Lytic transglycosylases mitigate periplasmic crowding by degrading soluble cell wall turnover products
Figures
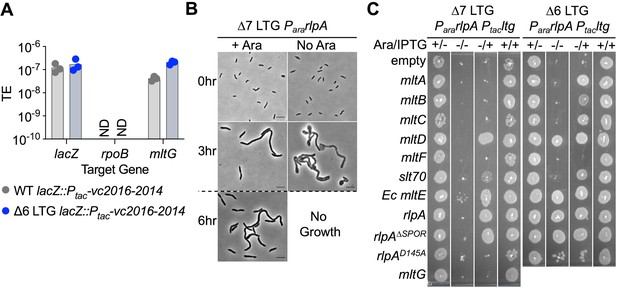
A single lytic transglycosylase (LTG) is necessary and sufficient for V. cholerae growth and envelope homeostasis.
(A) Trans expression of DNA synthesis genes vc2016-2014 permitted pAM299 disruption of native mltG locus. lacZ and rpoB were targeted as positive and negative controls for disruption, respectively. TE, transformation efficiency; ND, below limit of detection. Three biological replicates are shown. (B) RlpA was depleted from the WT, ∆6, and ∆7 LTG backgrounds by placing its native promoter under control of arabinose induction and growing from a 10–3 overnight culture dilution into 5 mL LB ± 0.4% arabinose (ara) at 37°C with shaking for 3 hr, back-diluting 10–3 into fresh media, and incubating for another 3 hr. Cells were imaged on LB agarose pads. Scale bars = 5 μm. Dotted line indicates 10–3 back-dilution. (C) Arabinose-dependent RlpA depletion in ∆6 and ∆7 LTG backgrounds was rescued with isopropyl-β-D-1-thiolgalactopyranoside (IPTG)-inducible LTGs by growing cultures in LB ± ara (0.4%) and ±IPTG (200 μM) in 96-well plates at 37°C without shaking for 3 hr, back-diluting 10–3 into fresh media, incubating another 3 hr, and spotting directly onto the same media + kan50. Plates were incubated at 30°C for 24 hr before imaging. Complete plating efficiencies associated with panels (B) and (C) can be found in Figure 1—figure supplement 2 and Figure 1—figure supplement 4, respectively. Images are representative of three biological replicates.
-
Figure 1—source data 1
Raw and uncropped mCherry Western blots.
- https://cdn.elifesciences.org/articles/73178/elife-73178-fig1-data1-v2.zip
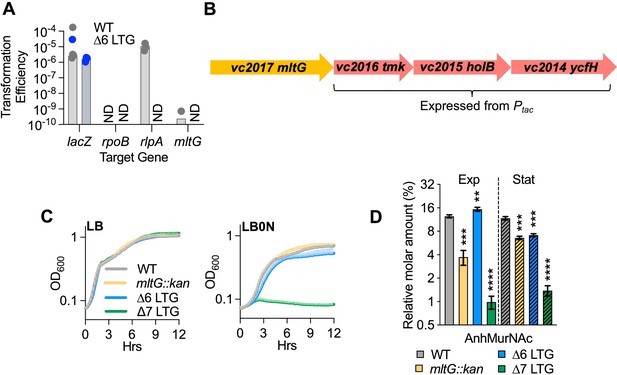
MltG is dispensable for growth of ∆6 lytic transglycosylase (LTG).
