Regulation of positive and negative selection and TCR signaling during thymic T cell development by capicua
Figures
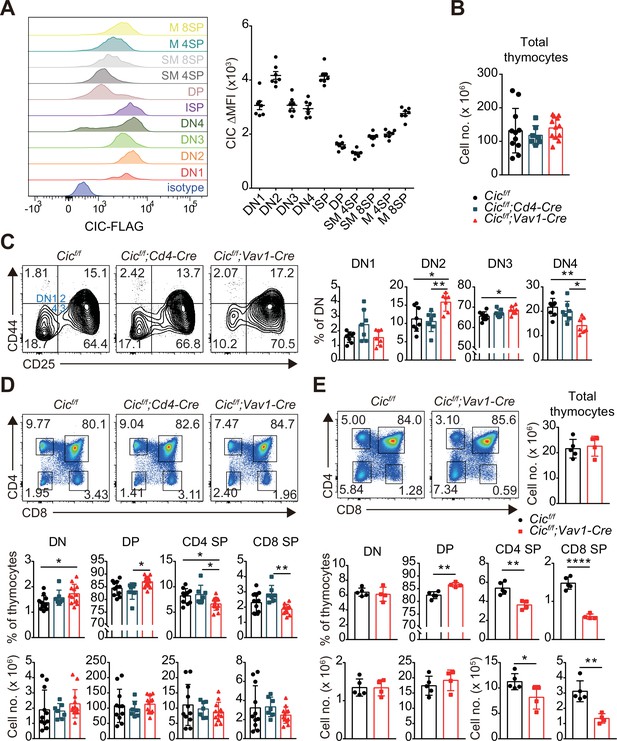
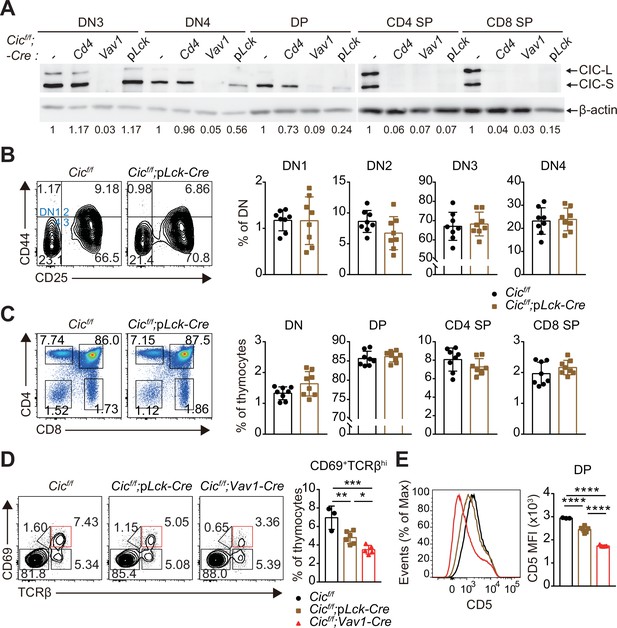
Altered T cell development in Cicf/f;Vav1-Cre mice.
(A) Capicua (CIC) protein levels in thymic T cell subsets. Thymocytes of CicFLAG/FLAG mice (N = 7) were subjected to flow cytometry using anti-FLAG antibody. Representative histograms of CIC-FLAG expression are shown in the left panel for each cell population. The difference in mean fluorescence intensity (ΔMFI) of the CIC-FLAG signal was calculated by subtraction of the MFI value of the isotype control from that obtained by anti-FLAG antibody staining. DN1: CD4-CD8-CD44hiCD25lo, DN2: CD4-CD8-CD44hiCD25hi, DN3: CD4-CD8-CD44loCD25hi, DN4: CD4-CD8-CD44loCD25lo, ISP: immature CD8+ single positive cells (CD4-CD8+TCRβloCD24hi), DP: CD4+CD8+, SM: semi-mature (CD69+TCRβhi), and M: mature (CD69-TCRβhi). Lineage (CD11b, CD11c, CD19, NK1.1, Gr-1, γδTCR, and TER119)-negative (Lin-)-gated cells were analyzed for DN cell populations. (B–D) Flow cytometric analysis of thymocytes from 7-week-old Cicf/f, Cicf/f;Cd4-Cre, and Cicf/f;Vav1-Cre mice. (B) Total numbers of thymocytes for each genotype. N = 11, 7, and 12 for Cicf/f, Cicf/f;Cd4-Cre, and Cicf/f;Vav1-Cre mice, respectively. (C) Proportions of DN1-4 subsets for each genotype. N = 8, 7, and seven for Cicf/f, Cicf/f;Cd4-Cre, and Cicf/f;Vav1-Cre mice, respectively. Lin--gated cells were used for analysis of DN cell populations. (D) Frequencies and numbers of DN, DP, CD4+ SP, and CD8+ SP cells for each genotype. N = 11, 7, and 12 for Cicf/f, Cicf/f;Cd4-Cre, and Cicf/f;Vav1-Cre mice, respectively. (E) Flow cytometric analysis of thymocytes isolated from 1-week-old Cicf/f and Cicf/f;Vav1-Cre mice using CD4 and CD8 markers. Total thymocyte numbers, and the frequencies and numbers of DN, DP, CD4+ SP, and CD8+ SP subsets in mice of each genotype, as well as representative plots are presented. N = 5 and 4 for Cicf/f and Cicf/f;Vav1-Cre mice, respectively. Data are representative of two independent experiments. Bar graphs represent the mean and SEM. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. One-way ANOVA with Tukey’s multiple comparison test (B–D) and unpaired two-tailed Student’s t-test (E) were used to calculate the corresponding p values. See also Figure 1—source data 1.
-
Figure 1—source data 1
Raw data for Figure 1.
- https://cdn.elifesciences.org/articles/71769/elife-71769-fig1-data1-v2.xlsx
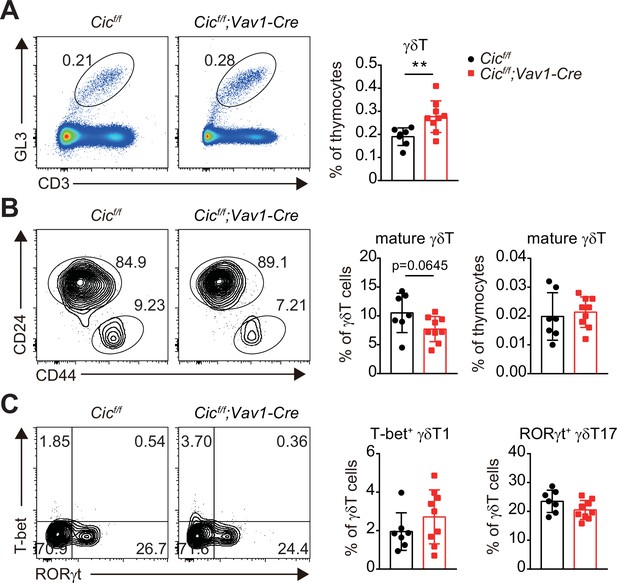
Analysis of thymic γδT cells in Cicf/f;Vav1-Cre mice.
