Crotamiton derivative JM03 extends lifespan and improves oxidative and hypertonic stress resistance in Caenorhabditis elegans via inhibiting OSM-9
Figures
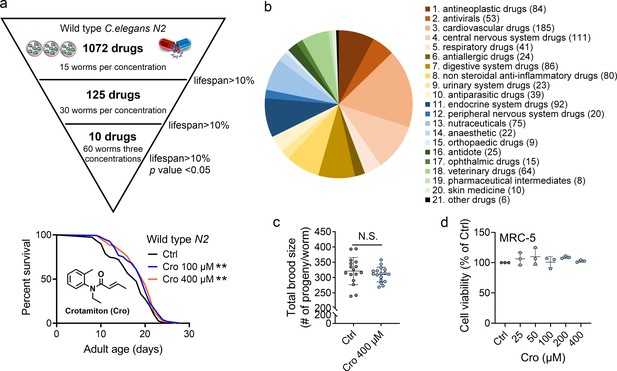
Crotamiton extends the lifespan of Caenorhabditis elegans.
(a) Phenotypic screening led to the discovery of crotamiton as a hit compound for prolonging the lifespan in wild type (N2) worms. Data were compared using the Log-rank test. p = 0.0032 for Cro 100 μM. p = 0.0011 for Cro 400 μM. (b) The pie charts show the distribution of approved drug combinations per disease area in this phenotypic screening study. (c) The total brood size of crotamiton-treated N2 worms. Control n = 16 and crotamiton n = 17. (d) The viability of crotamiton-treated MRC-5 cells. (c) Data have been represented as the mean ± SD, and comparisons are made using Student t-test. (d) Data have been represented as the mean ± SD, and comparisons are made using one-way ANOVA test. The graphics represent a compilation of at least three independent experiments. ** p < 0.01.
-
Figure 1—source data 1
Lifespan data in the 1st round screening.
- https://cdn.elifesciences.org/articles/72410/elife-72410-fig1-data1-v1.xlsx
-
Figure 1—source data 2
Lifespan data in the 2nd and 3rd round screening.
- https://cdn.elifesciences.org/articles/72410/elife-72410-fig1-data2-v1.xlsx
-
Figure 1—source data 3
Lifespan data for (a), brood size for (c), and cell viability for (d).
- https://cdn.elifesciences.org/articles/72410/elife-72410-fig1-data3-v1.xlsx

The anti-scabies drugs permethrin and benzyl benzoate failed to extend the lifespan in Caenorhabditis elegans.
(a) Permethrin failed to extend the lifespan of C. elegans. (b) Benzyl benzoate failed to extend the lifespan of C. elegans. (a–b) Data were compared using the Log-rank. * p < 0.05.
-
Figure 1—figure supplement 1—source data 1
Lifespan data of worms treated with permethrin or benzyl benzoatee.
- https://cdn.elifesciences.org/articles/72410/elife-72410-fig1-figsupp1-data1-v1.xlsx

Crotamiton and JM03 did not reduce the bacterial growth at 400 μM concentration.
(a) Crotamiton did not reduce the bacterial growth at 400 μM concentration. (b) JM03 did not reduce the bacterial growth at 400 μM concentration. Data represented as the mean ± SD, and comparisons were made using Student t-test. The graphics represent a compilation of at least three independent experiments. * p < 0.05.
-
Figure 1—figure supplement 2—source data 1
OD600 for OP50 culture measured every 12 hr in 72 hr.
