Systematic investigation of the link between enzyme catalysis and cold adaptation
Figures
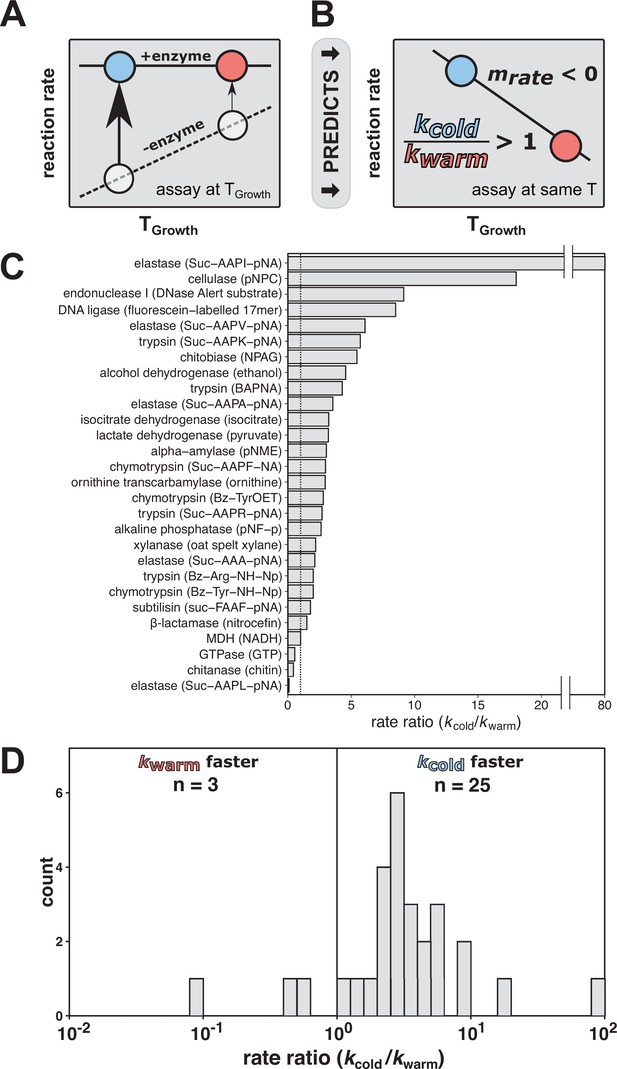
The rate compensation model of cold adaptation predicts that cold-adapted enzymes exhibit greater catalysis and are faster at a common temperature than their warm-adapted counterparts.
(A) According to the rate compensation model of cold adaptation, a cold-adapted variant (blue circle) has larger rate enhancement than a warm-adapted variant (red circle). The dashed line represents the uncatalyzed reaction, the solid line represents the catalyzed reaction, and the arrows represent the rate enhancement at the respective organism TGrowth. (B) When variants are assayed at a common temperature, rate compensation predicts a faster reaction for the enzyme from the cold-adapted organism, corresponding to a rate ratio (kcold/kwarm) of greater than one and a negative slope of rate vs. TGrowth (mrate). (C, D) Rate comparisons of warm-adapted and cold-adapted enzyme variants made at identical temperatures from cold adaptation literature spanning indicated reactions with substrate specified in parentheses (Collins and Gerday, 2017; Feller and Gerday, 1997; Siddiqui and Cavicchioli, 2006; Smalås et al., 2000). The black vertical lines represent no rate enhancement change with temperature (i.e., rate ratio = 1).
-
Figure 1—source data 1
Rate comparisons of warm-adapted and cold-adapted enzyme variants made at identical temperatures from cold adaptation literature.