(A) Flow cytometric analysis of γδT cells in the thymus of 7-week-old Cicf/f and Cicf/f;Vav1-Cre mice. Representative plots (left) and the frequency of total γδT cells (right) are shown. GL3: γδTCR. (B) Flow cytometric analysis of γδT cells using CD24 and CD44 markers. Frequencies of mature CD24loCD44hi γδT cells in total thymic γδT cells (middle) and total thymocytes (right) are shown. (C) Flow cytometric anaylsis of γδT cells for T-bet and RORγt expression. Frequencies of thymic T-bet+ type 1 (γδT1) and RORγt+ type 17 γδT (γδT17) cells are presented. N = 7 and 9 for Cicf/f and Cicf/f;Vav1-Cre mice, respectively. Data from two independent experiments were pooled. Bar graphs represent the mean and SEM. **p < 0.01. Unpaired two-tailed Student’s t-test was used to calculate the corresponding p values.
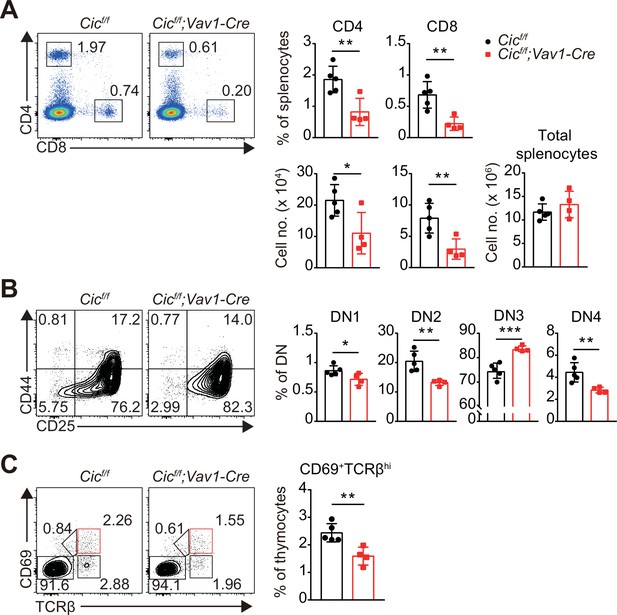
Analysis of splenic and thymic T cell subsets in 1-week-old Cicf/f;Vav1-Cre mice.
(A) Flow cytometric analysis of splenocytes from 1-week-old Cicf/f and Cicf/f;Vav1-Cre mice. Representative plots of CD4 and CD8 expression in splenocytes, frequency and number of CD4+ and CD8+ T cells, and total splenocyte numbers are shown. (B and C) Flow cytometric analysis of thymocytes from 1-week-old Cicf/f and Cicf/f;Vav1-Cre mice for proportions of (B) double-negative 1–4 (DN1-4) subsets and (C) CD69+TCRβhi cells. Lin--gated cells were analyzed to determine the frequency of DN subsets. N = 5 and 4 for Cicf/f and Cicf/f;Vav1-Cre mice, respectively. Data from two independent experiments are shown. Bar graphs represent the mean and SEM. *p < 0.05, **p < 0.01, and ***p < 0.001. Unpaired two-tailed Student’s t-test was used to calculate the corresponding p values.
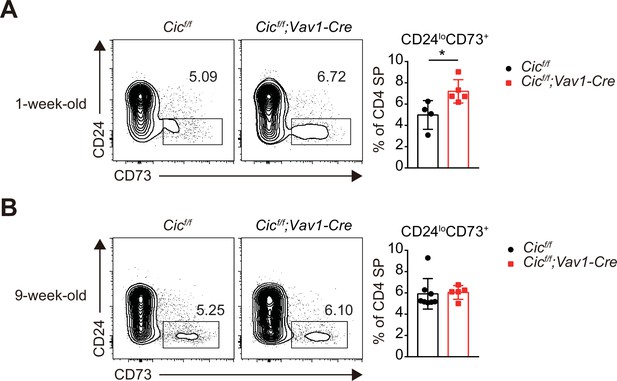
Flow cytometric analysis of recirculating CD4+ T cells in the thymus of Cicf/f;Vav1-Cre mice.
(A and B) Thymocytes from (A) 1-week-old Cicf/f (N = 4) and Cicf/f;Vav1-Cre (N = 5) and (B) 9-week-old Cicf/f (N = 8) and Cicf/f;Vav1-Cre (N = 5) mice were analyzed for recirculating CD4+ single-positive (SP) cells using CD24 and CD73 markers. Representative plots (left) and the frequency of CD24loCD73+CD4+ SP cells (right) are shown. Data are representative of two independent experiments. Bar graphs represent the mean and SEM. *p < 0.05. Unpaired two-tailed Student’s t-test was used to calculate the corresponding p values.
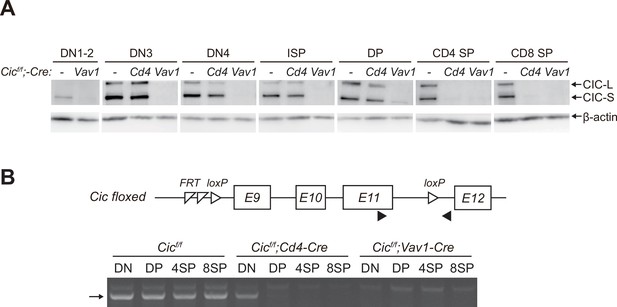
Stable capicua (CIC) expression in double-positive (DP) cells of Cicf/f;Cd4-Cre mice.
(A) Western blotting for detection of CIC levels in double-negative 1–2 (DN1-2), DN3, DN4, immature CD8+ single-positive (ISP), double-positive (DP), CD4+ SP, and CD8+ SP cells from Cicf/f, Cicf/f;Cd4-Cre, and Cicf/f;Vav1-Cre mice. Lin--gated DN1-2, DN3, DN4, ISP, DP, CD4+ SP, and CD8+ SP (TCRβhi) cells were sorted from mice of each genotype. (B) PCR analysis of Cic knock-out efficiency in Lin--gated DN, DP, CD4+ SP, and CD8+ SP (TCRβhi) cells from Cicf/f, Cicf/f;Cd4-Cre, and Cicf/f;Vav1-Cre mice. Genomic DNA was extracted from sorted DN, DP, CD4+ SP, and CD8+ SP cells and subjected to PCR amplification of a part of the floxed Cic allele. Upper panel, schematic of the Cic floxed allele. Arrowheads indicate the primers used for PCR. Lower panel, representative agarose gel image of PCR products. The arrow indicates the PCR products corresponding to the amplified part of the floxed Cic allele. See also Figure 2—source data 1.
-
Figure 2—source data 1
Original and labelled files for western blot and PCR gel images.
- https://cdn.elifesciences.org/articles/71769/elife-71769-fig2-data1-v2.zip
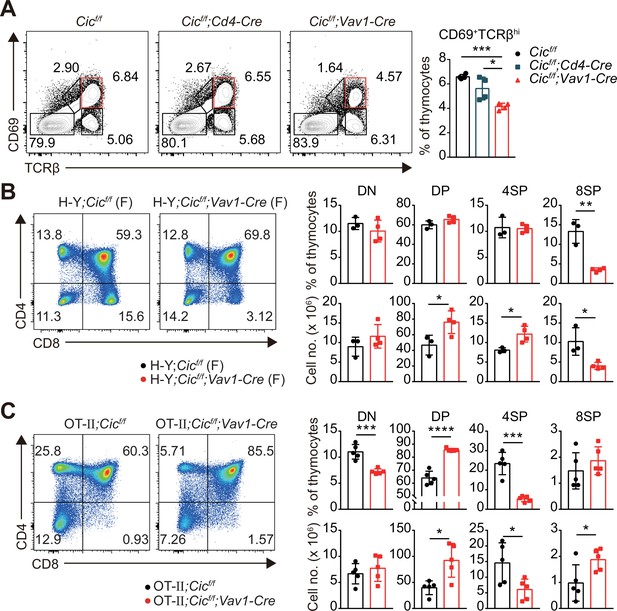
Defective positive selection in the absence of capicua (CIC).
