Novel multicellular prokaryote discovered next to an underground stream
Figures
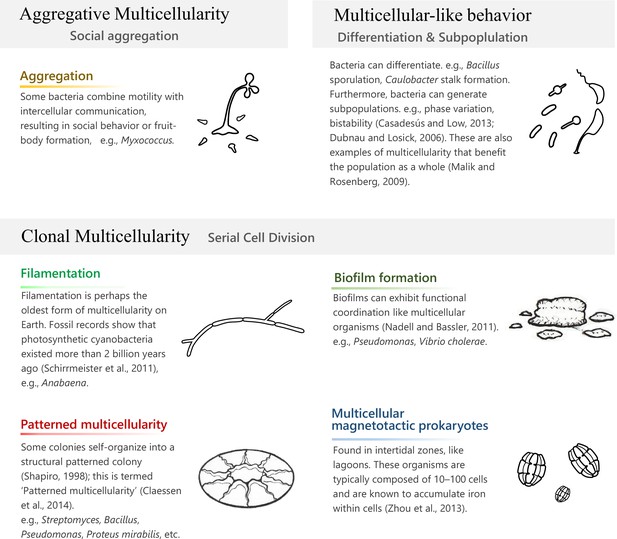
Prokaryotic multicellularity.
‘Clonal multicellularity’, ‘aggregative multicellularity’ (Brunet and King, 2017), and ‘multicellular-like behavior’ (Malik and Rosenberg, 2009) describe modes of multicellularity that arose through distinguished hypothetical conditions.
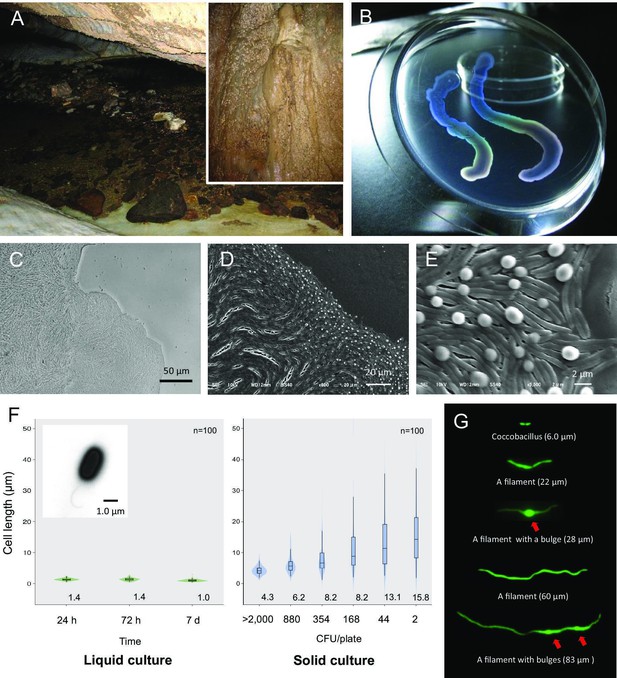
Liquid-crystal appearance and microstructure of a colony of Jeongeupia sacculi sp.nov.
HS-3, discovered in a limestone cave. (A) River in a cave showing the sampling site on the cave wall (inset). (B) Transparent and iridescent appearance of colonies. The edge of a colony growing on agar. Differential interference contrast (DIC) (C) and scanning electron microscopic (SEM) (D, E) images. Bulges of cells on the colony surface (E). (F) Cell length in liquid and solid cultures. The liquid cultures were monitored at different times. The solid cultures were inoculated at different colony densities (CFU/plate) and cultured for 2 days. The value given for each sample is the mean obtained from 100 cells. Inset: transmission electron microscopic (TEM) image of a liquid-cultured cell. (G) Typical cell morphology from a colony cultured on agar and observed by green fluorescent protein (GFP) fluorescence. Arrows: bulges of cells.

The isolation site of strain HS-3.