- https://cdn.elifesciences.org/articles/72884/elife-72884-fig1-data1-v1.csv
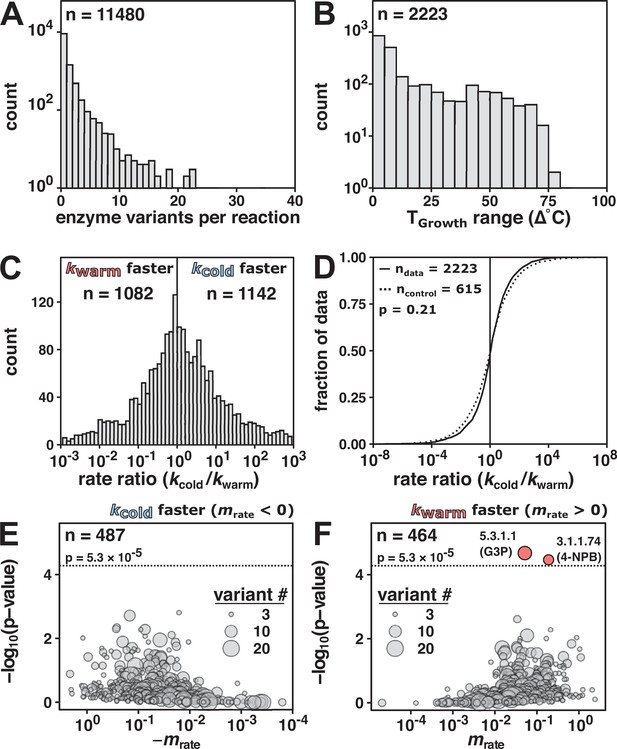
Enzyme rate constant data (kcat) do not indicate general rate compensation.
(A) Enzyme variants per reaction of wild-type enzyme kcat values (n = 11,480 reactions) matched to TGrowth. (B) Reactions with more than one enzyme variant (n = 2223 reactions). (C) Rate ratio distribution of the rate at the coldest TGrowth (kcold) divided by the rate of the variant from the warmest TGrowth (kwarm) (median = 1.1-fold, 95% CI [1.00, 1.22], n = 2223 reactions). Vertical line at rate ratio = 1. For clarity, only data with rate ratios between 10–3 and 103 are shown (>95% of the reactions). (D) Rate ratio (kcold/kwarm) data (solid line, n = 2223 from panel C) compared to fold change control distribution (same TGrowth; dashed line, median = 1.0-fold, 95% CI [0.89, 1.13], n = 615 reactions; p = 0.21, Mann–Whitney U test, two-sided). The black vertical line represents no rate enhancement change with temperature (i.e., rate ratio = 1). (E, F) The significance and magnitude of the linear fit of reaction rate as a function of TGrowth for negative slopes (E, n = 487) and positive slopes (F, n = 464) in log space. Enzyme Commission (E.C.) number and (substrate) indicated for reactions significantly associated with temperature (Bonferroni correction; p-value < 5.3 × 10–5, n = 951). Dotted horizontal lines at p = −log10(5.3 × 10–5). 5.3.1.1: triose-phosphate isomerase; G3P: glyceraldehyde 3-phosphate; 3.1.1.74: cutinase; 4-NPB: 4-nitrophenyl butyrate.
-
Figure 2—source code 1
Retrieval of updated BRENDA enzyme entries.
- https://cdn.elifesciences.org/articles/72884/elife-72884-fig2-code1-v1.zip
-
Figure 2—source code 2
Analysis of BRENDA enzyme rate constant entries.
- https://cdn.elifesciences.org/articles/72884/elife-72884-fig2-code2-v1.zip
-
Figure 2—source code 3
Control analysis of temperature-matched BRENDA enzyme rate constant entries.
- https://cdn.elifesciences.org/articles/72884/elife-72884-fig2-code3-v1.zip
-
Figure 2—source data 1
Downloaded BRENDA enzyme entries (July 2021).
- https://cdn.elifesciences.org/articles/72884/elife-72884-fig2-data1-v1.csv
-
Figure 2—source data 2
Analyzed BRENDA enzyme entries.
- https://cdn.elifesciences.org/articles/72884/elife-72884-fig2-data2-v1.csv
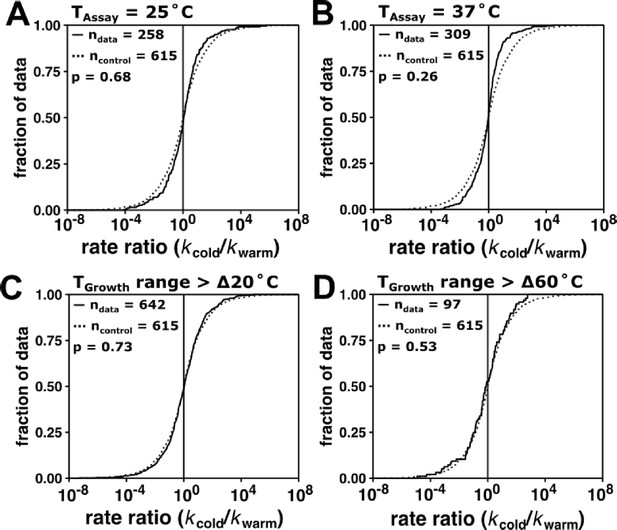
Specifying assay temperature and organism optimal growth temperature range per reaction does not alter conclusions.
