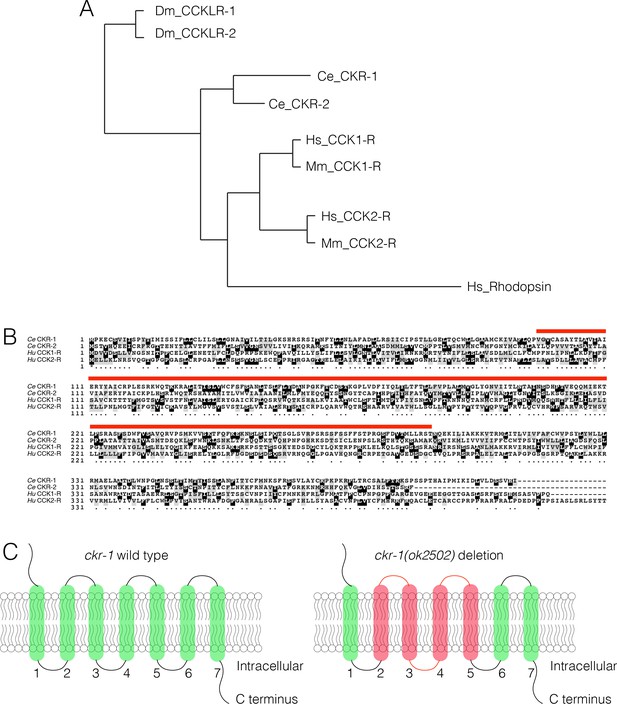
CKR-1 and CKR-2 GPCRs share similarity with vertebrate CCK GPCRs.
(A) Dendrogram (generated using Phylogeny,fr Dereeper et al., 2008). showing the predicted relationship between Drosophila (Dm_CCKLR-1/2), C. elegans (Ce_CKR-1/2), mouse (Mm), and human (Hs) CCK1/2R GPCRs. (B) Boxshade alignment of Caenorhabditis elegans CKR-1 and CKR-2 with Human CCK-1 and CCK-2 receptors. Black shading indicates identical amino acids, while gray shading indicates similar amino acids. Red bar indicates the amino acids removed by ckr-1(ok2502) deletion. (C) Schematic representation of CKR-1 GPCR membrane topology and domains affected by the ckr-1(ok2502) deletion (red shading).

NLP-12 peptides activate CKR-1 and CKR-2 in vitro.
NLP-12–1 and NLP-12–2 elicit Ca2+ responses in cells expressing CKR-1 or CKR-2, but not in cells transfected with an empty pcDNA3.1 vector. Bar graphs indicate the ratio of total Ca2+ response of CHO cells expressing CKR-1, CKR-2 or pcDNA3.1 empty vector, challenged with 10 µM of NLP-12 peptides (n=7), BSA (negative control, n=5), or ATP (positive control, n=5). Ratio of total Ca2+ response is calculated as peptide-evoked response normalized to the total Ca2+ response. Data were analyzed by two-way ANOVA; ****p<0.0001; ns, not significant (p>0.05).
-
Figure 1—figure supplement 2—source data 1
Source data for in vitro controls (ratio of total calcium response).
- https://cdn.elifesciences.org/articles/71747/elife-71747-fig1-figsupp2-data1-v2.xlsx
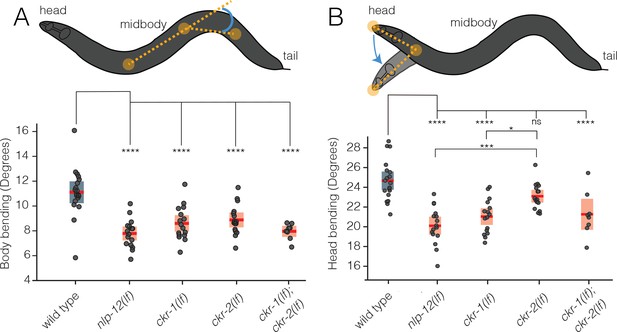
CKR-1 and CKR-2 differentially regulate head and body bending during basal locomotion.
Schematics showing body bending (A) and head bending (B) angles (solid orange circles indicate the vertices and measured angle in blue) quantified during single worm track analyses of movement (5 min) in the presence of food. Each data point in the scatterplots represents the average body or head bend angle for a single animal from analysis of 5 min of locomotion. Horizontal red bar indicates mean, shading indicates SEM for wild-type (blue) and mutants (orange). ****p<0.0001, ***p<0.001, *p<0.05, ns, not significant. ANOVA with Holms-Sidak post hoc test. wild-type: n=19, nlp-12(ok335): n=16, ckr-1(ok2502): n=16, ckr-2(tm3082): n=16, ckr-1(ok2502);ckr-2(tm3082): n=8.
-
Figure 2—source data 1
Source data for body bending measurements during single worm tracking of basal locomotion (Figure 2A).
- https://cdn.elifesciences.org/articles/71747/elife-71747-fig2-data1-v2.xlsx
-
Figure 2—source data 2
Source data for head bending measurements during single worm tracking of basal locomotion (Figure 2B).
- https://cdn.elifesciences.org/articles/71747/elife-71747-fig2-data2-v2.xlsx
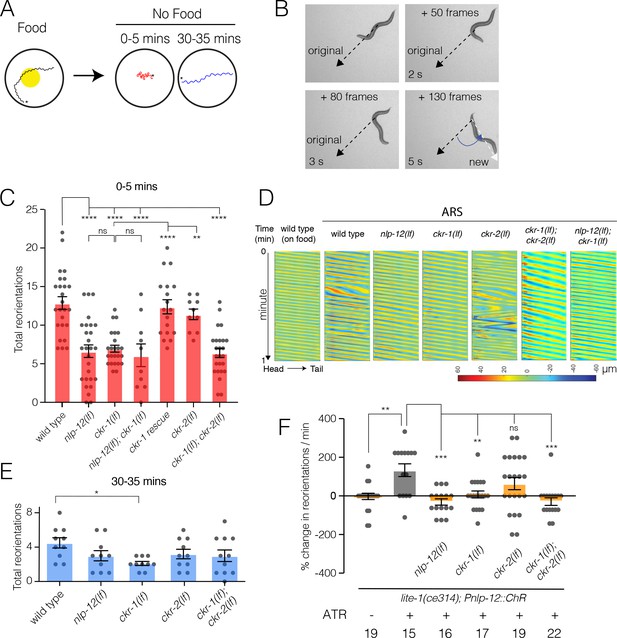
NLP-12/CCK food search responses are mediated through the GPCR CKR-1.