- https://cdn.elifesciences.org/articles/72410/elife-72410-fig1-figsupp2-data1-v1.xlsx
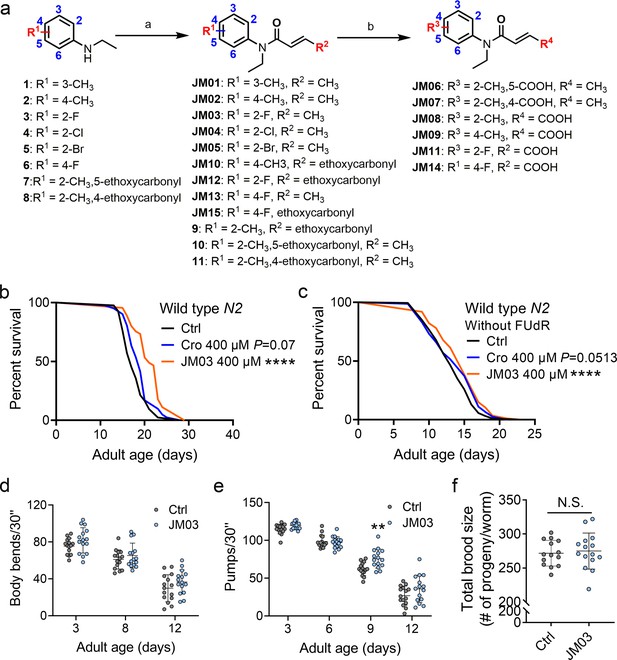
JM03 has better lifespan extension activity in Caenorhabditis elegans.
(a) Synthesis of compounds JM01−JM15. Reagents and conditions: a. Acryloyl chloride derivatives, K2CO3, CH2Cl2, 0 °C to rt; b. 1 M NaOH (aq.), CH3OH, rt. (b) JM03 prolonged lifespan in wild-type worms. p-values by Log-rank test. p = 0.0737 for Cro 400 μM. p < 0.0001 for JM03 400 μM. (c) JM03 prolonged lifespan in wild-type worms without FUdR treatment. p-values by Log-rank test. p = 0.0513 for Cro 400 μM. p = 0.0003 for JM03 400 μM. (d) The mobility of JM03-treated N2 worms by analyzing the body bend rate at days 3, 8, and 12. Control n = 15 and JM03 n = 15. (e) The pharyngeal pumping rate of JM03-treated N2 worms. Control n = 15 and JM03 n = 15 at days 3, 6, 9, and 12. p-values by two-way AVOVA. p = 0.0015 for 9 days. (f) The total brood size of JM03-treated N2 worms. Control n = 14 and JM03 n = 15. (b–c) Data are compared using the Log-rank test. (d-e) Data have been represented as the mean ± SD, and comparisons are made using two-way AVOVA. (f) Data have been represented as the mean ± SD, and comparisons are made using Student t-test. The graphics represent a compilation of at least three independent experiments. ** p < 0.01, **** p < 0.0001.
-
Figure 2—source data 1
Lifespan data for (b); number of body bends for (c); number of pumps in 30″ for (d); brood size for (e) and cell viability for (f).
- https://cdn.elifesciences.org/articles/72410/elife-72410-fig2-data1-v1.xlsx

The structures and mean percentage of lifespan extension by crotamiton derivatives.
-
Figure 2—figure supplement 1—source data 1
Lifespan data of crotamiton derivatives.
- https://cdn.elifesciences.org/articles/72410/elife-72410-fig2-figsupp1-data1-v1.xlsx

The structures and mean percentage of lifespan extension by crotamiton derivatives.
-
Figure 2—figure supplement 2—source data 1
Lifespan data of crotamiton derivatives.
- https://cdn.elifesciences.org/articles/72410/elife-72410-fig2-figsupp2-data1-v1.xlsx
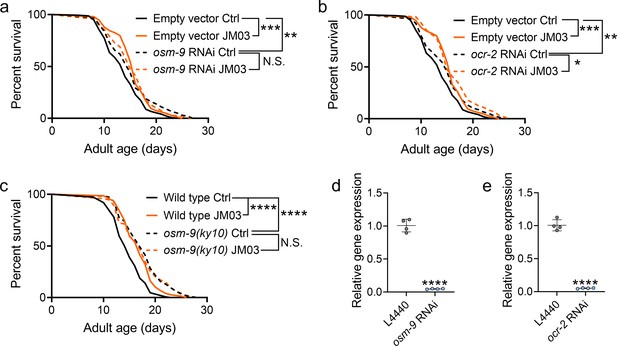
JM03-induced lifespan extension depends on OSM-9.