(A) Distribution of kcat rate ratio values including only measurements made at 25°C and (B) 37°C. (C) Distribution of rate ratios with TGrowth range >∆20°C and (D) TGrowth range >60°C. Reported p-values from two-sided Mann–Whitney U test comparing filtered data (solid line) and the control data (dotted line, see Materials and methods).
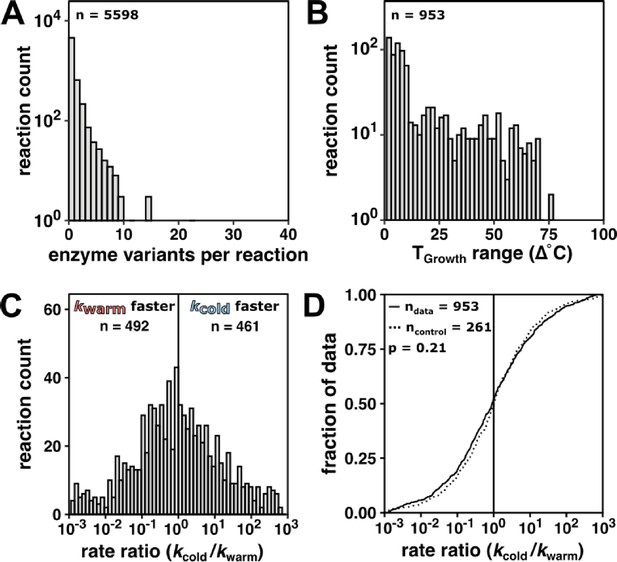
Enzyme rate constant data for kcat/KM do not indicate rate compensation, supporting the conclusions from the kcat analysis in the main text.
(A) Variants per reaction of wild-type enzyme kcat values (n = 5598 reactions) matched to TGrowth. (B) Number of reactions spanning the specified TGrowth range (n = 953 reactions with >1 variant). (C) kcat/KM rate ratio (kcold/kwarm) distribution (median = 0.93-fold, 95% CI [0.78, 1.12], n = 953 reactions). Gray vertical line at rate ratio = 1. (D) kcat/KM rate ratio (kcold/kwarm) data (black line, n = 953 reactions) with kcat/KM rate ratio control (gray line, median = 1.00-fold, 95% CI [0.82, 1.21], n = 307 reactions) determined in the same way as the kcat rate ratio control in the main text (see Materials and methods) (p = 0.80, Mann–Whitney U test, two-sided). For clarity, only data with rate ratios between 10–3 and 103 are shown, representing >90% rate ratio data in (C) and >83% of rate ratio control values in (D). Black vertical line represents no rate change with temperature (i.e., rate ratio = 1).
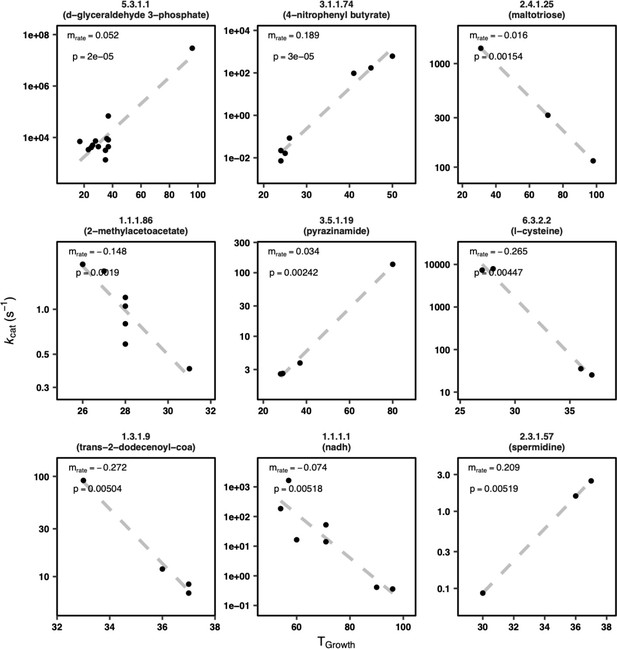
Example mrate plots (9 of 951 reactions shown).