(A) Transformation efficiencies represent the strepR and kanR CFU divided by total viable strepR CFU recovered from a single mating. StreptomycinR (StrepR) V. cholerae WT and ∆6 LTG strains were mated with a strepS E. coli MFD donor strain carrying the pAM299 suicide vector carrying kanamycin resistance (KanR) and targeting positive control gene lacZ, negative control gene rpoB, rlpA, or mltG. (B) The genetic region downstream of mltG encodes essential DNA synthesis genes. (C) Mean growth curves for mltG::kan strains in WT Ptacvc2016-2014 and ∆6 LTG Ptacvc2016-2014 backgrounds diluted 1:100 from overnight cultures into LB or LB0N and grown at 37°C. (D) Relative molar abundance of all anhMurNAc-containing muropeptide species in sacculus. Overnight cultures of WT, mltG::kan, ∆6 LTG, and ∆6 LTG mltG::kan (∆7 LTG) in lacZ::Ptacvc2016-2014 backgrounds were diluted 1:100 into LB + 200 μM IPTG and samples collected for HPLC analysis at OD600 0.3 (Exp, solid bars) and OD600 1.2 (Stat, striped bars). Complete muropeptide profiles can be found in Supplementary file 1. Error bars represent standard deviation from three biological replicates. Mutants were compared to the WT within each growth phase with an unpaired t-test. **p<0.01, ***p<0.001, ****p<0.0001.
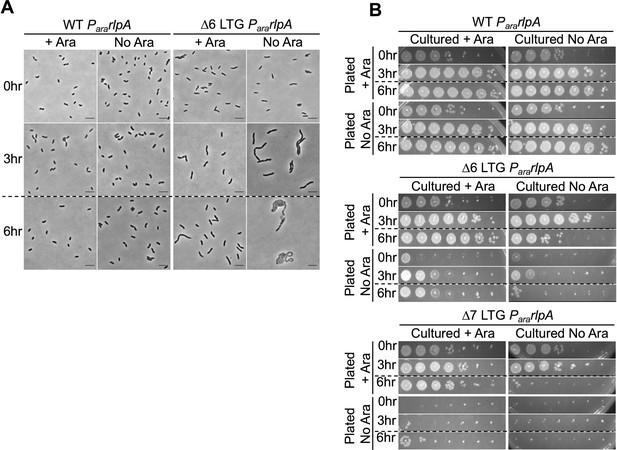
RlpA depletion in the ∆6 and ∆7 lytic transglycosylase (LTG) backgrounds.
Native RlpA was depleted from the WT, ∆6, and ∆7 LTG backgrounds as described in Figure 1B, (A) imaged on LB agarose pads and (B) spot plated in 10-fold serial dilutions (100–10–6) onto LB + kan50 ± 0.4% arabinose and incubated at 30°C for 24 hr before imaging.
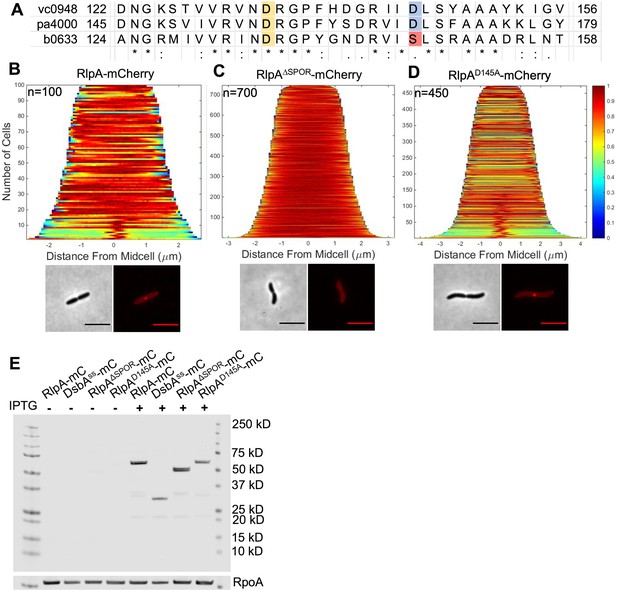
RlpA active site architecture and localization.