(a) JM03 failed to extend the lifespan of osm-9 RNAi worms. p-values by Log-rank test. p = 0.0002 between Empty vector Ctrl and Empty vector JM03. p = 0.0028 between Empty vector Ctrl and osm-9 RNAi Ctrl. (b) JM03 extended the lifespan of ocr-2 RNAi worms. p-values by Log-rank test. p = 0.0002 between Empty vector Ctrl and Empty vector JM03. p = 0.0084 between Empty vector Ctrl and ocr-2 RNAi Ctrl. p = 0.0259 between ocr-2 RNAi Ctrl and ocr-2 RNAi JM03. (c) JM03 failed to extend the lifespan of osm-9(ky10) mutants. p-values by Log-rank test. p < 0.0001 for wild-type JM03 and osm-9(ky10) Ctrl. (d) The transcriptional level of osm-9 decreased after RNAi treatment. p-values by Student t-test. p < 0.0001 for osm-9 RNAi. (e) The transcriptional level of ocr-2 decreased after RNAi treatment. p-values by Student t-test. p < 0.0001 for ocr-2 RNAi. (a–c) Data are compared using the Log-rank test. (d-e) Data have been represented as the mean ± SD, and comparisons are made using Student t-test. The graphics represent a compilation of at least three independent experiments. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001. N.S., not significant.
-
Figure 3—source data 1
Lifespan data for (a–c) and relative gene expression for (d, e).
- https://cdn.elifesciences.org/articles/72410/elife-72410-fig3-data1-v1.xlsx
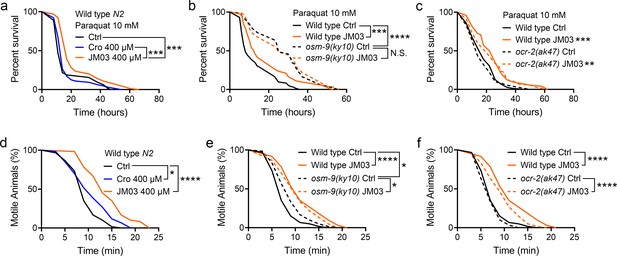
OSM-9 inhibition induced by JM03 has beneficial effect for Caenorhabditis elegans lifespan under oxidative and hypertonic stress conditions.
(a) JM03 extended the lifespan of wild-type (N2) worms under paraquat-induced oxidative stress condition. p-values by Log-rank test. p = 0.0001 between Ctrl and JM03 400 μM. p = 0.0002 between Cro 400 μM and JM03 400 μM. (b) JM03 failed to extend the lifespan of osm-9(ky10) mutants under oxidative stress condition. p-values by Log-rank test. p = 0.0009 between Wild-type Ctrl and Wild-type JM03. p < 0.0001 between Wild-type Ctrl and osm-9(ky10) Ctrl. (c) JM03 extended the lifespan of ocr-2(ak47) mutants under oxidative stress condition. p-values by Log-rank test. p = 0.0005 between Wild-type Ctrl and Wild-type JM03. p = 0.0024 between ocr-2(ak47) Ctrl and ocr-2(ak47) JM03. (d) JM03 significantly reduced the paralysis for wild-type worms under NaCl-induced hypertonic stress condition. p-values by Log-rank test. p = 0.0171 between Ctrl and Cro 400 μM. p < 0.0001 between Ctrl and JM03 400 μM. (e) JM03 reduced the responsiveness for osm-9(ky10) mutants under hypertonic stress condition. p-values by Log-rank test. p < 0.0001 between Wild-type Ctrl and Wild-type JM03. p = 0.0128 between Wild-type Ctrl and osm-9(ky10) Ctrl. p = 0.0254 between osm-9(ky10) Ctrl and osm-9(ky10) JM03. (f) JM03 significantly reduced the paralysis for ocr-2(ak47) mutants similar to wild-type worms under hypertonic stress condition. p-values by Log-rank test. p < 0.0001 between Wild-type Ctrl and Wild-type JM03. p < 0.0001 between ocr-2(ak47) Ctrl and ocr-2(ak47) JM03. (a–f) Data are compared using the Log-rank test. The graphics represent a compilation of at least three independent experiments. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001. N.S., not significant.