-
Figure 3—source data 1
Raw data for Figure 3.
- https://cdn.elifesciences.org/articles/71769/elife-71769-fig3-data1-v2.xlsx
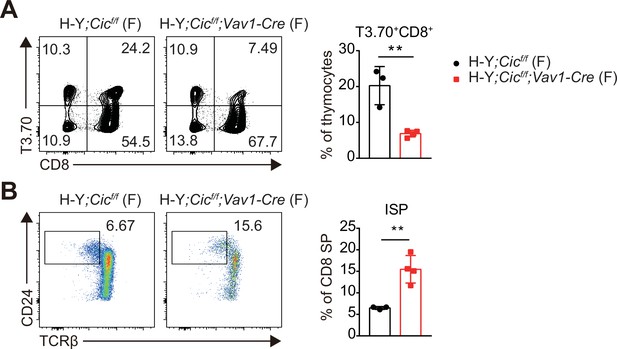
Analysis of H-Y TCR transgenic CD8+ T and immature CD8+ single positive (ISP) cells in female H-Y mice.
(A) Flow cytometric analysis of thymocytes from female H-Y;Cicf/f (N = 3) and H-Y;Cicf/f;Vav1-Cre (N = 4) mice for H-Y TCR (T3.70) and CD8 expression. Frequencies of H-Y TCR+ CD8+ cells among total thymocytes are presented. (B) Flow cytometric analysis of thymocytes from female H-Y;Cicf/f (N = 3) and H-Y;Cicf/f;Vav1-Cre (N = 4) mice for ISP cell populations. Frequencies of CD24hiTCRβlo ISP cells among CD8+ SP cells are presented. Data are representative of two independent experiments. Bar graphs represent the mean and SEM. **p < 0.01. Unpaired two-tailed Student’s t-test was used to calculate the corresponding p values.
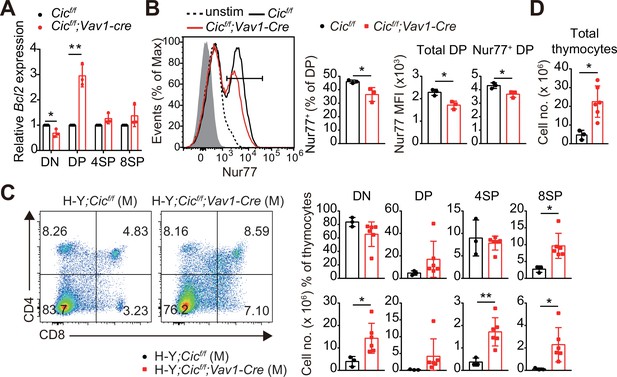
Defective negative selection in the absence of capicua (CIC).
(A) qRT-PCR quantification of Bcl2 expression in double-negative (DN), double-positive (DP), CD4+ single-positive (4SP), and CD8+ SP (8SP, TCRβhi) cells of Cicf/f and Cicf/f;Vav1-Cre mice. N = 3 for each group. (B) Flow cytometric analysis of Nur77 expression in DP thymocytes from Cicf/f and Cicf/f;Vav1-Cre mice. Freshly isolated thymocytes were treated with plate-coated anti-CD3 (5 μg/ml) and anti-CD28 (10 μg/ml) antibodies for 2 hr and subsequently subjected to flow cytometry. Representative histograms for Nur77 expression in DP thymocytes of Cicf/f (black line) and Cicf/f;Vav1-Cre (red line) mice overlaid with isotype (gray shaded) and unstimulated (dotted line) control histograms (left), the frequency of Nur77+ DP cells (middle), and Nur77-derived mean fluorescence intensities (MFIs) of total and Nur77+ DP cells (right) are presented. N = 3 for each genotype. Data are representative of two independent experiments. (C and D) Thymocytes from male H-Y;Cicf/f (N = 3) and H-Y;Cicf/f;Vav1-Cre (N = 6) mice were analyzed for CD4 and CD8 expression. (C) Representative flow cytometry plots (left) and frequencies (top, right) and numbers (bottom, right) of DN, DP, CD4+ SP (4SP), and CD8+ SP (8SP) cells are shown. (D) Total thymocyte numbers. Data are representative of two independent experiments. Bar graphs represent the mean and SEM. *p < 0.05 and **p < 0.01. Unpaired two-tailed Student’s t-test was used to calculate the corresponding p values. See also Figure 4—source data 1.
-
Figure 4—source data 1
Raw data for Figure 4.
- https://cdn.elifesciences.org/articles/71769/elife-71769-fig4-data1-v2.xlsx
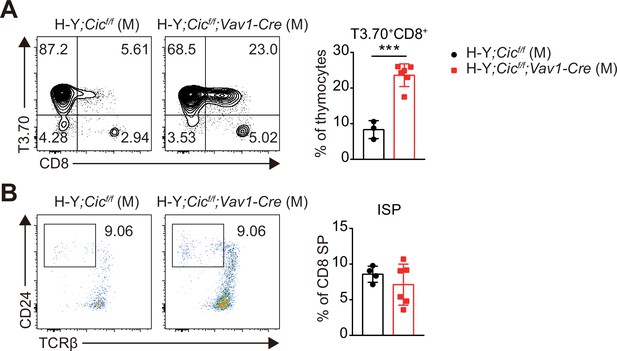
Analysis of H-Y TCR transgenic CD8+ T and immature CD8+ single positive (ISP) cells in male H-Y mice.
(A) Flow cytometric analysis of thymocytes from male H-Y;Cicf/f (N = 3) and H-Y;Cicf/f;Vav1-Cre (N = 6) mice for H-Y TCR (T3.70) and CD8 expression. Frequencies of H-Y TCR+ CD8+ cells among total thymocytes are presented. (B) Flow cytometric analysis of thymocytes from male H-Y;Cicf/f (N = 3) and H-Y;Cicf/f;Vav1-Cre (N = 6) mice for ISP cell populations. Frequencies of CD24hiTCRβlo ISP cells among CD8+ SP cells are presented. Data are representative of two independent experiments. Bar graphs represent the mean and SEM. ***p < 0.001. Unpaired two-tailed Student’s t-test was used to calculate the corresponding p values.
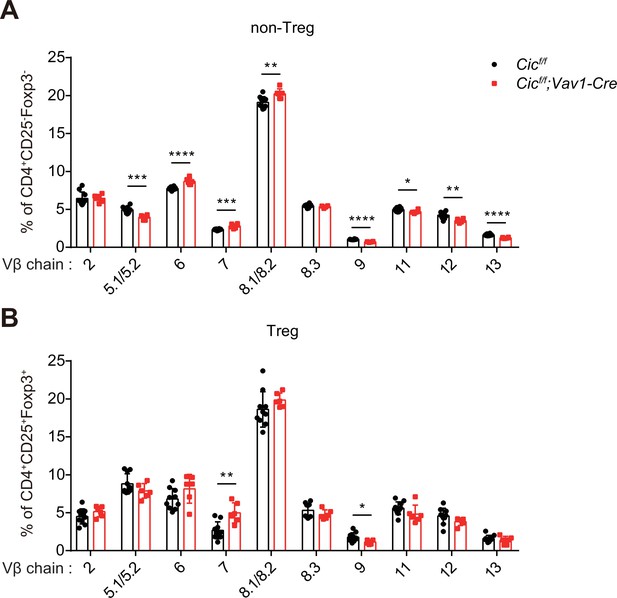
Analysis of TCR Vβ chains in CIC-deficient CD4+ SP thymocytes.