(A) Clustal Omega alignment of a relevant portion of lyase domain from V. cholerae RlpA (vc0948), P. aeruginosa RlpA (pa4000), and E. coli RlpA (b0633). Two Asp residues are implicated in PaRlpA activity: PaRlpA D156 aligns with VcRlpA D133 and EcRlpA D135, and PaRlpA D168 aligns with VcRlpA D145 and EcRlpA S147. WT V. cholerae carrying (B) pHL100 rlpA-mCherry (C) rlpA∆SPOR-mCherry or (D) rlpAD145A-mCherry was grown in M9 + 0.2% glucose at 30°C for 2 hr, induced with 1 mM isopropyl-β-D-1-thiolgalactopyranoside (IPTG), and imaged on M9 + 0.2% glucose agarose pads at OD600 ~ 0.2. Demographs were generated using Oufti. Panels (B–D) are representative of two biological replicates. Scale bars = 5 μm. (E) mCherry (mC) fusion proteins were detected with mCherry antibody, with mCherry fused to a DsbA signal sequence serving as a soluble mCherry size standard. RpoA loading control detected with RpoA antibody. Raw and unedited blots can be found in Figure 1—source data 1.
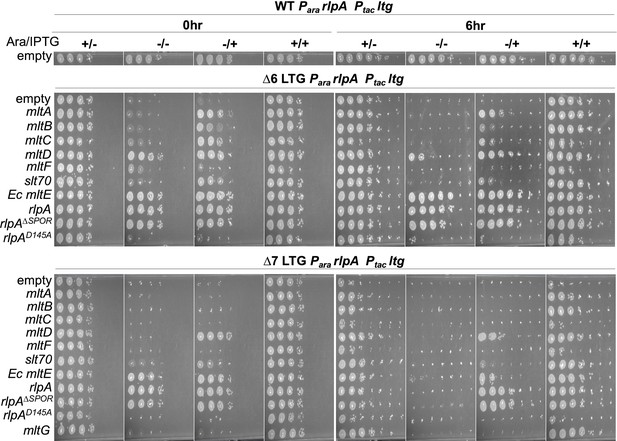
Lytic transglycosylases (LTGs) have variable ability to sustain growth.
Native RlpA depletion in ∆6 and ∆7 LTG backgrounds was rescued by single LTGs as described in Figure 1B, spot plated in 10-fold serial dilutions (from 100 to 10–6) at 0 hr and 6 hr, then incubated at 30°C for 24 hr before imaging. Images are representative of three biological replicates.
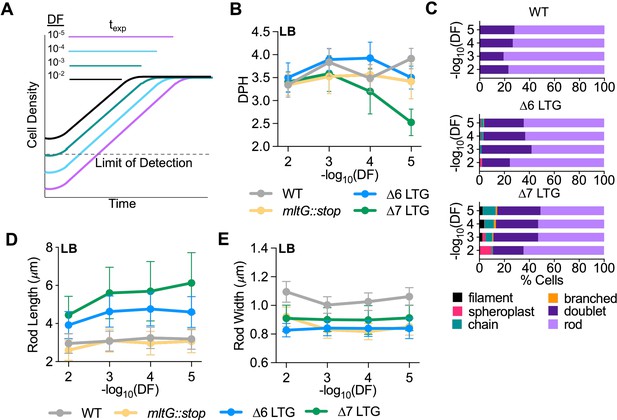
Lytic transglycosylase (LTG) insufficiency causes cumulative growth and morphology defects.
(A) Schema describing relationship between dilution factor (DF) of saturated cultures into fresh media and time spent in exponential growth (texp). (B) Mass doubling times (doublings per hour [DPH]) from growth curves performed in LB inoculated with 10-fold serial dilutions of saturated overnight cultures. Values were calculated from growth curves shown in Figure 2—figure supplement 2. Error bars represent standard deviation of the mean, n ≥ 3. (C) Relative abundance of cell morphologies from cultures at OD600 0.3 from panel (C). n > 500 cells. Definition criteria and images are shown in Figure 2—figure supplement 1. Mean length (D) and width (E) as a function of DF of rod cells from panel (C). n > 500 rods.