(A) Schematic of the food search assay indicating the time intervals when reorientations were scored. Wild-type animals increase reorientations during the first 5 min (0–5 min) after removal from food (local search) and reduce reorientations during dispersal (30–35 min). Asterisks (*) indicate the position of worm at the start of recording. (B) Frame grabs showing worm position and posture prior to, during and after reorientation. Angle (blue) between the black (original trajectory) and white (new trajectory) dashed lines indicates the change in trajectory. Frame numbers and time points indicated are relative to the first image in each sequence, which represents the start point (frame 0, time 0 s) when the reorientation event began, and the last frame was when the reorientation was completed. Trajectory changes were scored as reorientations if changes in trajectory were greater than 50°. (C) Quantification of reorientations during 0–5 min following removal from food for the genotypes indicated. Rescue refers to transgenic expression of wild-type ckr-1 in ckr-1 mutants. Bars represent mean ± SEM. ****p<0.0001, **p<0.01, ns, not significant, ANOVA with Holms-Sidak post hoc test. wild-type: n=25, nlp-12(ok335): n=27, ckr-1(ok2502): n=24, nlp-12(ok335);ckr-1(ok2502): n=10, ckr-1 rescue: n=18, ckr-2(tm3082): n=10, ckr-1(ok2502);ckr-2(tm3082): n=25. (D) Representative body curvature kymographs for worm locomotion during basal locomotion and area restricted searching (ARS). Head to tail orientation along the horizontal axis in each kymograph is left to right as indicated for wild type. Time is indicated along the vertical axis from 0 min to 1 min. (E) Total number of reorientations during an interval of 30–35 min following removal from food for the genotypes as shown. Each bar represents mean ± SEM. *p<0.05, ANOVA with Holms-Sidak post hoc test. wild-type: n=10, nlp-12(ok335): n=10, ckr-1(ok2502): n=10, ckr-2(tm3082): n=10, ckr-1(ok2502);ckr-2(tm3082): n=11. (F) Trajectory changes (reorientations) scored in response to photostimulation of DVA. Percent change in the number of high angle turns elicited during 1 min of blue light exposure compared to prestimulus (no blue light). Bars represent mean ± SEM. ***p<0.001, **p<0.01, ns, not significant, compared to +ATR control, ANOVA with Holms-Sidak post hoc test. ATR, all-trans retinal.
-
Figure 3—source data 1
Source data for reorientations quantified during area restricted search (0–5 min off food, Figure 3C).
- https://cdn.elifesciences.org/articles/71747/elife-71747-fig3-data1-v2.xlsx
-
Figure 3—source data 2
Source data for reorientations quantified during dispersal (30–35 min off food, Figure 3E).
- https://cdn.elifesciences.org/articles/71747/elife-71747-fig3-data2-v2.xlsx
-
Figure 3—source data 3
Source data for % change in reorientations from mean quantified for DVA photostimulation (Figure 3F).
- https://cdn.elifesciences.org/articles/71747/elife-71747-fig3-data3-v2.xlsx
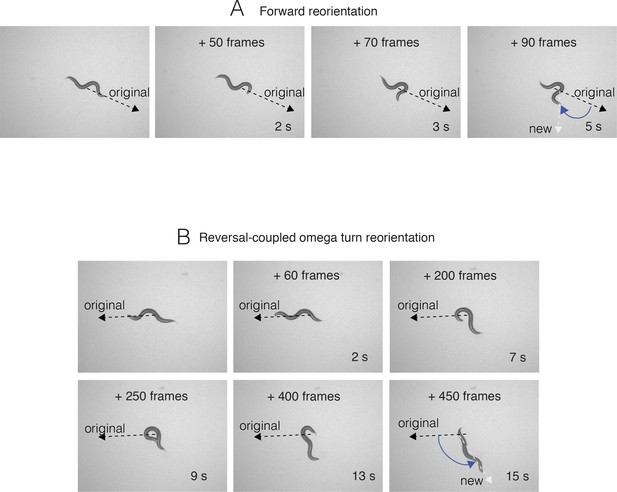
Sequential snapshots of frames from a representative reorientation, for forward reorientations (A) and reversal-coupled omega turn mediated reorientations (B).
Frame #s and time points are indicated in each panel. Frame numbers and time points indicated are relative to the first image in each sequence, which represents the start point (frame 0, time 0 s) when the reorientation event began, and the last frame was when the reorientation was completed. Black dashed line shows the original trajectory, and white dashed line the new trajectory upon completion of the reorientation. Blue angle shows the measured change in trajectory (degrees).

NLP-12 signaling through CKR-1 promotes forward reorientations.
(A) Quantification of reorientations during ARS (0–5 min following removal from food) compare to animals on food. Note that the increased number of forward and reversal coupled reorientations. Bars represent mean ± SEM. ****p<0.0001, ***p<0.001, Student’s t-test. wild-type on food: n=9, wild-type ARS: n=8. (B) Quantification of reorientations during ARS (0–5 min following removal from food) for the genotypes indicated. Note that the number of forward reorientations during ARS are significantly decreased in nlp-12(ok335) and ckr-1(ok2502) animals. However, reversal coupled reorientations are unaffected. Bars represent mean ± SEM. **p<0.01, ANOVA with Holms-Sidak post hoc test. wild-type: n=14, nlp-129(ok335): n=13, ckr-1(ok2502): n=9. ARS, area-restricted searching.
-
Figure 3—figure supplement 2—source data 1
Source data for reorientations quantified on food and during area restricted search (0–5 min off food, Figure 3—figure supplement 2A).
- https://cdn.elifesciences.org/articles/71747/elife-71747-fig3-figsupp2-data1-v2.xlsx
-
Figure 3—figure supplement 2—source data 2
Source data for reorientations quantified during area restricted search (0–5 min off food, Figure 3—figure supplement 2B).