Reactions with the rate constants of constituent variants in order of mrate p-value (for all reactions shown, p < 5.2 × 10–3). mrate is the slope of log10(kcat) vs. TGrowth. Note different scales for the axes. 5.3.1.1: triose-phosphate isomerase; 3.1.174: cutinase; 2.4.1.25: 4-alpha-glucanotransferase; 1.1.1.86: ketol-acid reductoisomerase; 3.5.1.19: nicotinamidase; 6.3.2.2: glutamate-cysteine ligase; 1.3.1.9: enoyl-[acyl-carrier-protein] reductase (NADH); 1.1.1.1: alcohol dehydrogenase; 2.3.1.57: diamine N-acetyltransferase.
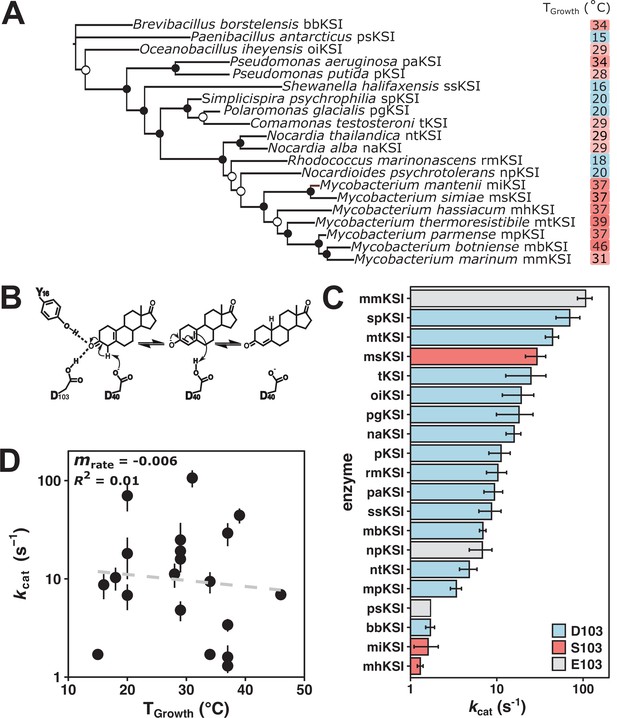
Ketosteroid isomerase (KSI) rate enhancements do not indicate rate compensation.
(A) Unrooted maximum likelihood phylogenetic tree of KSI variants. Closed circles represent bootstrap values of >70%; open circles represent bootstrap values of 40–70%. (B) The mechanism of isomerization of the steroid 5 (10)-estrene-3,17-dione by KSI. 5 (10)-EST was used to allow direct measurement of the rate-limiting chemical step kcat (Pollack et al., 1986). (C) Activity of KSI variants (kcat) at a common assay temperature of 25°C. Error bars represent standard deviation of at least two different experimental replicates varying [E] at least 5-fold. KSI variants with D103 are represented in blue, S103 in red, and E103 in gray (Pseudomonas putida numbering throughout). (D) Activity (log10(kcat)) of KSI variants at a common assay temperature (25°C) vs. organism growth temperature (TGrowth) (n = 20, mrate = –0.006, R2 = 0.01, p = 0.02).
-
Figure 3—source data 1
Ketosteroid isomerase (KSI) origins and organism growth temperatures.
a (Engqvist, 2018). b Alternatively reported to grow optimally at 65°C (Schröder et al., 1997). For consistency, curated values from Engqvist, 2018, are used in this work.
- https://cdn.elifesciences.org/articles/72884/elife-72884-fig3-data1-v1.docx
-
Figure 3—source data 2
Kinetic measurement of ketosteroid isomerases (KSIs) at 25°C with substrate 5 (10)-estrene-3,17-dione.
a (Engqvist, 2018). b Reported assay temperatures are the average of at least three measurements per experiment. c aAverage ± standard deviation from two to nine independent experiments with enzyme concentration varied by at least 5-fold. Values measured with substrate concentrations from 9 to 600 µM. Value of kcat/KM are less than 107 M–1 s–1 and thus unlikely to be limited by substrate binding. Reported assay temperatures are the average of at least three measurements per experiment.