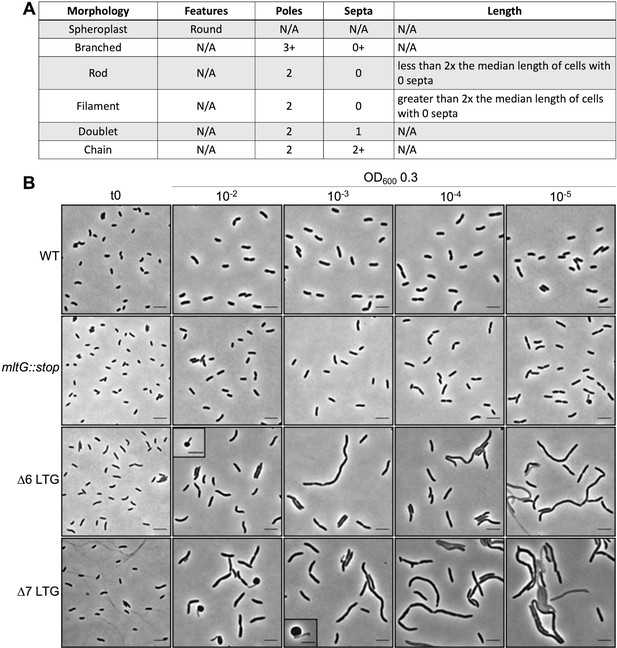
Definitions of morphology defect categories.
(A) Cell morphology classification criteria. (B) Representative micrographs of cell morphologies where each cell type depicted accounts for >1% of population as sampled from cultures grown in LB to OD600 0.3 from 10-fold serial dilutions of overnight cultures. Scale bars = 5 μm.
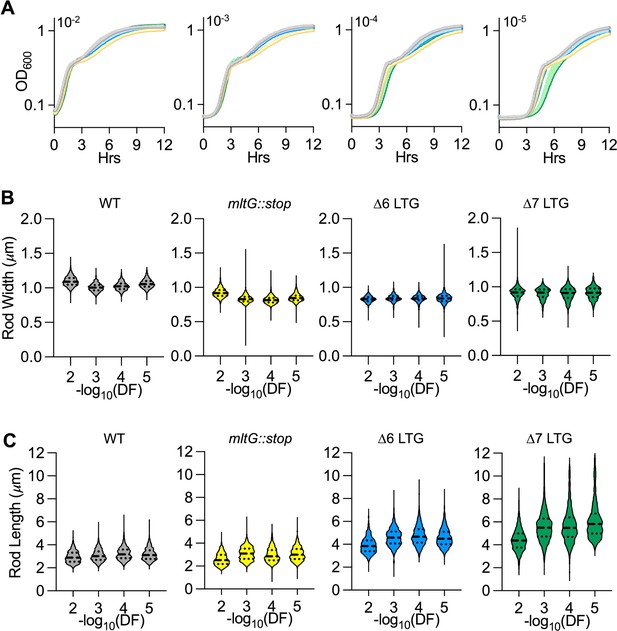
Quantitative growth and morphology of lytic transglycosylase (LTG)-deficient mutants in LB during extended exponential phase.
(A) Growth curves were performed in LB at 37°C inoculated with 10-fold serial dilutions of saturated overnight cultures. Violin plots of raw (B) mean width or (C) length of single rods. n > 500 single rods. Error bars represent standard deviation of the mean, n ≥ 3. DF, dilution factor.