- https://cdn.elifesciences.org/articles/71747/elife-71747-fig3-figsupp2-data2-v2.xlsx
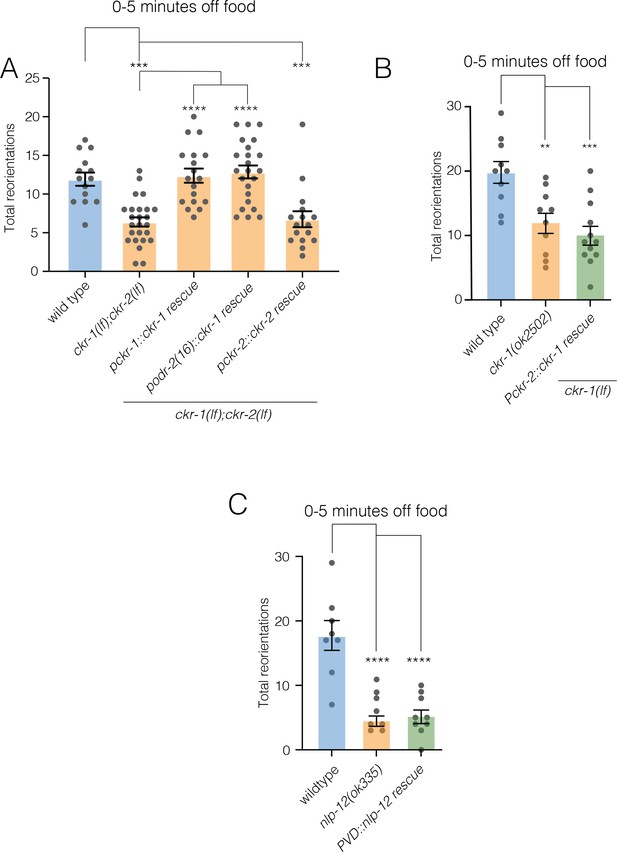
NLP-12 released from DVA acts selectively through CKR-1 to promote reorientations.
(A) Quantification of reorientations during ARS (0–5 min following removal from food) for the genotypes indicated. Rescue refers to transgenic expression of wild-type ckr-1 or ckr-2 in ckr-1(ok2502);ckr-2(tm3082) mutants. Bars represent mean ± SEM. ****p<0.0001, ***p<0.001, ANOVA with Holms-Sidak post hoc test. wild-type: n=14, ckr-1(ok2502);ckr-2(tm3082): n=25, Pckr-1::ckr-1 rescue: n=18, Podr-2(16)::ckr-1 rescue: n=23, Pckr-2::ckr-2 rescue: n=16. (B) Quantification of reorientations during 0–5 min following removal from food for the genotypes indicated. Note that the expression of ckr-1 under the ckr-2 promoter does not rescue reorientations during ARS in ckr-1(ok2502) animals. Bars represent mean ± SEM. ***p<0.001, **p<0.01, ANOVA with Holms-Sidak post hoc test. wild-type: n=10, ckr-1(ok2502): n=10, Pckr-2::ckr-1 rescue: n=12. (C) Quantification of reorientations during 0–5 min following removal from food for the genotypes indicated. Note that the expression of nlp-12 under the PVD specific promoter (ser-2prom3) does not rescue reorientations during ARS in nlp-12(ok335) animals. Bars represent mean ± SEM. ****p<0.0001, ANOVA with Holms-Sidak post hoc test. wild-type: n=8, nlp-12(ok335): n=8, Pser-2prom3::nlp-12 rescue: n=9. ARS, area-restricted searching.
-
Figure 3—figure supplement 3—source data 1
Source data for reorientations quantified during area restricted search (0–5 min off food, Figure 3—figure supplement 3A).
- https://cdn.elifesciences.org/articles/71747/elife-71747-fig3-figsupp3-data1-v2.xlsx
-
Figure 3—figure supplement 3—source data 2
Source data for reorientations quantified during area restricted search (0–5 min off food, Figure 3—figure supplement 3B).
- https://cdn.elifesciences.org/articles/71747/elife-71747-fig3-figsupp3-data2-v2.xlsx
-
Figure 3—figure supplement 3—source data 3
Source data for reorientations quantified during area restricted search (0–5 min off food, Figure 3—figure supplement 3C).
- https://cdn.elifesciences.org/articles/71747/elife-71747-fig3-figsupp3-data3-v2.xlsx
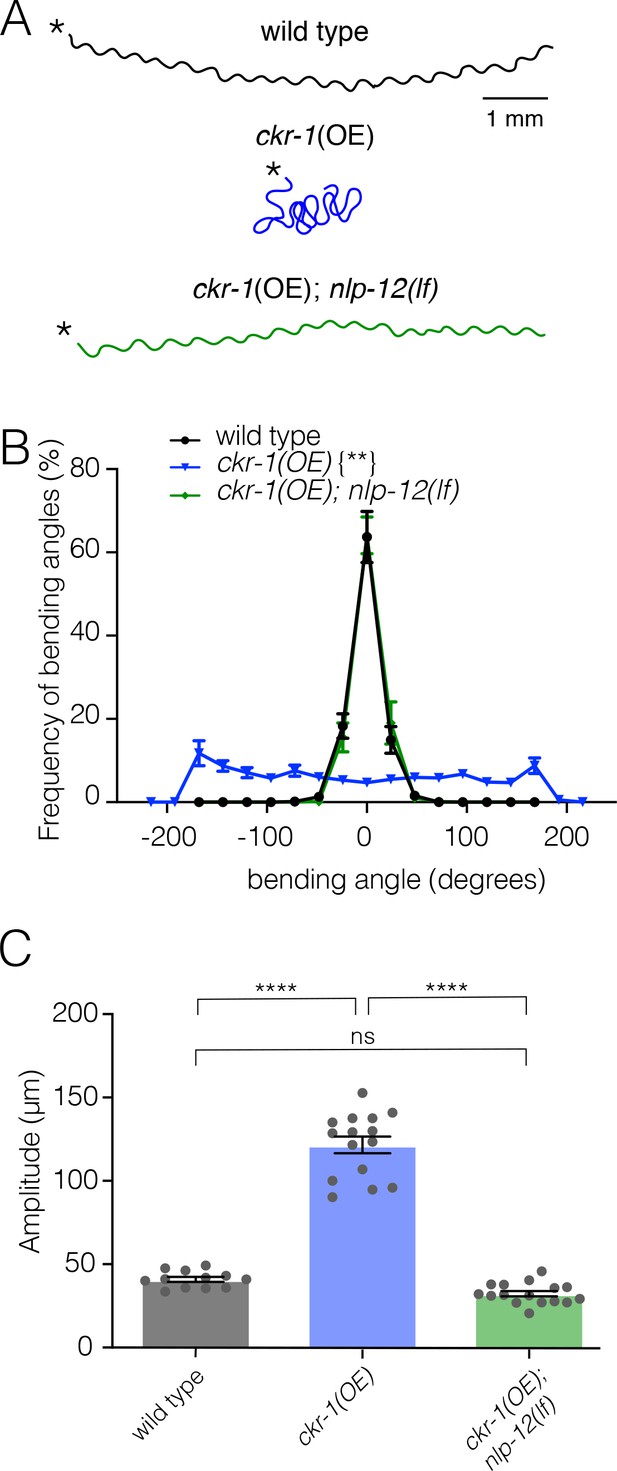
Elevated CKR-1 signaling enhances bending angle and amplitude in an nlp-12 dependent manner.