(A and B) Thymocytes from + –9 week-old Cicf/f and Cicf/f;Vav1-Cre mice were analyzed for the expression of 10 different types of TCR Vβ chains by flow cytometry. (A) CD4+CD25-Foxp3- non-Treg and (B) CD4+CD25+Foxp3+ Treg cells were gated; and cells expressing each TCR Vβ chain were analyzed. N = 10 and 6 for Cicf/f and Cicf/f;Vav1-Cre, respectively. Bar graphs represent the mean and SEM. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. Unpaired two-tailed Student’s t-test was used to calculate the corresponding p values.
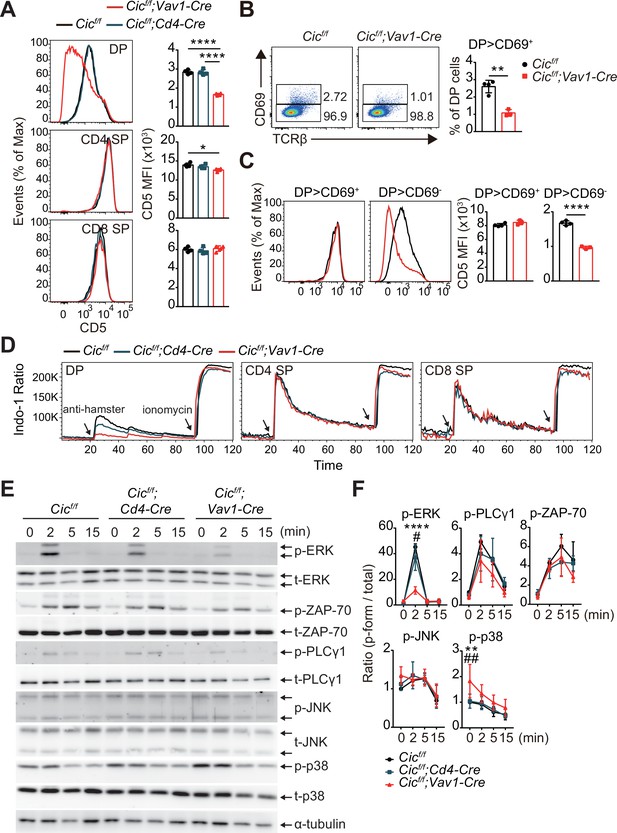
Attenuated TCR signaling in capicua (CIC)-deficient double-positive (DP) thymocytes.
(A) Thymocytes from 7-week-old Cicf/f, Cicf/f;Cd4-Cre, and Cicf/f;Vav1-Cre mice were FACS-gated into the DP, CD4+ single-positive (SP), and CD8+ SP (TCRβhi) cell population, and the CD5 mean fluorescence intensity (MFI) was measured. Representative histograms of CD5 expression in each cell population (left) and calculated CD5 MFI values (right) are shown (N = 4 per group). (B and C) DP cells from Cicf/f (N = 4) and Cicf/f;Vav1-Cre (N = 3) mice were analyzed for CD69 and CD5 expression. (B) Representative flow cytometry plots (left) and frequencies (right) of CD69+ DP cells are shown. (C) Representative histograms of CD5 expression in CD69+ or CD69- DP cells (left) and CD5 MFI values in each cell population (right) are shown. (D) TCR stimulation-induced Ca2+ influx in DP, CD4+ SP, and CD8+ SP (TCRβhi) thymocytes from 7-week-old Cicf/f, Cicf/f;Cd4-Cre, and Cicf/f;Vav1-Cre mice. Data are representative of at least three independent experiments. (E and F) Western blot analysis of TCR cascade component activation in DP thymocytes from 7-week-old Cicf/f, Cicf/f;Cd4-Cre, and Cicf/f;Vav1-Cre mice. Sorted DP cells were stimulated with soluble anti-CD3 and anti-CD4 antibodies for the times indicated. (E) Representative western blot images are shown. (F) Signal densities were measured using ImageJ software and are presented as ratios of phosphorylated to total forms for each protein (N = 3). Graphs represent the mean and SEM. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. #p < 0.05 and ##p < 0.01 (comparison between Cicf/f;Cd4-Cre and Cicf/f;Vav1-Cre mice). One-way (A) or two-way (F) ANOVA with Tukey’s multiple comparison test and unpaired two-tailed Student’s t-test (B and C) were used to calculate the corresponding p values. See also Figure 5—source data 1, Figure 5—source data 2.
-
Figure 5—source data 1
Raw data for Figure 5.
- https://cdn.elifesciences.org/articles/71769/elife-71769-fig5-data1-v2.xlsx
-
Figure 5—source data 2
Original and labeled files for western blot images.
- https://cdn.elifesciences.org/articles/71769/elife-71769-fig5-data2-v2.zip
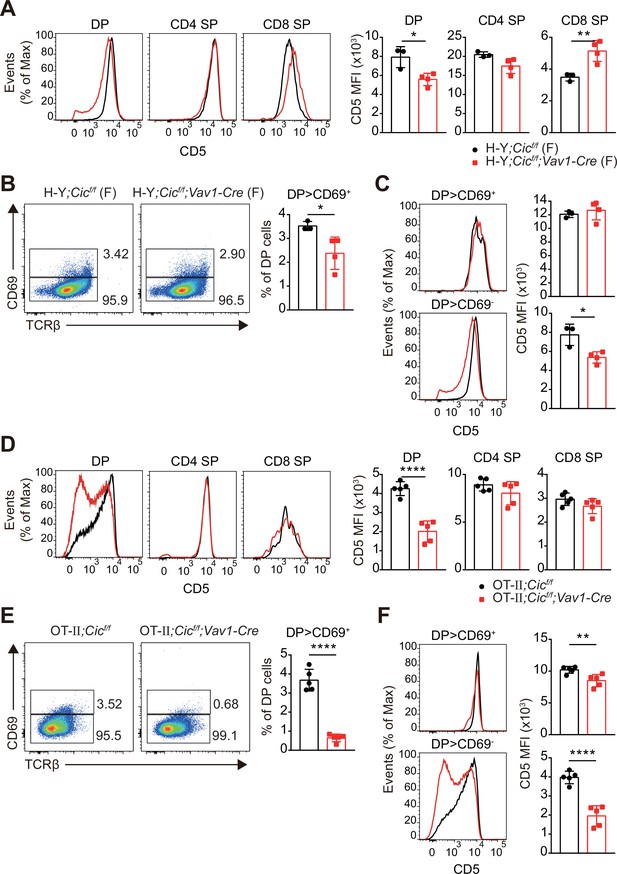
Analysis of CD5 expression in TCR transgenic thymocytes.