- https://cdn.elifesciences.org/articles/72884/elife-72884-fig3-data2-v1.pdf
-
Figure 3—source data 3
Kinetic measurement of ketosteroid isomerases (KSIs) at 15°C with substrate 5 (10)-estrene-3,17-dione.
a (Engqvist, 2018). b Reported assay temperatures are the average of at least three measurements per experiment. c Average ± standard deviation from two to four independent experiments with enzyme concentration varied by at least 2-fold. Values measured with substrate concentrations from 9 to 600 µM.
- https://cdn.elifesciences.org/articles/72884/elife-72884-fig3-data3-v1.pdf
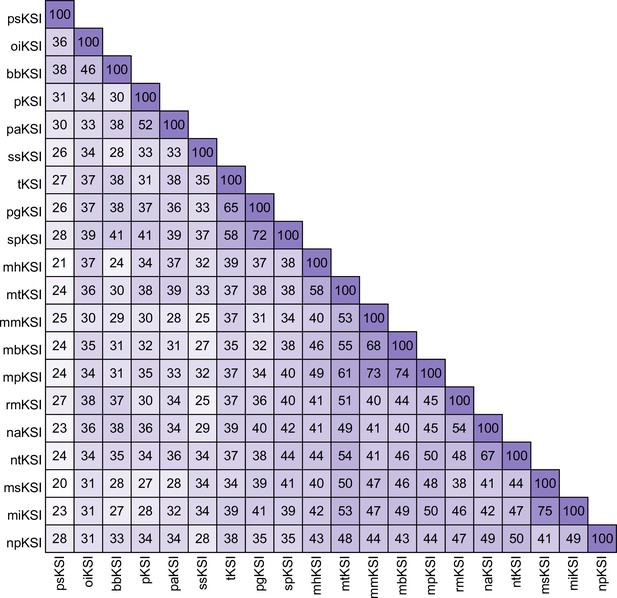
ketosteroid isomerase (KSI) variant similarity.
The primary sequence variation of each KSI variant ranges from 20% to 75% amino acid identity.
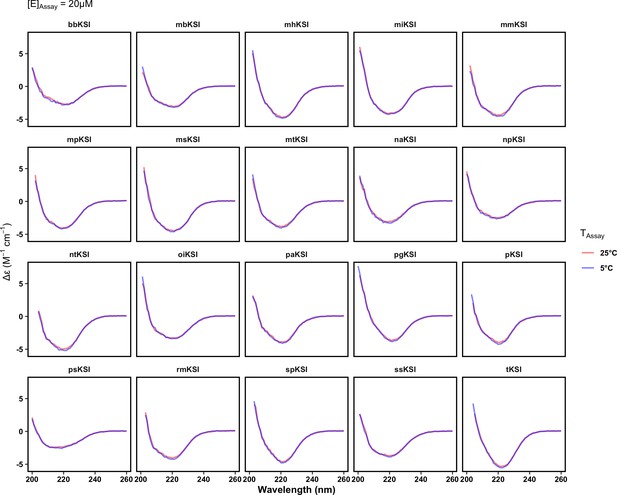
ketosteroid isomerase (KSI) variant circular dichroism (CD) spectra are indistinguishable at cold and warm temperature.
Far ultraviolet CD spectra at 5°C (blue) and 25°C (red) are indistinguishable. Measurements for each variant were made at an enzyme concentration of 20 µM.
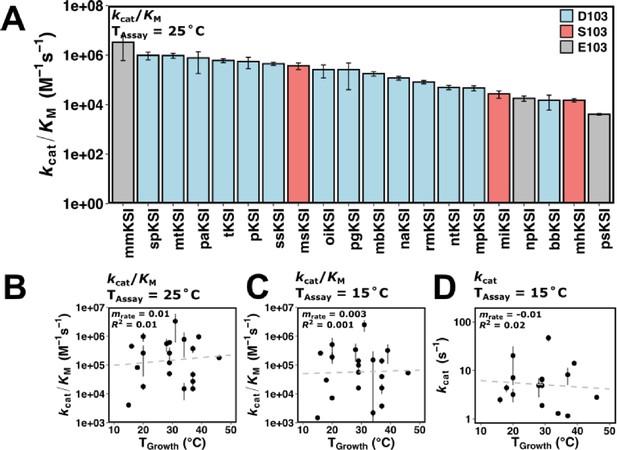
Ketosteroid isomerase (KSI) rate enhancements vary with organism growth temperature in kcat and in kcat/KM.