Images of the site where the bacterium was collected in a limestone cave: (A) Map of Japan. Arrowhead: Kyushu Island. (Inset) Arrowhead: Hirao karst plateau; (B) Aerial photograph of the sampling site in the Hirao karst plateau, the whole area of which measures 6.5 km from north to south and 2.5 km from east to west, and has an altitude of between 300 and 700 m. Arrowhead: Seiryu limestone cave. The aerial image was obtained from Geospatial Information Authority of Japan; (C) Photograph of Hirao karst plateau; (D) Cave wall (sampling site); (E) Entrance viewed from inside the cave; (F) Entrance from outside the cave; (G–I) River flowing in the cave. The air temperature inside the cave was around 15°C. The pH of the water on the wall was 8.5.

Phylogenetic analysis and genome information of HS-3.
(A) Phylogenetic tree based on 16S rRNA gene sequence. The tree represents a cluster in the Neisseriaceae family (shadowed area) and some relatives isolated from similar oligotrophic environments such as tufa, another form of limestone, as indicated by blue arrows. The closest relatives of strain HS-3T are Jeongeupia naejangsanensis KCTC 66233T (FJ669217) and Jeongeupia chitinilytica JchiT (NR_109420) with similarity values of 98.2% and 97.8%, respectively. (B) Diagrams of the genome and the plasmid. The plasmid seems to be a low copy number one for which it has not been experimentally detected by a commercial extraction kit.
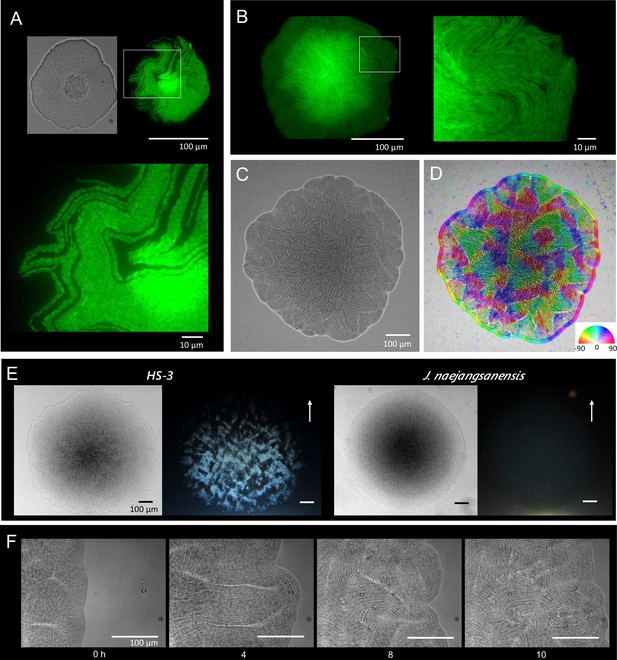
Colony morphogenesis and liquid-crystal-like ordering.
(A) A microcolony showing a single layer of cells. Differential interference contrast (DIC) (top left) and green fluorescent protein (GFP) (top right) images and an enlarged image (bottom) of the area indicated by the white box. (B) A young colony (larger than A) with a single layer of cells at the edge. Enlarged image of the area indicated by the white box (right). (C) Anisotropic pattern of the bottom cell layer of a single colony. (D) The color mapping to show the orientation of cells. The anisotropic pattern is a typical characteristics in a nematic phase (Doostmohammadi et al., 2018). (E) Anisotropic optical texture of colonies of HS-3 (left) and Jeongeupia naejangsanensis (right), the close relative strain. DIC images (left) and diagonally illuminated images (right). Arrows indicate the direction of light illumination. (F) Time-lapse images of liquid-crystal-like pattern formation on the edge of a growing colony. Time (hr) after chase is indicated. The original video is available as Video 1.

Emergence of analogous nematic patterns on the colony edge.
(A) The emergence of analogous nematic patterns at the colony edge. Two boxed areas are shown as examples on the right. The areas are chronologically aligned and show two types of topological defects characterized by two-dimensional nematic liquid crystals. (B) Time course images of the enlarged area indicated by a box on the colony edge. A key feature underlying the development of the unique cellular architecture is related to the cells adopting the two-dimensional, nematic liquid-crystal orientation (Doostmohammadi et al., 2018), in which cells are aligned tightly together to form the layered structure. Other bacteria, such as E. coli and B. subtilis, can produce nematic features, but only for short periods when the cells are allowed to grow two dimensionally (Dell’Arciprete et al., 2018; Yaman et al., 2019). In these species, collisions between the expanding layer of cells results in edge instability and buckling of the two-dimensional nematic order. Conversely, HS-3 filamentous cells have the remarkable ability to sustain an ordered structure.