(A) Representative movement trajectories of wild-type (black), ckr-1(OE) (blue) and ckr-1(OE); nlp-12(lf) (green) animals for 30 s on NGM agar plates seeded with OP50 bacteria. ckr-1(OE) refers to high copy expression of the wild-type ckr-1 genomic locus (ufEx802). Note the increased frequency of high angle turns and convoluted track for ckr-1(OE). These movement phenotypes are reversed by nlp-12 deletion. Scale bar, 1 mm. (B) Frequency distribution of body bending angles (mean ± SEM) during forward runs (30 s) on plates thinly seeded with OP50 bacteria. Kolmogorov-Smirnov test: wild-type versus ckr-1(OE)**, ckr-1(OE) versus ckr-1(OE); nlp-12(ok335)**, wild-type versus ckr-1(OE); nlp-12(ok335) ns. **p<0.01, ns, not significant. wild-type: n=8, ckr-1(OE): n=10, and ckr-1(OE);nlp-12(lf): n=10. (C) Comparison of the average body bend amplitude for the indicated genotypes. Bars represent mean ± SEM. ****p<0.0001, ns, not significant, ANOVA with Holms-Sidak post hoc test. wild-type: n=12, ckr-1(OE): n=15, ckr-1(OE);nlp-12(ok335): n=16. NGM, nematode growth media.
-
Figure 4—source data 1
Source data for frequency of bending angles (Figure 4B).
- https://cdn.elifesciences.org/articles/71747/elife-71747-fig4-data1-v2.xlsx
-
Figure 4—source data 2
Source data for body bending amplitude (Figure 4C).
- https://cdn.elifesciences.org/articles/71747/elife-71747-fig4-data2-v2.xlsx
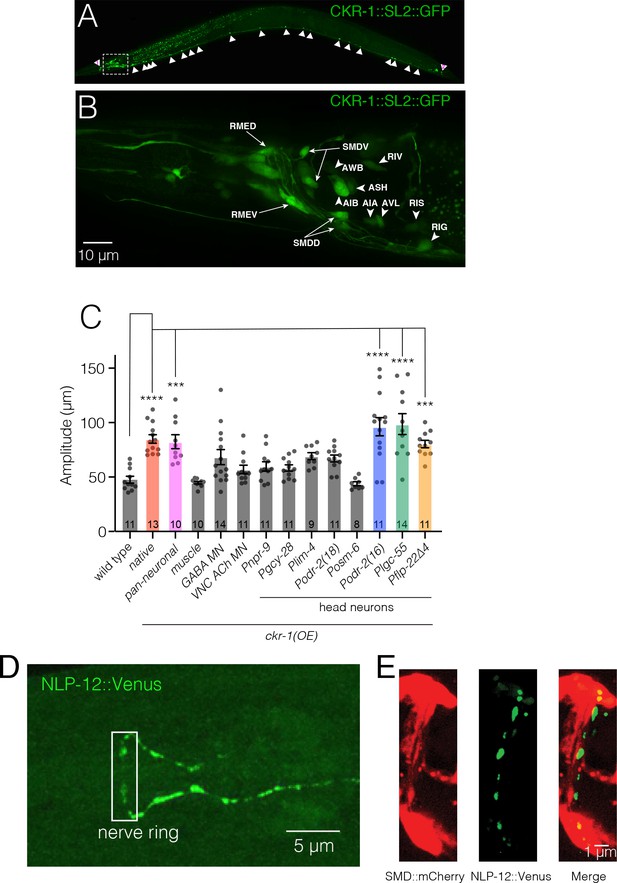
ckr-1 functions in the SMD head motor neurons to modulate body bending.
(A) Confocal maximum intensity projection of adult expressing the Pckr-1::ckr-1::SL2::GFP reporter. Note that the expression in multiple head neurons (white box) and a subset of ventral nerve cord motor neurons (white arrowheads). (B) Confocal maximum intensity projection of the head region of adult expressing the Pckr-1::ckr-1::SL2::GFP reporter. Scale bar, 10 μm. See Figure 5—figure supplement 1 and Supplementary file 2 for additional expression information. (C) Quantification of average body bend amplitudes (mean ± SEM) for ckr-1 overexpression in the indicated cell types. Promoters used for listed cell types: pan-neuronal Prgef-1, muscle Pmyo-3, GABA motor neurons Punc-47, cholinergic ventral cord motor neurons Punc-17β. See Supplementary file 3 for details about cellular expression of promoters used for head neurons. ****p<0.0001, ***p<0.001, ANOVA with Holms-Sidak’s post hoc test. Numbers within bars indicate n for each genotype. (D) Confocal maximum intensity projection of the nerve ring region of a transgenic animal expressing Pnlp-12::NLP-12::Venus. Note the high levels of NLP-12::Venus in the nerve ring. White box indicates approximate nerve ring region where close localization of NLP-12 clusters to SMD processes has been shown in panel (E). Scale bar, 5 µm. (E) Confocal maximum intensity projection of the nerve ring region of a transgenic animal expressing Pnlp-12::NLP-12::Venus (DVA) and Pflp-22∆4::mCherry (SMD). Note the close localization of NLP-12::Venus dense core vesicle clusters to the SMD process. Scale bar, 1 µm.
-
Figure 5—source data 1
Source data for body bending amplitude (Figure 5C).