(A) Rate of KSI variants (kcat/KM) at a common assay temperature (TAssay) of 25°C. KSI variants with D103 are represented in blue, S103 in red, and E103 in gray (Pseudomonas putida numbering). (B) Rates (kcat/KM) of KSI variants at 25°C assay temperature (TAssay) vs. organism growth temperature (TGrowth) (n = 20, mrate = 0.01, R2 = 0.01, p = 4 × 10–7). (C) Rates (kcat/KM) of KSI variants at 15°C assay temperature (TAssay) vs. organism growth temperature (TGrowth) (n = 20, mrate = 0.003, R2 = 0.001, p = 3 × 10–7). (D) Rates (kcat) of KSI variants at 15°C assay temperature (TAssay) vs. organism growth temperature (TGrowth) (n = 20, mrate = –0.01, R2 = 0.02, p = –0.11). Error bars represent standard deviation of at least two different experimental measurements varying [E] at least 5-fold (25°C) or 2-fold (15°C).
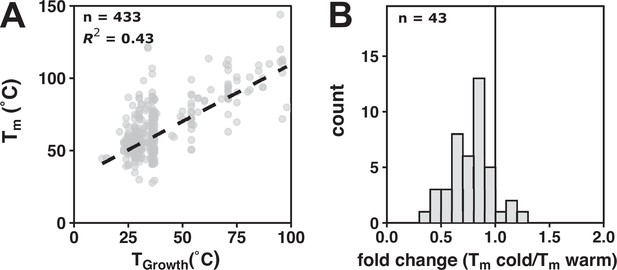
Protein stability data display stability compensation.
(A) Wild-type Tm stability data from ProThermDB as a function of organism TGrowth. Dashed black line represents a linear fit (n = 433, R2 = 0.43). (B) Fold change (Tm cold/Tm warm) of wild-type protein variants (n = 43, median = 0.81, 95% [0.70, 0.85]). The black vertical line represents no change (i.e., fold change = 1).
-
Figure 4—source data 1
ProThermDB wild-type protein stability entries.
- https://cdn.elifesciences.org/articles/72884/elife-72884-fig4-data1-v1.csv
-
Figure 4—source data 2
ProThermDB wild-type protein stability entries of protein families with more than one reported variant.
- https://cdn.elifesciences.org/articles/72884/elife-72884-fig4-data2-v1.csv
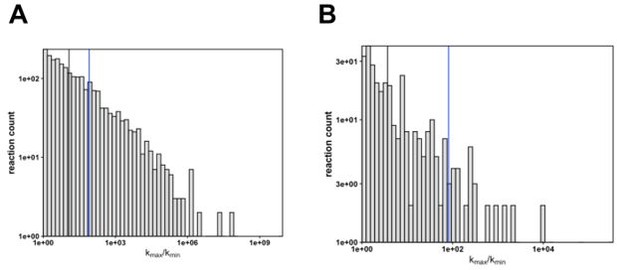
Differences in rate constants between enzyme variants.
(A) The fold change in kcat (kmax/kmin) of enzyme reactions (n = 2223). The black line indicates the fold change median value (median kmax/kmin = 11.8) and the blue line indicates the fold change value of KSI (kmax/kmin = 82). (B) The fold change in kcat (kmax/kmin) of enzyme reactions measured at 37°C (n = 319). The black line indicates the fold change median value (median kmax/kmin = 3.7) and the blue line indicates the fold change value of KSI (kmax/kmin = 82); while the median is much smaller there remain a number of enzymes with fold change values similar or larger than the KSIs.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Escherichia coli) | BL21(DE3) | Sigma-Aldrich | CMC0016 | Electrocompetent cells for protein expression |
| Chemical compound, drug | 5 (10)-estrene-3,17-dione | Steraloids | E4500-000 | Ketosteroid isomerase reaction substrate |
Additional files
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/72884/elife-72884-transrepform1-v1.docx
-
Supplementary file 1
Overview of proposed molecular models of cold adaptation.
- https://cdn.elifesciences.org/articles/72884/elife-72884-supp1-v1.docx
-
Supplementary file 2
Ketosteroid isomerase (KSI) sequences.
- https://cdn.elifesciences.org/articles/72884/elife-72884-supp2-v1.docx