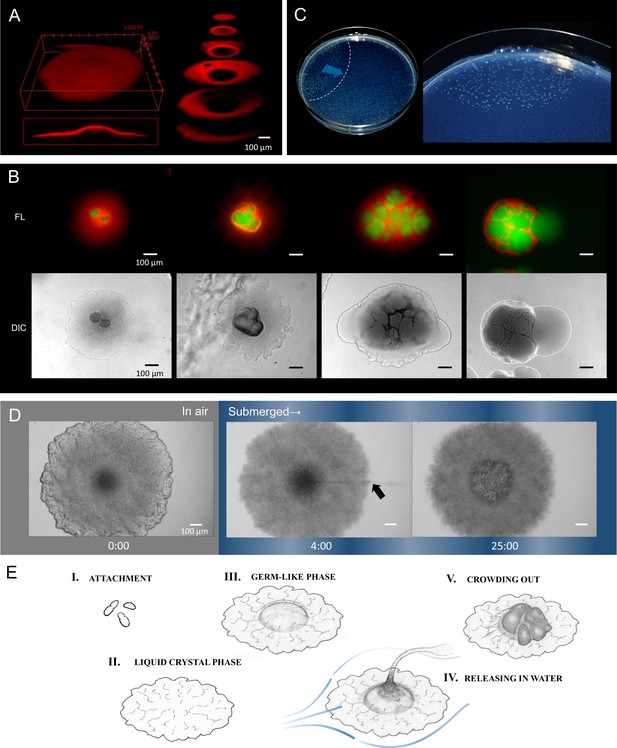
Internally growing cells and crowding out.
(A) Three-dimensional image of a 4-day-old colony reconstructed from 100 Z-axis slices with a total height of 168 µm. The whole Nile-red image (left) and its cross-sections (right). (B) Five-day-old, mature colonies showing internal growth at the center of the colony. Two right panels show the ‘crowding out’. (C) Colony-crowding influences adjacent colonies by inducing a ‘chain reaction’. The direction of the chain reaction and its front line are indicated by the blue arrow and the dotted line, respectively. More details of a crowding colony are shown in Video 2. (D) Time-lapse images of a mature colony in air (left panel) and after being submerged in water (middle and right panels). The arrow indicates the waterborne coccobacillus cells released from the colony. The colonies were grown for 5 days on agar before the time-lapse analysis. The time (min) after chase is shown. Bars = 100 µm. The original video is available as Video 3. (E) Illustration of the potential multicellular life-cycle of HS-3. (I) Coccobacillus cells attached to a new solid surface. (II) The cells proliferate to some extent, generating a liquid-crystal texture. (III) The center area becomes raised due to clustering of internal cells, leading to the germ-like phase. (IV) A waterflow releases coccobacilli. (V) In the absence of a waterflow, outward growth of the internal coccobacillus cells results in ‘crowding out’.

Reversibility between filament and coccobacillus cells.
(A) HS-3 crowds out coccobacillus cells from mature colonies. Streaking of the daughter coccobacillus cells on a new plate showed that the coccobacilli could reproduce the original colony morphology with filamentous cells. Phase contrast images (top left) and a differential interference contrast (DIC) image (top right), and scanning electron microscopic (SEM) images (second row). (B) The filamentous cells were selected from a single 2-day-old colony, suspended in phosphate-buffered saline (PBS), and inoculated on a new agar plate. The mature filament cells that survived physical damage during inoculation grew by dividing on agar. The microscopic observation was performed without a cover slip. The time after chase is shown. Bars = 50 µm.
Videos
The edge of a colony.
Emergence of coccobacillus cells.
Waterborne coccobacillus cells from a colony.
Additional files
-
Supplementary file 1
Characteristics that distinguish strain HS-3 from its close phylogenetic relatives.
- https://cdn.elifesciences.org/articles/71920/elife-71920-supp1-v1.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/71920/elife-71920-transrepform1-v1.docx