- https://cdn.elifesciences.org/articles/71747/elife-71747-fig5-data1-v2.xlsx
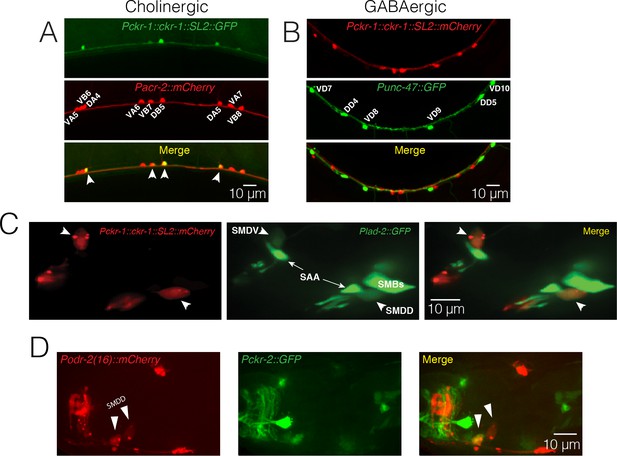
Neuronal expression of CKR-1 and CKR-2.
(A) Confocal maximum intensity projections of a segment of the ventral nerve cord of a transgenic animal co-expressing Pckr-1::ckr-1::SL2::GFP and the cholinergic reporter Pacr-2::mCherry. ckr-1 is expressed in the DA and DB motor neurons in the ventral nerve cord. Anterior is to the left in all panels. Scale bar, 10 μm. (B) Confocal maximum intensity projections of a segment of the ventral nerve cord of a transgenic animal co-expressing Pckr-1::ckr-1::SL2::mCherry and the GABAergic reporter Punc-47::GFP. (C) Confocal maximum intensity projections of optical sections with SMD fluorescence (GFP) from the head region of a transgenic animal expressing ckr-1::SL2::mCherry (left panel) together with Plad-2::GFP (middle panel). White arrowheads denote the SMD cell bodies in all cases. Note the colocalization of the red and green fluorescence exclusively in the SMD neurons (merge right panel). (D) Confocal maximum intensity projections of optical sections with SMD fluorescence (mCherry) from the head region of a transgenic animal co-expressing Podr-2(16)::mCherry (left panel), and Pckr-2::GFP (middle panel). Note weak ckr-2 expression in a single SMDD neuron (merge, right panel).
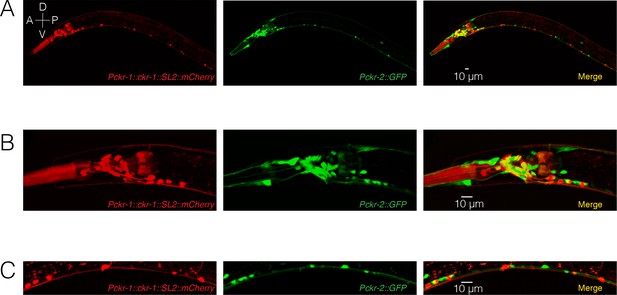
CKR-1 and CKR-2 expression are largely non-overlapping.
Confocal maximum intensity projections of transgenic worm expressing Pckr-1::ckr-1::SL2::mCherry and Pckr-2::GFP. (A) ckr-1 and ckr-2 expression in the entire worm. Both ckr-1 and ckr-2 are highly expressed in head neurons and ventral nerve cord motor neurons. However, there is very little overlap between the expression of ckr-1 and ckr-2. (B) Magnified view of ckr-1 and ckr-2 expression in the head region. (C) Magnified view of ckr-1 and ckr-2 expression in the ventral nerve cord. Scale bar, 10 µm.
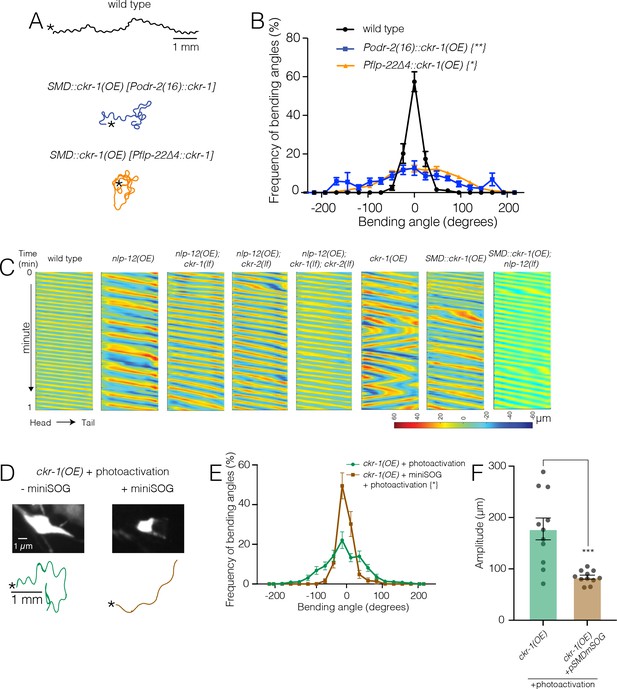
Ablation of SMD motor neurons abolishes the effects of ckr-1 overexpression.
(A) Representative tracks (1 min) for indicated genotypes. Asterisks indicate the position of animal at the beginning of recordings. Note that the increased reorientations and body bending depth in the tracks with cell-specific ckr-1 overexpression. Scale bar, 1 mm. (B) Average body bending angle distribution (mean ± SEM) for the indicated genotypes. High level expression of ckr-1 in SMDs using the odr-2(16) or flp-22∆4 promoters increases bending angle. Kolmogorov-Smirnov test: wild-type versus Podr-2(16)::ckr-1(OE)**, wild-type versus Pflp-22∆4::ckr-1(OE)*, **p<0.01, *p<0.05. wild-type n=9 (black circles), Podr-2(16)::ckr-1(OE): n=9 (blue squares), Pflp-22∆4::ckr-1(OE): n=11 (orange triangles). (C) Representative body curvature kymographs for worm locomotion during basal locomotion for indicated genotypes. Head to tail orientation along the horizontal axis in each kymograph is left to right as indicated for wild-type. Time is indicated along the vertical axis from 0 min to 1 min. (D) Top, representative fluorescent images of SMD motor neuron in ckr-1(OE) animals without (left) or with (right) miniSOG expression 16 hr following photoactivation. Bottom, representative 30 s track for control ckr-1(OE) (−miniSOG, left) animal or SMD ablated ckr-1(OE) (+miniSOG, right) animal 16 hr after photostimulation. Scale bar, 1 µm. (E) Average body bending angle distribution (mean ± SEM) for control ckr-1(OE) (green circles, n=11) and SMD ablated ckr-1(OE) (brown squares, n=11) animals. SMD ablation reduces the frequency of large bending angles produced by ckr-1(OE). Kolmogorov-Smirnov test: *p<0.05. (F) Comparison of average body bending amplitude for control ckr-1(OE) (n=11) and SMD ablated ckr-1(OE) (n=11). SMD ablation significantly reduces the enhanced body bending amplitude observed by ckr-1(OE). Bars represent mean ± SEM. ***p<0.001, Student’s t-test.