-
Figure 4—source data 1
lifespan data for (a–c) and relative gene expression data for (d, e).
- https://cdn.elifesciences.org/articles/72410/elife-72410-fig4-data1-v1.xlsx
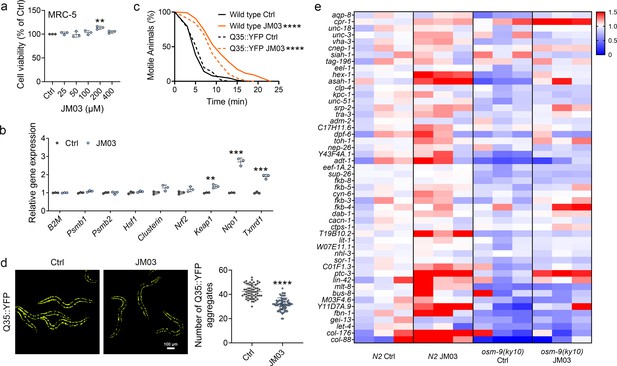
OSM-9 inhibition induced by JM03 has beneficial effect for Caenorhabditis elegans lifespan under hypertonic stress conditions.
(a) The viability of JM03-treated MRC-5 cells. p-values by one-way ANOVA test. p = 0.0036 for JM03 200 μM. (b) JM03 significantly increased the transcriptional expression of proteostasis related genes in MRC-5 cells. p-values by Student t-test. p = 0.0041 for Keap1. p = 0.0001 for Nqo1. p = 0.0003 for Txnrd1. (c) JM03 significantly reduced the paralysis for Q35::YFP worms similar to wild-type worms under hypertonic stress condition. p-values by Log-rank test. p < 0.0001 between Wild-type Ctrl and Wild-type JM03. p < 0.0001 between Q35::YFP Ctrl and Q35::YFP JM03. (d) JM03 significantly reduced the Q35::YFP aggregation. p-values by Student t-test. p < 0.0001 for JM03 treatment. (e) Putative proteostasis genes differentially upregulated by JM03 treatment in N2 control worms but not osm-9(ky10) worms by transcriptome analysis. (a) Data have been represented as the mean ± SD, and comparisons are made using one-way ANOVA test. (b, d) Data have been represented as the mean ± SD, and comparisons are made using Student t-test. (c) Data are compared using the Log-rank test. The graphics represent a compilation of at least three independent experiments. ** p < 0.01, *** p < 0.001, **** p < 0.0001.
-
Figure 5—source data 1
Cell viability for (a); relative gene expression data for (b); lifespan data for (c); number of Q35::YFP aggregates of each worm for (d); RNA sequencing data of proteostasis-related genes for (e).
- https://cdn.elifesciences.org/articles/72410/elife-72410-fig5-data1-v1.xlsx

The principal component analysis (PCA) of the transcriptome analysis by RNA-sequencing.
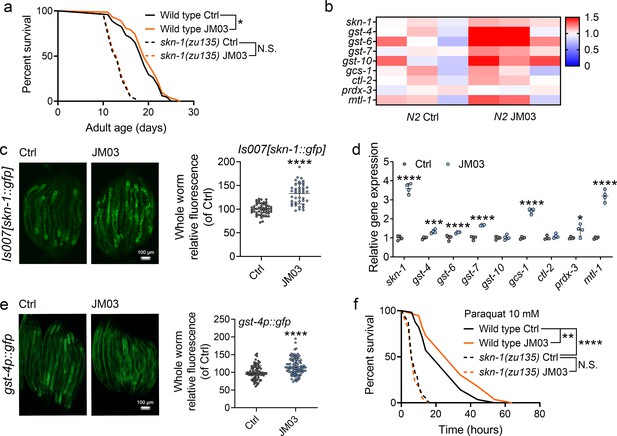
JM03-induced lifespan extension through SKN-1 pathway.