Thymocytes from (A–C) female H-Y;Cicf/f (N = 3) and H-Y;Cicf/f;Vav1-Cre (N = 4) mice and (D–F) OT-II;Cicf/f (N = 5) and OT-II;Cicf/f;Vav1-Cre (N = 5) mice were analyzed for CD69 and CD5 expression. (A and D) Representative histograms of CD5 expression in double-positive (DP), CD4+ single-positive (SP), and CD8+ SP (TCRβhi) cells (left), and CD5 mean fluorescence intensities (MFIs) of each cell population (right) are shown. (B and E) Representative flow cytometry plots (left) and frequencies of CD69+ cells among DP cells (right) are shown. (C and F) Representative histograms of CD5 expression in CD69+-gated (top) or CD69--gated (bottom) DP cells are shown. CD5 MFI was calculated for each gated cell population (right). Bar graphs represent the mean and SEM. *p < 0.05, **p < 0.01, and ****p < 0.0001. Unpaired two-tailed Student’s t-tests were used to calculate p values.
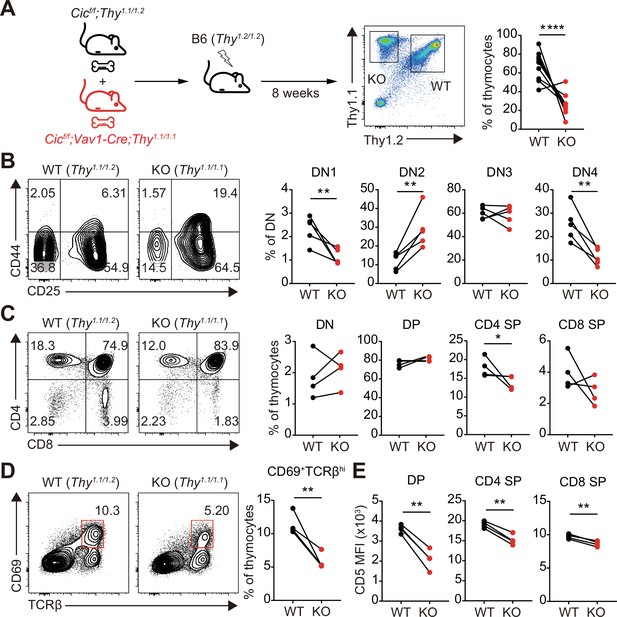
Altered T cell development and TCR intensity in Cicf/f;Vav1-Cre mice are caused by CIC loss in T cells.
(A) Schematic of the generation and analysis of mixed bone marrow (BM) chimeric mice. Equal numbers of BM cells from Cicf/f;Thy1.1/1.2 (WT) and Cicf/f;Vav1-Cre;Thy1.1/1.1 (KO) mice were mixed and transferred to irradiated B6 (Thy1.2/1.2) recipient mice. Representative FACS plot showing the thymocytes of different origin (left) and their frequencies (right) are presented (N = 10). (B–D) Flow cytometric analysis of thymocytes from mixed BM chimeras (N = 4) for the frequencies of (B) double-negative (DN) subsets based on CD44 and CD25 expression, (C) DN, double-positive (DP), CD4+ single-positive (SP), and CD8+ SP cells, and (D) post-positive selection subsets (CD69+TCRβhi). The CD69+TCRβhi cell population is highlighted by the red box in the flow cytometry plots in (D). (E) Flow cytometric analysis of surface expression levels of CD5 in DP, CD4+ SP, and CD8+ SP (TCRβhi) thymocytes derived from WT and KO BM cells in the same BM chimeric mice (N = 4). Data are representative of two independent experiments. Graphs represent the mean and SEM. *p < 0.05, **p < 0.01, and ****p < 0.0001. Unpaired two-tailed Student’s t-test was used to calculate the corresponding p values. See also Figure 6—source data 1.
-
Figure 6—source data 1
Raw data for Figure 6.
- https://cdn.elifesciences.org/articles/71769/elife-71769-fig6-data1-v2.xlsx
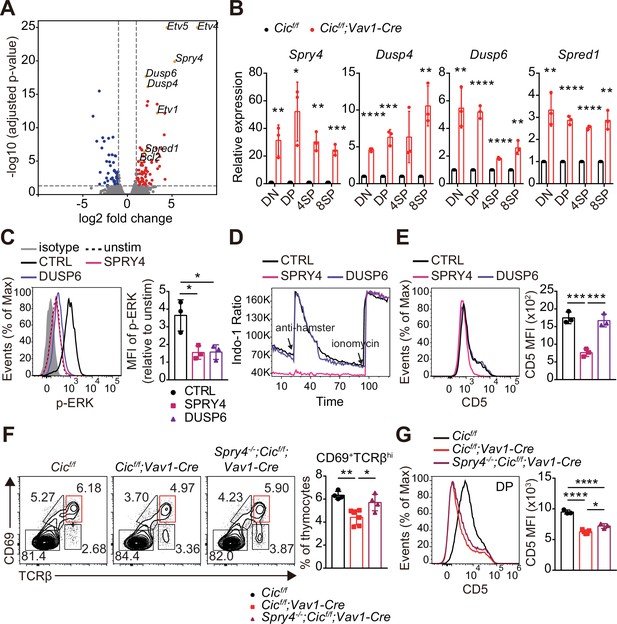
Identification of capicua (CIC) target genes regulating TCR signaling in double-positive (DP) thymocytes.
(A) Volcano plot showing differentially expressed genes (DEGs) in CIC-deficient DP thymocytes (fold change, > 2; adjusted P value, < 0.05). CIC target genes and Bcl2 are indicated at the corresponding dots. (B) qRT-PCR quantification of Spry4, Dusp4, Dusp6, and Spred1 expression in double-negative (DN), DP, CD4+ single-positive (4SP), and CD8+ SP (8SP) thymocytes from Cicf/f and Cicf/f;Vav1-Cre mice. N = 3 for each group. (C–E) Effects of SPRY4 and DUSP6 overexpression on TCR signaling in DP cells. Thymocytes were infected with retroviruses co-expressing GFP and either SPRY4 or DUSP6, and subjected to flow cytometry for (C) ERK activation, (D) Ca2+ influx, and (E) CD5 expression in GFP+ DP thymocytes. Three independent experiments were performed. (F) Thymocytes from 7-week-old Cicf/f, Cicf/f;Vav1-Cre, and Spry4-/-;Cicf/f;Vav1-Cre mice were analyzed for surface expression of CD69 and TCRβ. Representative FACS plots (left) and the frequency of CD69+TCRβhi cells (right) are shown. The CD69+TCRβhi cell population is highlighted by the red box in the FACS plots. N = 4, 6, and four for Cicf/f, Cicf/f;Vav1-Cre, and Spry4-/-;Cicf/f;Vav1-Cre mice, respectively. (G) CD5 levels in DP thymocytes from mice used in (F). Representative FACS plots (left) and CD5 mean fluorescence intensities (MFIs; right) are shown. N = 3, 5, and three for Cicf/f, Cicf/f;Vav1-Cre, and Spry4-/-;Cicf/f;Vav1-Cre mice, respectively. Bar graphs represent the mean and SEM. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. Unpaired two-tailed Student’s t-test (B) and one-way ANOVA with Tukey’s multiple comparison test (C, E, F and G) were used to calculate the corresponding p values. See also Figure 7—source data 1.
-
Figure 7—source data 1
Raw data for Figure 7.