-
Figure 6—source data 1
Source data for frequency of bending angles (Figure 6B).
- https://cdn.elifesciences.org/articles/71747/elife-71747-fig6-data1-v2.xlsx
-
Figure 6—source data 2
Source data for frequency of bending angles (Figure 6E).
- https://cdn.elifesciences.org/articles/71747/elife-71747-fig6-data2-v2.xlsx
-
Figure 6—source data 3
Source data for bending amplitude (Figure 6F).
- https://cdn.elifesciences.org/articles/71747/elife-71747-fig6-data3-v2.xlsx
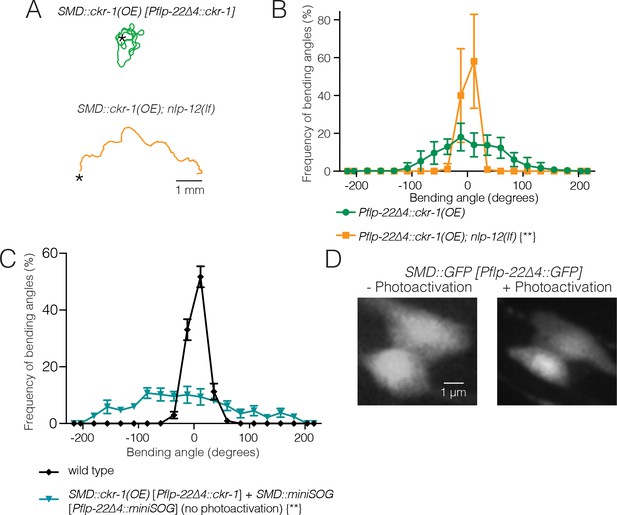
Effects of ckr-1(OE) are dependent on NLP-12 and miniSOG expression alone does not alter SMD morphology or behavior.
(A) Representative tracks (30 s) for transgenic animals with high levels of cell-specific ckr-1 overexpression (Pflp-22∆4::ckr-1) in wild-type (top) or nlp-12 deletion background (bottom). Asterisks indicate the position of animals at the beginning of recording. Scale bar, 1 mm. (B) Average bending angle distribution (mean ± SEM) for SMD-specific ckr-1(OE) in wild-type (green circles) or nlp-12(lf) background (orange squares). n=8 for each group. Kolmogorov-Smirnov test. **p<0.01. (C) Average body bending angle distribution (mean ± SEM) for pSMD::ckr-1(OE) animals expressing miniSOG in SMDs (Pflp-22∆4::miniSOG), but not subjected to photoactivation (control, blue triangles) compared to wild type (black diamonds). n=7 for each group. Kolmogorov-Smirnov test. **p<0.01. (D) Single confocal slices of GFP-labeled SMD neurons, following photoactivation (right) compared to control (-photoactivation, left), in transgenic animals without miniSOG expression. Photoactivation protocol does not alter SMD neuron morphology in the absence of miniSOG expression. Scale bar, 1 µm.
-
Figure 6—figure supplement 1—source data 1
Source data for frequency of bending angles (Figure 6—figure supplement 1B).
- https://cdn.elifesciences.org/articles/71747/elife-71747-fig6-figsupp1-data1-v2.xlsx
-
Figure 6—figure supplement 1—source data 2
Source data for frequency of bending angles (Figure 6—figure supplement 1C).
- https://cdn.elifesciences.org/articles/71747/elife-71747-fig6-figsupp1-data2-v2.xlsx
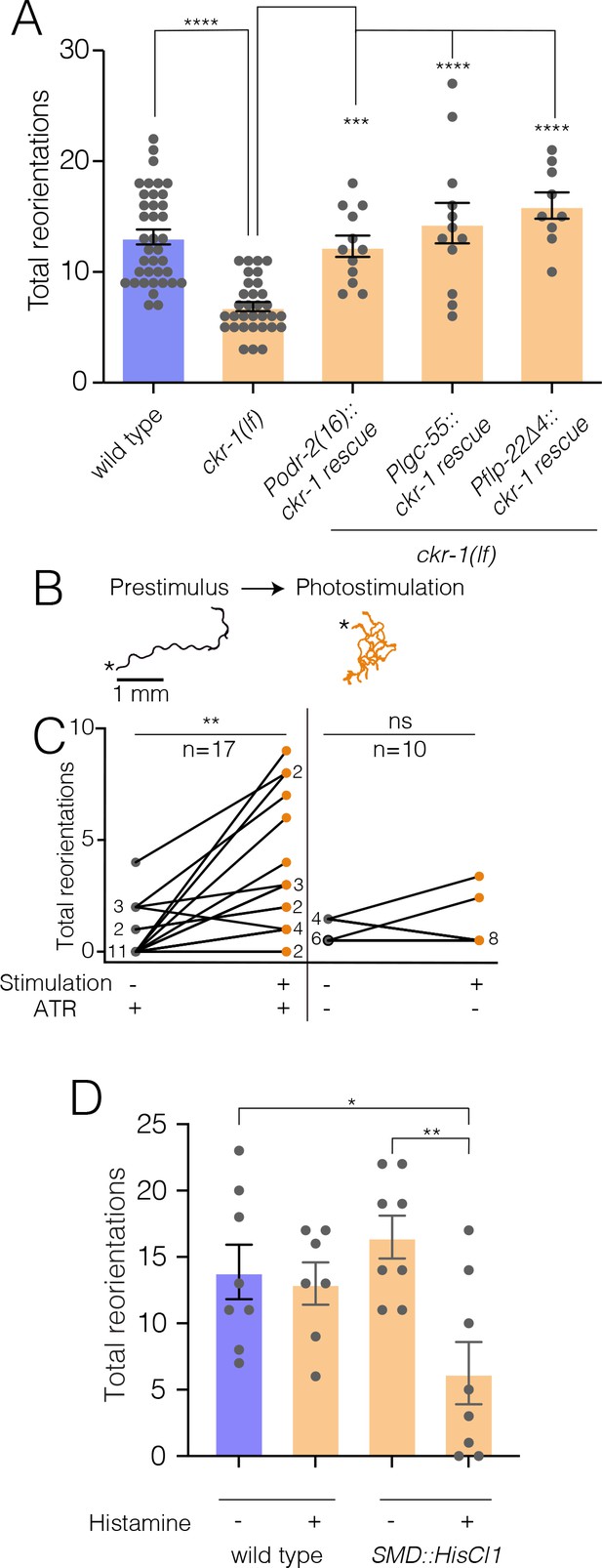
NLP-12/CKR-1 excitation of the SMD neurons promotes reorientations.