(a) JM03 treatment failed to extend the lifespan of skn-1(zu135) mutants. p-values by Log-rank test. p = 0.0569 between Wild-type Ctrl and Wild-type JM03. (b) Skn-1 and its targets genes upregulated by JM03 in wild-type worms by transcriptome analysis. (c) JM03 significantly increased the fluorescence intensity of skn-1::gfp. Scale bar = 100 µm. p-values by Student t-test. p < 0.0001 for JM03 treatment. (d) JM03 significantly increased the transcriptional expression of skn-1 and skn-1 regulated genes. p-values by Student t-test. p < 0.0001 for skn-1. p = 0.0006 for gst-4. p < 0.0001 for gst-6. p < 0.0001 for gst-7. p < 0.0001 for gcs-1. p = 0.0424 for prdx-3. p < 0.0001 for mtl-1. (e) JM03 significantly upregulated the fluorescence intensity of gst-4p::gfp. Scale bar = 100 µm. p-values by Student t-test. p < 0.0001 for JM03 treatment. (f) JM03 treatment failed to extend the lifespan of osm-9(ky10) and skn-1(zu135) mutants under oxidative stress condition. p-values by Log-rank test. p = 0.0059 between Wild-type Ctrl and Wild-type JM03. p < 0.0001 between Wild-type Ctrl and skn-1(zu135) Ctrl. (a, f) Data are compared using the Log-rank test. The graphics represent a compilation of at least three independent experiments. (c–e) Data have been represented as the mean ± SD, and comparisons are made using Student t-test. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001. N.S., not significant.
-
Figure 6—source data 1
Lifespan data for (a); RNA-sequencing data of Skn-1 and its targets genes for (b); mean fluorescence intensity of each worm for (c); relative gene expression data for (d); mean fluorescence intensity of each worm for (e); lifespan of each worm in paraquat for (f).
- https://cdn.elifesciences.org/articles/72410/elife-72410-fig6-data1-v1.xlsx

JM03 treatment extended the lifespan of daf-16(mu86) mutants.
Data are compared using the Log-rank test. The graphics represent a compilation of at least three independent experiments. ****p < 0.0001.
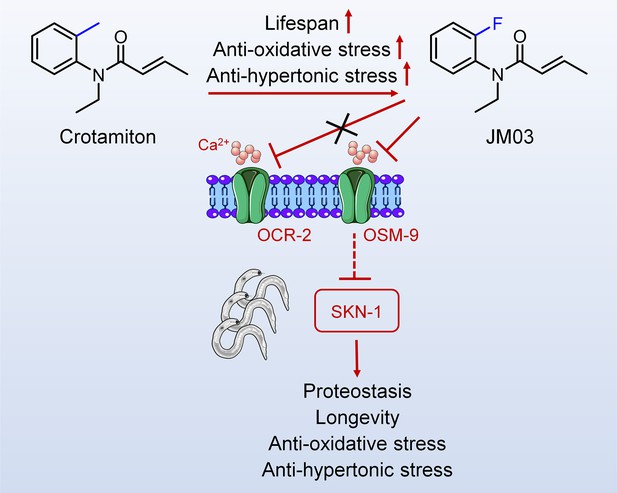
Schematic diagram of the mechanism of action of JM03 for regulating the lifespan, anti-oxidative and anti-hypertonic stress ability in Caenorhabditis elegans.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Commercial assay or kit | GMyc-PCR Mycoplasma Test Kit | Yeasen | Cat# 40,601ES10 | |
| Commercial assay or kit | cell counting kit-8 solution | Targetmol | Cat# C0005 | |
| Commercial assay or kit | Trizol Reagent | Beyotime | Cat# R0016 | |
| Commercial assay or kit | Total RNA Kit II | Omega | Cat# R6934-01 | |
| Commercial assay or kit | Hifair II 1st Strand cDNA Synthesis SuperMix | Yeasen | Cat# 11,120ES60 | |
| Commercial assay or kit | Hieff qPCR SYBR Green Master Mix | Yeasen | Cat# 11,201ES08 | |