- https://cdn.elifesciences.org/articles/71769/elife-71769-fig7-data1-v2.xlsx
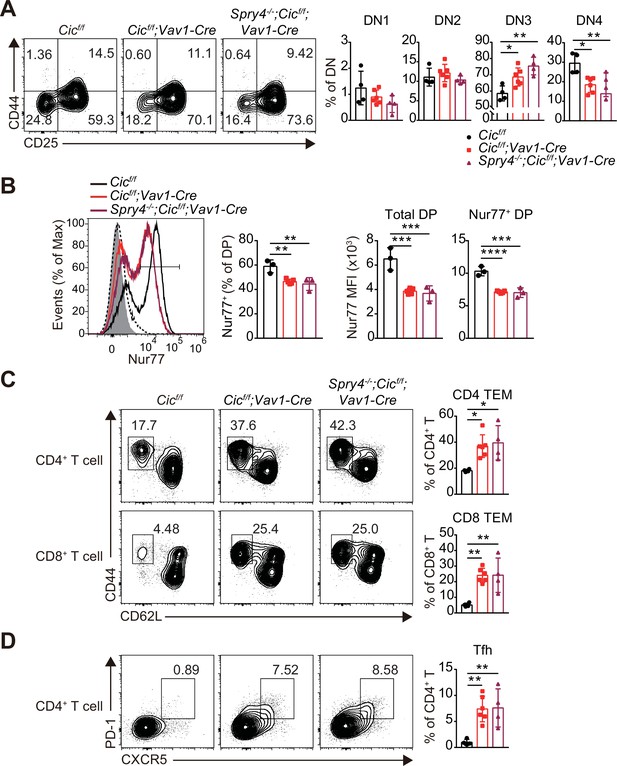
Analysis of T cell subsets in the thymus and spleen of Spry4-/-;Cicf/f;Vav1-Cre mice.
(A) Flow cytometric analysis of thymocytes from 7-week-old Cicf/f (N = 4), Cicf/f;Vav1-Cre (N = 6), and Spry4-/-;Cicf/f;Vav1-Cre (N = 4) mice for the frequency of DN1–four subsets. (B) FACS analysis of Nur77 expression in double-positive (DP) thymocytes from 7-week-old Cicf/f (N = 3), Cicf/f;Vav1-Cre (N = 5), and Spry4-/-;Cicf/f;Vav1-Cre (N = 3) mice. Freshly isolated thymocytes were treated with plate-coated anti-CD3 (5 μg/ml) and anti-CD28 (10 μg/ml) antibodies for 2 hr and subsequently subjected to flow cytometry. Representative histograms of Nur77 expression in DP thymocytes from Cicf/f (black line), Cicf/f;Vav1-Cre (red line), and Spry4-/-;Cicf/f;Vav1-Cre (burgundy line) mice overlaid with isotype (gray shaded) and unstimulated (dotted line) control histograms (left), the frequency of Nur77+ DP cells (middle), and Nur77 mean fluorescence intensities (MFIs) of total and Nur77+ DP cells (right) are presented. (C and D) Flow cytometric analysis of the frequency of (C) CD44hiCD62Llo effector memory T (TEM) and (D) Tfh cells in the spleen of 7-week-old Cicf/f (N = 4), Cicf/f;Vav1-Cre (N = 6), and Spry4-/-;Cicf/f;Vav1-Cre (N = 4) mice. Bar graphs represent the mean and SEM. *p < 0.05, **p < 0.01, and ***p < 0.001. One-way ANOVA with Tukey’s multiple comparison test was used to calculate the corresponding p values.
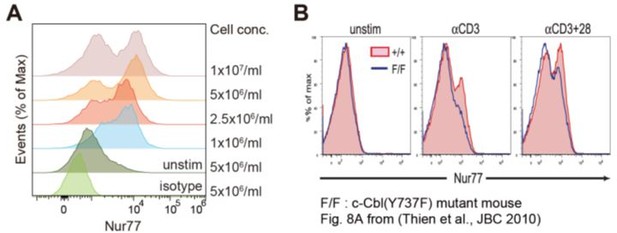
Histograms of Nur77 expression in double-positive (DP) thymocytes (A) Flow cytometric analysis of Nur77 expression in DP thymocytes.
Various concentrations of freshly isolated thymocytes were incubated with plate-coated anti-CD3 (5 μg/ml) and anti-CD28 (10 μg/ml) antibodies for 2 h, followed by staining of surface markers (CD4, CD8, and TCRβ) and intracellular staining of Nur77. Representative histograms are presented. (B) Independent experiment showing two-peaked histograms of Nur77-expressing cells. Adapted from Thien et al. (2010).
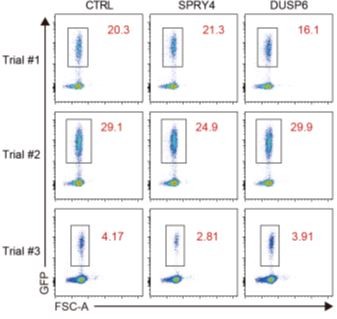
Efficiency of retroviral transduction of thymocytes.
FACS plots showing the frequency of GFP+ thymocytes transduced with retrovirus co-expressing GFP and control, SPRY4, or DUSP6. Three independent experiments were performed.
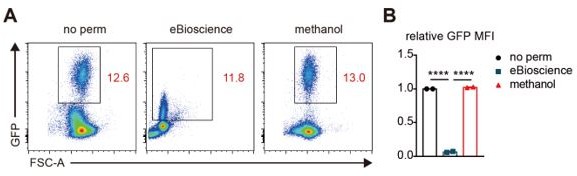
Comparison of the extent of preserved GFP signal after cell permeabilization by different methods.