Total reorientations measured during 0–5 min following removal from food for the genotypes indicated. ckr-1 rescue refers to expression of wild-type ckr-1 (5 ng/µl) in ckr-1(ok2502) animals using the indicated promoters. Bars represent mean ± SEM. ****p<0.0001, ***p<0.001 ANOVA with Holms-Sidak post hoc test. wild-type: n=38, ckr-1(lf): n=32, Podr-2(16)::ckr-1 rescue: n=12, Plgc-55::ckr-1 rescue: n=12, Pflp-22(∆4)::ckr-1 rescue: n=9. (B) Representative tracks (1 min) on thinly seeded NGM agar plates prior to (left) and during photostimulation (right) for transgenic animals expressing Podr-2(16)::Chrimson. Scale bar, 1 mm. Asterisks (*) indicate the position of worm at the start of recording. (C) Left, quantification of reorientations for individual animals over 1 min durations prior to (prestimulus) and during photostimulation (+ATR). Right, quantification of reorientations for individual animals prior to and during photostimulation in control animals (−ATR). Black circles, reorientations during prestimulus. Orange circles, reorientations during photostimulation. Numbers adjacent to circles indicate number of overlapping data points. **p<0.01, ns, not significant. Paired t-test. ATR, all-trans retinal. (D) Quantification of reorientations for wild-type and transgenic animals, (Pflp-22∆4::His-Cl1::SL2::GFP), in the presence and absence of histamine. Note reduced reorientations with SMD silencing in transgenics (+histamine). **p<0.01, *p<0.05, ANOVA with Holms-Sidak post hoc test. wild-type: −Histamine: n=8, +Histamine: n=7, pSMD::HisCl1::SL2::GFP: −Histamine: n=8, +Histamine: n=8. NGM, nematode growth media.
-
Figure 7—source data 1
Source data for reorientations quantified during area restricted search (0–5 min off food, Figure 7A).
- https://cdn.elifesciences.org/articles/71747/elife-71747-fig7-data1-v2.xlsx
-
Figure 7—source data 2
Source data for reorientations quantified during SMD photostimulation (Figure 7C).
- https://cdn.elifesciences.org/articles/71747/elife-71747-fig7-data2-v2.xlsx
-
Figure 7—source data 3
Source data for reorientations quantified during area restricted search upon SMD silencing (0–5 min off food, Figure 7D).
- https://cdn.elifesciences.org/articles/71747/elife-71747-fig7-data3-v2.xlsx

SMD activation modestly impacts body bending.
(A) Average body bending angle distribution (mean ± SEM) plotted for wild-type control animals (soid black circles, n=8) and Pflp-22∆4::ckr-1 (solid orange squares, n=8). Low level (5 ng/µl) cell-specific expression of ckr-1 in SMDs in wild type did not alter body bending. Kolmogorov-Smirnov test not significant. (B) Photostimulation of SMDs modestly increases body bending amplitude. **p<0.01, paired Student’s t-test. Black circles, reorientations during prestimulus. Orange circles, reorientations during photostimulation.
-
Figure 7—figure supplement 1—source data 1
Source data for frequency of bending angles (Figure 7—figure supplement 1A).
- https://cdn.elifesciences.org/articles/71747/elife-71747-fig7-figsupp1-data1-v2.xlsx
-
Figure 7—figure supplement 1—source data 2
Source data for body bending amplitude quantified during SMD photostimulation (Figure 7—figure supplement 1B).
- https://cdn.elifesciences.org/articles/71747/elife-71747-fig7-figsupp1-data2-v2.xlsx
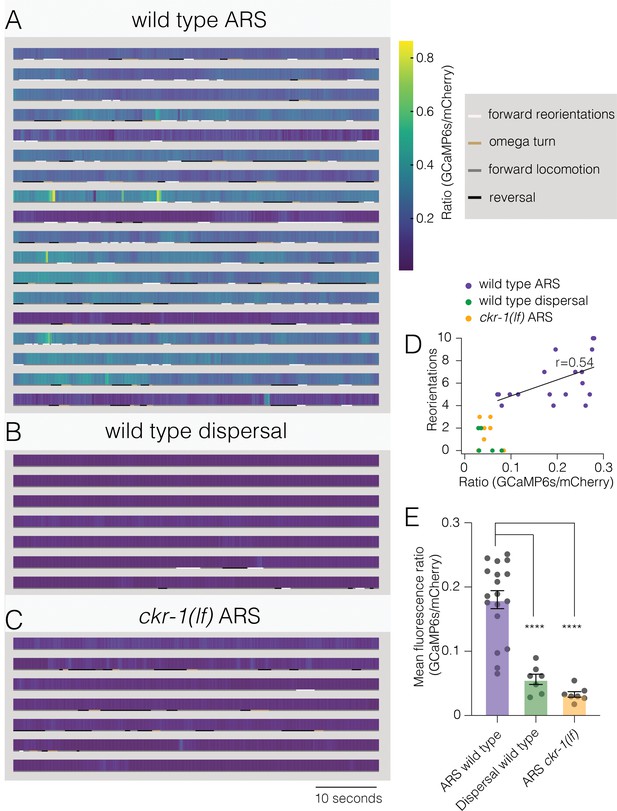
Elevated activity in SMD motor neurons during ARS promotes reorientations.