| Cell line (Homo-sapiens) | MRC-5 cells | National Collection of Authenticated Cell Cultures | Cat# SCSP-5040 | |
| Strain, strain background (Escherichia coli) | OP50 | Caenorhabditis Genetics Center | ||
| Strain, strain background (Escherichia coli) | HT115 | Caenorhabditis Genetics Center | ||
| Strain, strain background (Caenorhabditis elegans) | N2 | Caenorhabditis Genetics Center | WormBase ID: WBStrain00000001 | wild type |
| Strain, strain background (Caenorhabditis elegans) | CX4544 | Caenorhabditis Genetics Center | WormBase ID: WBStrain00005264 | Genotype: ocr-2(ak47) IV |
| Strain, strain background (Caenorhabditis elegans) | CX10 | Caenorhabditis Genetics Center | WormBase ID: WBStrain00005214 | Genotype: osm-9(ky10) IV |
| Strain, strain background (Caenorhabditis elegans) | AM140 | Caenorhabditis Genetics Center | WormBase ID: WBStrain00000182 | Genotype: rmIs132 [unc-54p::Q35::YFP] |
| Strain, strain background (Caenorhabditis elegans) | LG333 | Caenorhabditis Genetics Center | WormBase ID: WBStrain00024182 | Genotype: skn-1(zu135) (IV)/nT1[qIs51] (IV;V); ldIs7 [skn-1b/c::gfp] |
| Strain, strain background (Caenorhabditis elegans) | CF1038 | Caenorhabditis Genetics Center | WormBase ID: WBStrain00004840 | Genotype: daf-16(mu86) (I) |
| Strain, strain background (Caenorhabditis elegans) | CL2166 | Caenorhabditis Genetics Center | WormBase ID: WBStrain00005102 | Genotype: dvIs19 [gst-4p::gfp::NLS] (III) |
| Strain, strain background (Caenorhabditis elegans) | EU31 | Caenorhabditis Genetics Center | WormBase ID: WBStrain00007251 | Genotype: skn-1(zu135) (IV)/ nT1[unc-?(n754);let-?] (IV;V) |
| Sequence-based reagent | B2M-F | This paper | PCR primers | ACTGAATTCACCCCCACTGA |
| Sequence-based reagent | B2M-R | This paper | PCR primers | AAGCAAGCAAGCAGAATTTGG |
| Sequence-based reagent | Psmb1-F | This paper | PCR primers | GGATGCAGCGGTTTTCATGG |
| Sequence-based reagent | Psmb1-R | This paper | PCR primers | AATTGCCCCCGTAGTCATGG |
| Sequence-based reagent | Psmb2-F | This paper | PCR primers | CTGCTCCGCCCTCCATTAAC |
| Sequence-based reagent | Psmb2-R | This paper | PCR primers | GCCAAGCATGGAGTAGAACG |
| Sequence-based reagent | Hsf-1-F | This paper | PCR primers | TATGGCTTCCGGAAAGTGGT |
| Sequence-based reagent | Hsf-1-R | This paper | PCR primers | GGAACTCCGTGTCGTCTCTC |
| Sequence-based reagent | Clusterin-F | This paper | PCR primers | AAACGAAGAGCGCAAGACAC |
| Sequence-based reagent | Clusterin-R | This paper | PCR primers | TGTTTCAGGCAGGGCTTACA |
| Sequence-based reagent | Nrf2-F | This paper | PCR primers | CATCCAGTCAGAAACCAGTGG |
| Sequence-based reagent | Nrf2-R | This paper | PCR primers | GCAGTCATCAAAGTACAAAGCAT |
| Sequence-based reagent | Keap1-F | This paper | PCR primers | AGTTCATGGCCCACAAGGTG |