(A) Flow cytometric analysis of GFP expression in live thymocytes infected with GFP-expressing retrovirus. Representative FACS plots are presented. No perm: without permeabilization, eBioscience: the Foxp3 staining buffer set, and methanol: cold methanol. (B) Relative expression levels of GFP in thymocytes before and after permeabilization with the Foxp3 staining buffer set (eBioscience) or cold methanol. N=2 for each group.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Mus musculus) | C57BL/6 J | The Jackson Laboratory | RRID:IMSR_JAX:000664 | |
| Strain, strain background (M. musculus) | B6.Cg-Commd10Tg(Vav1-icre)A2Kio/J | The Jackson Laboratory | RRID:IMSR_JAX:008610 | |
| Strain, strain background (M. musculus) | B6.Cg-Tg(Cd4-cre)1Cwi/BfluJ | The Jackson Laboratory | RRID:IMSR_JAX:022071 | |
| Strain, strain background (M. musculus) | B6NTac.Cg-Tg(Lck-cre)1Cwi/Mmnc | Lee et al., 2001 | RRID:MMRRC_037396-UNC | |
| Strain, strain background (M. musculus) | Cicfloxed | Lu et al., 2017; Park et al., 2017 | RRID:IMSR_JAX:030555 | |
| Strain, strain background (M. musculus) | CicFLAG/FLAG | Park et al., 2019 | PMID:30810242 | |
| Strain, strain background (M. musculus) | B6.Cg-Tg(TcraH-Y,TcrbH-Y)71Vbo | Kisielow et al., 1988 | RRID:MGI:3588781 | |
| Strain, strain background (M. musculus) | B6.Cg-Tg(TcraTcrb)425Cbn/J | The Jackson Laboratory | RRID:IMSR_JAX:004194 | |
| Strain, strain background (M. musculus) | B6.Cg-Foxp3tm2Tch/J | The Jackson Laboratory | RRID:IMSR_JAX:006772 | |
| Strain, strain background (M. musculus) | Spry4-/- | This paper | N/A | generated using Spry4tm1a(KOMP)Mbp embryonic stem cells obtained from the UC Davis KOMP repository. |
| Cell line (Homo-sapiens) | Platinum-E (Plat-E) Retroviral Packaging Cell Line | Cell Biolabs | Cat# RV-101, RRID:CVCL_B488 | |
| Antibody | Anti-mouse CD3ε (Armenian Hamster monoclonal) | BioLegendTonbo Biosciences | Cat# 100329, RRID:AB_1877171Cat# 50–0031, RRID:AB_2621730 | FC (1:300) |
| Antibody | Anti-mouse CD4 (Rat monoclonal) | BiolegendBD Biosciences | Cat# 100552, RRID:AB_2563053Cat# 562891, RRID:AB_2737870 | FC (1:300) |
| Antibody | Anti-mouse CD5 (Rat monoclonal) | Biolegend eBioscience | Cat# 100625, RRID:AB_2563928Cat# 45-0051-80, RRID:AB_914332 | FC (1:300) |
| Antibody | Anti-mouse CD8α (Rat monoclonal) | BioLegend | Cat# 100723, RRID:AB_389304Cat# 100721, RRID:AB_312760 | FC (1:300) |
| Antibody | Anti-mouse CD11b (Rat monoclonal) | BioLegend | Cat# 101211, RRID:AB_312794 | FC (1:300) |
| Antibody | Anti-mouse CD11c (Armenian Hamster monoclonal) | BioLegend | Cat# 117309, RRID:AB_313778 | FC (1:300) |
| Antibody | Anti-mouse CD19 (Rat monoclonal) | BD Biosciences | Cat# 561738, RRID:AB_10893995 | FC (1:300) |
| Antibody | Anti-mouse CD24 (Rat monoclonal) | BioLegend | Cat# 101819, RRID:AB_572010 | FC (1:300) |
| Antibody | Anti-mouse CD25 (Rat monoclonal) | Tonbo Biosciences | Cat# 75–0251, RRID:AB_2621943 | FC (1:300) |
| Antibody | Anti-mouse CD44 (Rat monoclonal) | BD Biosciences | Cat# 553133, RRID:AB_2076224Cat# 561860, RRID:AB_10895375 | FC (1:300) |
| Antibody | Anti-mouse CD62L (Rat monoclonal) | BD Biosciences | Cat# 560516, RRID:AB_1645257 | FC (1:300) |
| Antibody | Anti-mouse CD69 (Armenian Hamster monoclonal) | BioLegend | Cat# 104513, RRID:AB_492844Cat# 104545, RRID:AB_2686969 | FC (1:300) |
| Antibody | Anti-mouse CD73 (Rat monoclonal) | Biolegend eBioscience | Cat# 127223, RRID:AB_2716102Cat# 12-0731-81, RRID:AB_763516 | FC (1:300) |
| Antibody | Anti-mouse CD90.1 (Mouse monoclonal) | eBioscience | Cat# 45-0900-82, RRID:AB_2573662 | FC (1:300) |
| Antibody | Anti-mouse CD90.2 (Rat monoclonal) | BioLegend | Cat# 140303, RRID:AB_10642686 | FC (1:300) |
| Antibody | Anti-mouse CXCR5 (Rat monoclonal) | BD Biosciences | Cat# 551960, RRID:AB_394301 | FC (1:100) |
| Antibody | Anti-mouse TCRβ (Armenian Hamster monoclonal) | Tonbo Biosciences | Cat# 35–5961, RRID:AB_2621723 | FC (1:300) |
| Antibody | Anti-mouse TCR Vβ2 (Rat monoclonal) | BioLegend | Cat# 127908, RRID:AB_1227784 | FC (1:300) |
| Antibody | Anti-mouse TCR Vβ5.1/5.2 (Mouse monoclonal) | BD Biosciences | Cat# 553189, RRID:AB_394697 | FC (1:300) |
| Antibody | Anti-mouse TCR Vβ6 (Rat monoclonal) | BioLegend | Cat# 140004, RRID:AB_10643583 | FC (1:300) |
| Antibody | Anti-mouse TCR Vβ7 (Rat monoclonal) | BioLegend | Cat# 118308, RRID:AB_893628 | FC (1:300) |
| Antibody | Anti-mouse TCR Vβ8.1/8.2 (Rat monoclonal) | BioLegend | Cat# 118408, RRID:AB_1134109 | FC (1:300) |
| Antibody | Anti-mouse TCR Vβ8.3 (Armenian Hamster monoclonal) | BD Biosciences | Cat# 553664, RRID:AB_394980 | FC (1:300) |
| Antibody | Anti-mouse TCR Vβ9 (Mouse monoclonal) | BioLegend | Cat# 139804, RRID:AB_10641563 | FC (1:300) |
| Antibody | Anti-mouse TCR Vβ11 (Rat monoclonal) | BioLegend | Cat# 125907, RRID:AB_1227781 | FC (1:300) |
| Antibody | Anti-mouse TCR Vβ12 (Mouse monoclonal) | BioLegend | Cat# 139704, RRID:AB_10639729 | FC (1:300) |
| Antibody | Anti-mouse TCR Vβ13 (Mouse monoclonal) | BioLegend | Cat# 140704, RRID:AB_10639945 | FC (1:300) |
| Antibody | Anti-mouse TCRγ/δ (Armenian Hamster monoclonal) | eBioscience | Cat# 17-5711-81, RRID:AB_842757 | FC (1:300) |
| Antibody | Anti-mouse NK-1.1 (Mouse monoclonal) | BioLegend | Cat# 108709, RRID:AB_313396 | FC (1:300) |
| Antibody | Anti-mouse TER-119 (Rat monoclonal) | eBioscience | Cat# 17-5921-81, RRID:AB_469472 | FC (1:300) |
| Antibody | Anti-mouse Gr-1 (Rat monoclonal) | eBioscience | Cat# 17-5931-81, RRID:AB_469475 | FC (1:300) |
| Antibody | Anti-mouse TCR H-Y (Mouse monoclonal) | eBioscience | Cat# 11-9930-81, RRID:AB_465452 | FC (1:300) |
| Antibody | Anti-mouse PD-1 (Rat monoclonal) | eBioscience | Cat# 11-9981-81, RRID:AB_465466 | FC (1:300) |
| Antibody | Anti-mouse Nur77 (Mouse monoclonal) | eBioscience | Cat# 12-5965-82, RRID:AB_1257209 | FC (1:100) |
| Antibody | Anti-T-bet (Mouse monoclonal) | eBioscience | Cat# 12-5825-80, RRID:AB_925762 | FC (1:100) |