(A–C) Representative heat maps showing activity of SMD neurons in transgenic animals (Pflp-22∆4::GCaMP6s::SL2::mCherry) during ARS (A) and dispersal (B) for wild type, and ARS for ckr-1(ok2502) (C). Each row represents one animal over a duration of 1 min. Corresponding behaviors (forward, reversal, omega turn, forward reorientation) are annotated by color-coded (as indicated in legend) horizontal bar below each heat map. The SMD GCaMP6s/mCherry fluorescence ratio is elevated during wild-type ARS, compared with either ckr-1(lf) ARS, and wild-type dispersal. (D) Number of reorientations plotted against mean SMD GCaMP6s/mCherry ratio for the individuals in (A–C). Black line indicates linear fit for wild-type ARS values, with Pearson’s correlation coefficient (r), *p=0.02. (E) Quantification of mean SMD fluorescence ratio (GCaMP6s/mCherry) during ARS or dispersal for the genotypes indicated. ****p<0.0001, ANOVA with Holms-Sidak post hoc test. ARS wild-type: n=18, ARS ckr-1(ok2502): n=7, Dispersal wild-type: n=7. ARS, area-restricted searching.
-
Figure 8—source data 1
Source data for GCaMP6s/mCherry ratio during SMD calcium imaging (Figure 8A–D).
- https://cdn.elifesciences.org/articles/71747/elife-71747-fig8-data1-v2.xlsx
-
Figure 8—source data 2
Source data for mean GCaMP6s/mCherry ratio during SMD calcium imaging (Figure 8E).
- https://cdn.elifesciences.org/articles/71747/elife-71747-fig8-data2-v2.xlsx

Representative calcium signals (GCaMP6s/mCherry ratio) for wild-type ARS, wild-type dispersal, and ck-1(lf) ARS.
Corresponding behaviors are annotated by shading as indicated.
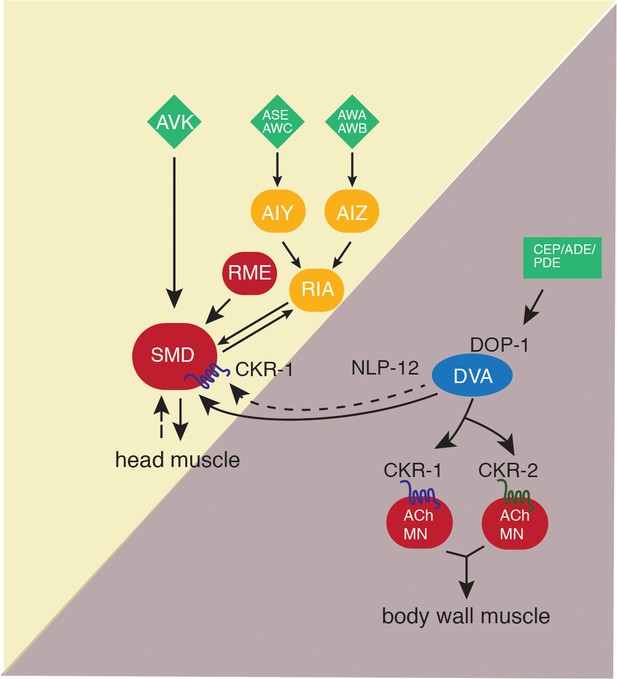
Proposed model for NLP-12 action through CKR-1 and CKR-2.
During basal locomotion, NLP-12 activation of CKR-1 and CKR2 GPCRs in ventral nerve cord motor neurons regulates body bending. During local searching, NLP-12 acts primarily through CKR-1 in SMD motor neurons to promote increased turning, trajectory changes and enhance body bending. Solid arrows indicate known synaptic connections, dotted arrows indicate extrasynaptic. Sensory neurons (green), head interneurons (orange), and motor neurons (red). Olfactory sensory neurons: AWA, AWB, AWC, and ASE.
Videos
Representative 20-s video showing locomotion on food of animal overexpressing nlp-12.
Video has been sped up 4×.
Representative 20-s video showing locomotion of wild-type animal during area restricted search (0–5 min off food).
Video has been sped up 4×.
Representative 20-s video showing locomotion of wild-type animal during dispersal (30–35 mi off food).
Video has been sped up 4×.
Representative 20-s video showing locomotion on food of animal overexpressing ckr-1.
Video has been sped up 4×.
Representative 20-s video showing locomotion on food of animal overexpressing ckr-1 in the SMD motor neurons.
Video has been sped up 4×.
Representative 20-s video showing locomotion on food of animal in the absence (left) and during SMD photostimulation (right).
Video has been sped up 4×.
Representative 20-s video showing simultaneous post hoc tracking of mCherry and GCaMP6s fluorescence for ratiometric calcium imaging analysis.
Video has been sped up 4×.
Representative 20-s video showing tracking locomotion of animal overexpressing nlp-12 in WormLab to analyze body bending.
Video has been sped up 4×.
Representative 20-s video showing single worm tracking of wild-type animal during basal locomotion on food to analyze body bending and head bending.
Video has been sped up 4×.
Additional files
-
Supplementary file 1
Stains generated/used in this work.
- https://cdn.elifesciences.org/articles/71747/elife-71747-supp1-v2.docx
-
Supplementary file 2
Identification (method of ID, marker and strain indicated for each neuron) to determine ckr-1 expressing neurons.
* Indicated strains were crossed into ufIs141 (Pckr-1::ckr-1::SL2::GFP) to generate strains to determine colocalization. #+ or – indicates presence or absence of ckr-1 expression in identified neuron. * Indicated strains were crossed into ufIs141 to generate strains to determine colocalization, #+ indicates ckr-1 expression, - indicates absence.
- https://cdn.elifesciences.org/articles/71747/elife-71747-supp2-v2.docx
-
Supplementary file 3
Promoters used in ckr-1(OE) screen (Figure 5C) indicating expression pattern.
**Bold indicates neurons where ckr-1 is expressed.
- https://cdn.elifesciences.org/articles/71747/elife-71747-supp3-v2.docx
-
Supplementary file 4
Plasmid constructs used in cell specific ckr-1(OE) screen or cell-specific rescue (Figures 5C and 7A).
For cell specific overexpression or rescue of ckr-1, ckr-1 minigene was expressed under indicated promoters. Entry vectors containing promoters recombined with destination vectors pRB12 or pRB13 for cell-specific overexpression or rescue of ckr-1.
- https://cdn.elifesciences.org/articles/71747/elife-71747-supp4-v2.docx
-
Supplementary file 5
Promoter lengths and primer information for promoters used.
- https://cdn.elifesciences.org/articles/71747/elife-71747-supp5-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/71747/elife-71747-transrepform1-v2.pdf