| Sequence-based reagent | Keap1-R | This paper | PCR primers | AATGGACACCACCTCCATGC |
| Sequence-based reagent | Nqo1-F | This paper | PCR primers | AGCAGACGCCCGAATTCAAA |
| Sequence-based reagent | Nqo1-R | This paper | PCR primers | AGAGGCTGCTTGGAGCAAAA |
| Sequence-based reagent | Txnrd1-F | This paper | PCR primers | TTGGAGTGCGCTGGATTTCT |
| Sequence-based reagent | Txnrd1-R | This paper | PCR primers | TTTGTTGGCCATGTCCTGGT |
| Sequence-based reagent | ama-1-F | This paper | PCR primers | TGGAACTCTGGAGTCACACC |
| Sequence-based reagent | ama-1-R | This paper | PCR primers | CATCCTCCTTCATTGAACGG |
| Sequence-based reagent | act-1-F | This paper | PCR primers | ATGTGTGACGACGAGGTTGC |
| Sequence-based reagent | act-1-R | This paper | PCR primers | ACTTGCGGTGAACGATGGATG |
| Sequence-based reagent | skn-1-F | This paper | PCR primers | ACAGTGCTTCTCTTCGGTAGC |
| Sequence-based reagent | skn-1-R | This paper | PCR primers | GAGACCCATTGGACGGTTGA |
| Sequence-based reagent | gst-4-F | This paper | PCR primers | TGCTCAATGTGCCTTACGAG |
| Sequence-based reagent | gst-4-R | This paper | PCR primers | AGTTTTTCCAGCGAGTCCAA |
| Sequence-based reagent | gst-6-F | This paper | PCR primers | TTTGGCAGTTGTTGAGGAG |
| Sequence-based reagent | gst-6-R | This paper | PCR primers | TGGGTAATCTGGACGGTTTG |
| Sequence-based reagent | gst-7-F | This paper | PCR primers | AGGACAACAGAATCCCAAAGG |
| Sequence-based reagent | gst-7-R | This paper | PCR primers | AGCAAATCCCATCTTCACCAT |
| Sequence-based reagent | gst-10-F | This paper | PCR primers | GTCTACCACGTTTTGGATGC |
| Sequence-based reagent | gst-10-R | This paper | PCR primers | ACTTTGTCGGCCTTTCTCTT |
| Sequence-based reagent | gcs-1-F | This paper | PCR primers | AATCGATTCCTTTGGAGACC |
| Sequence-based reagent | gcs-1-R | This paper | PCR primers | ATGTTTGCCTCGACAATGTT |
| Sequence-based reagent | ctl-1-F | This paper | PCR primers | GCGGATACCGTACTCGTGAT |
| Sequence-based reagent | ctl-1-R | This paper | PCR primers | GTGGCTGCTCGTAGTTGTGA |
| Sequence-based reagent | prdx-3-F | This paper | PCR primers | CTTGACTTCACCTTTGTATGCC |
| Sequence-based reagent | prdx-3-R | This paper | PCR primers | GGCGATCTTCTTGTTGAAATCA |
| Sequence-based reagent | mtl-1-F | This paper | PCR primers | CAAGTGTGACTGCAAAAACAAG |
| Sequence-based reagent | mtl-1-R | This paper | PCR primers | GCAGTACTTCTCACAACACTTG |
| Sequence-based reagent | osm-9-F | This paper | PCR primers | TTCGGTTGGATCAGGAAGGC |
| Sequence-based reagent | osm-9-R | This paper | PCR primers | GCTTGCTTTCTCTGACGTGC |
| Sequence-based reagent | ocr-2-F | This paper | PCR primers | ACTTGTAGATATGCATGGCGGT |
| Sequence-based reagent | ocr-2-R | This paper | PCR primers | CCAAGTCGTTCATTTCTTTCCTTA |