| Antibody | Anti-RORγt (Rat monoclonal) | eBioscience | Cat# 12-5825-80, RRID:AB_925762 | FC (1:100) |
| Antibody | Anti-Foxp3 (Rat monoclonal) | eBioscience | Cat# 17-5773-80, RRID:AB_469456 | FC (1:100) |
| Antibody | Anti-DYKDDDDK(flag) Tag antibody (Rat monoclonal) | BioLegend | Cat# 637309, RRID:AB_2563147 | FC (1:100) |
| Antibody | PE Donkey anti-rabbit IgG (min. x-reactivity) antibody (Donkey Polyclonal) | BioLegend | Cat# 406421, RRID:AB_2563484 | FC (1:100) |
| Antibody | Anti-CIC (Rabbit polyclonal) | Kim et al., 2015 | PMID:25653040 | WB (1:1000) |
| Antibody | PLCγ1 (D9H10) XP Rabbit mAb antibody (Rabbit monoclonal) | Cell Signaling Technology | Cat# 5690, RRID:AB_10691383 | WB (1:1000) |
| Antibody | Phospho-PLC 1 (Tyr783) antibody (Rabbit polyclonal) | Cell Signaling Technology | Cat# 2821, RRID:AB_330855 | WB (1:500) |
| Antibody | Zap-70 (99F2) Rabbit mAb antibody (Rabbit monoclonal) | Cell Signaling Technology | Cat# 2705, RRID:AB_2273231 | WB (1:1000) |
| Antibody | Phospho-Zap-70 (Tyr319)/Syk (Tyr352) antibody (Rabbit polyclonal) | Cell Signaling Technology | Cat# 2701, RRID:AB_331600 | WB (1:500) |
| Antibody | SAPK/JNK antibody (Rabbit polyclonal) | Cell Signaling Technology | Cat# 9252, RRID:AB_2250373 | WB (1:2000) |
| Antibody | Phospho-SAPK/JNK (Thr183/Tyr185) antibody (Rabbit polyclonal) | Cell Signaling Technology | Cat# 9251, RRID:AB_331659 | WB (1:1000) |
| Antibody | p44/42 MAPK (Erk1/2) antibody (Rabbit polyclonal) | Cell Signaling Technology | Cat# 9102, RRID:AB_330744 | WB (1:2000) |
| Antibody | Phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) antibody (Rabbit monoclonal) | Cell Signaling Technology | Cat# 4370, RRID:AB_2315112 | WB (1:1000)FC (1:100) |
| Antibody | Anti-p38 MAPK antibody (Rabbit polyclonal) | Cell Signaling Technology | Cat# 9212, RRID:AB_330713 | WB (1:2000) |
| Antibody | Phospho-p38 MAPK (Thr180/ Tyr182) antibody (Rabbit polyclonal) | Cell Signaling Technology | Cat# 9211, RRID:AB_331641 | WB (1:1000) |
| Antibody | β-Actin Antibody (Mouse monoclonal) | Santa Cruz Biotechnology | Cat# sc-47778, RRID:AB_626632 | WB (1:1000) |
| Antibody | α-Tubulin antibody (A-6) (Mouse monoclonal) | Santa Cruz Biotechnology | Cat# sc-398103, RRID:AB_2832217 | WB (1:1000) |
| Antibody | Purified NA/LE Hamster Anti-Mouse CD3e (Armenian Hamster monoclonal) | BD Biosciences | Cat# 553057, RRID:AB_394590 | (10 µg/mL) |
| Antibody | AffiniPure Goat Anti-Armenian Hamster IgG (H + L) (Goat polyclonal) | Jackson ImmunoResearch Labs | Cat# 127-005-160, RRID:AB_2338972 | (25 µg/mL) |
| Recombinant DNA reagent | pMIGR1-GFP (plasmid) | Pear et al., 1998 | RRID:Addgene_27490 | |
| Recombinant DNA reagent | pMIGR1-SPRY4-GFP (plasmid) | This paper | N/A | CDS of mouse Spry4 was inserted into pMIGR1-GFP. |
| Recombinant DNA reagent | pMIGR1-DUSP6-GFP (plasmid) | This paper | N/A | CDS of mouse Dusp6 was inserted into pMIGR1-GFP. |
| Recombinant DNA reagent | pCL-Eco (plasmid) | Naviaux et al., 1996 | RRID:Addgene_12371 | Retrovirus packaging vector |
| Sequence-based reagent | primers used in qRT-PCR | This paper | See Supplementary file 3 for sequence information. | |
| Peptide, recombinant protein | Streptavidin | Southern Biotech | Cat# 7100–01 | |
| Peptide, recombinant protein | Streptavidin | eBioscienceBD Biosciences | Cat# 45-4317-80, RRID:AB_10260035Cat# 554067, RRID:AB_10050396 | FC (1:100) |
| Commercial assay or kit | Fixable Viability Dye eFluor 780 | eBioscience | Cat# 65-0865-14 | |
| Commercial assay or kit | Ghost Dye Violet 510 | Tonbo Biosciences | Cat# 13–0870 | |
| Commercial assay or kit | Foxp3/ Transcription Factor Staining Buffer Set | eBioscience | Cat# 00-5523-00 | |
| Commercial assay or kit | BD Cytofix Fixation Buffer | BD Biosciences | Cat# 554655, RRID:AB_2869005 | |
| Commercial assay or kit | FuGENE HD Transfection Reagent | Promega | Cat# E2311 | |
| Commercial assay or kit | RiboEx | GeneAll | Cat# 301–002 | |
| Commercial assay or kit | GoScript Reverse Transcriptase Kit | Promega | Cat# A5001 | |
| Commercial assay or kit | SYBR Green Realtime PCR Master Mix | TOYOBO | Cat# TOQPK-201 | |
| Commercial assay or kit | EasySep Mouse Streptavidin RapidSpheres Isolation Kit | Stem Cell Technologies | Cat# 19,860 | |
| Commercial assay or kit | BCA Protein Assay Kit | Pierce | Cat# 23,225 | |
| Commercial assay or kit | Clarity Western ECL Substrate | Bio-Rad | Cat# 1705061 | |
| Commercial assay or kit | SuperSignal West Dura Extended Duration Substrate | Thermo Scientific | Cat# 34,076 | |
| Chemical compound, drug | Indo-1, AM, cell permeant | Invitrogen | Cat# I1203 | |
| Chemical compound, drug | Ionomycin from Streptomyces conglobatus | Sigma-Aldrich | Cat# I9657-1MG | |
| Chemical compound, drug | Hexadimethrine bromide | Sigma-Aldrich | Cat# H9268-10G | |
| Software, algorithm | FlowJo | Tree Star Inc. | RRID:SCR_008520, https://www.flowjo.com/solutions/flowjo | |
| Software, algorithm | ImageJ | NIH | RRID:SCR_003070, https://imagej.nih.gov/ij/ | |
| Software, algorithm | GraphPad Prism 7 | GraphPad Software | RRID:SCR_002798, https://www.graphpad.com/scientific- software/prism/ |
Additional files
-
Supplementary file 1
List of Differentially expressed genes (DEGs) in Cic-deficient DP thymocytes.
- https://cdn.elifesciences.org/articles/71769/elife-71769-supp1-v2.docx
-
Supplementary file 2
Gene Ontology (GO) analysis of DEGs in Cic-deficient DP thymocytes.
- https://cdn.elifesciences.org/articles/71769/elife-71769-supp2-v2.docx
-
Supplementary file 3
Oligonucleotide sequences used for qRT-PCR.
- https://cdn.elifesciences.org/articles/71769/elife-71769-supp3-v2.docx
-
Supplementary file 4
Flow cytometry gating strategy.
- https://cdn.elifesciences.org/articles/71769/elife-71769-supp4-v2.pdf
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/71769/elife-71769-transrepform1-v2.docx