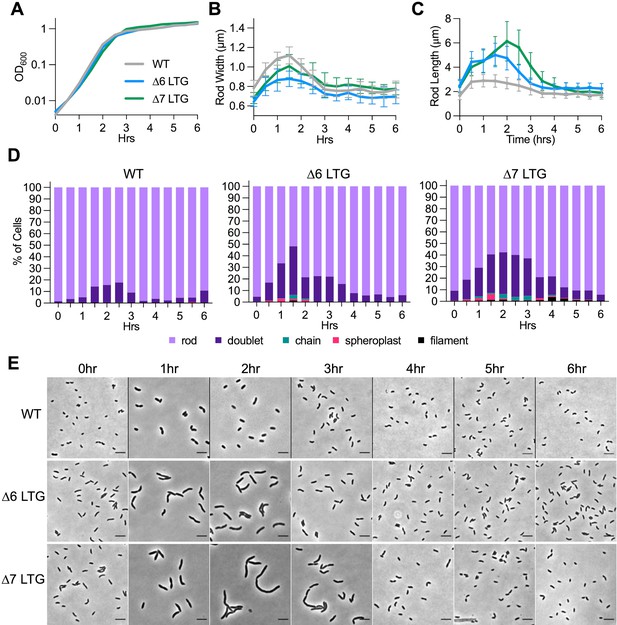
Morphology of lytic transglycosylase (LTG)-deficient mutants in LB is growth-phase dependent.
Saturated overnight cultures were diluted 1/1000 into LB at 37°C and periodically sampled for (A) OD600 and imaged on LB agarose pads for (B) mean width, (C) length, and (D) cell morphology as exhibited in (E) representative micrographs. Error bars in (B) and (C) represent standard deviation of the mean, n > 100 single rods. n > 200 cells for (D). Scale bars = 5 μm.
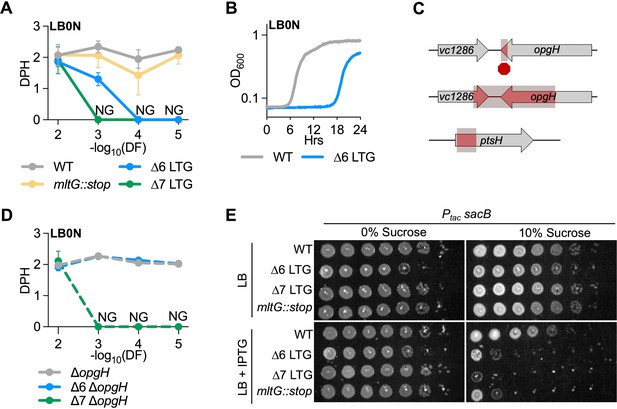
Lytic transglycosylase (LTG) mutants are hypersensitive to low osmolarity and accumulation of periplasmic polymers.
(A) Mass doublings (doublings per hour [DPH]) upon increasing dilutions (dilution factor [DF]) during growth in LB0N. Values were calculated from growth curves in Figure 3—figure supplement 1. Error bars represent standard deviation of the mean, n ≥ 3. NG, no growth. (B) Representative growth curve in low-salt LB (LB0N) showing late-growing spontaneous suppressor in ∆6 LTG. (C) Whole-genome sequencing of ∆6 LB0N suppressor mutations identifies a premature stop, resulting in a 6% 3′ truncation of opgH; a deletion resulting in a 56% 3′ truncation of opgH and 36% 3′ truncation of vc1286; and a deletion of the 5′ end of ptsH. (D) Validation of low osmolarity growth defect suppression by an opgH mutation. Shown are mass doublings (DPH) upon increasing dilutions (DF) during growth in LB0N. Values were calculated from growth curves shown in Figure 3—figure supplement 1. (E) Saturated overnight cultures harboring isopropyl-β-D-1-thiolgalactopyranoside (IPTG)-inducible sacB were 10-fold serially diluted and plated on LB + kan50 ± 200 μM IPTG ± 10% sucrose, incubated at 30°C, and imaged 24 hr before. Representative of three biological replicates. Empty vector and LB0N controls are shown in Figure 3—figure supplement 4.
-
Figure 3—source data 1
Raw growth curve data for single lytic transglycosylase (LTG) mutants and LTG complementation in LTG-deficient mutants.
Raw OD600 measurements for growth curves performed in LB and low-salt LB (LB0N) with and without induction of trans-complementation by single LTGs in LTG-deficient mutant backgrounds, as well as growth curves in LB and LB0N of single LTG-deficient mutants, are presented. n ≥ 3.
- https://cdn.elifesciences.org/articles/73178/elife-73178-fig3-data1-v2.xlsx
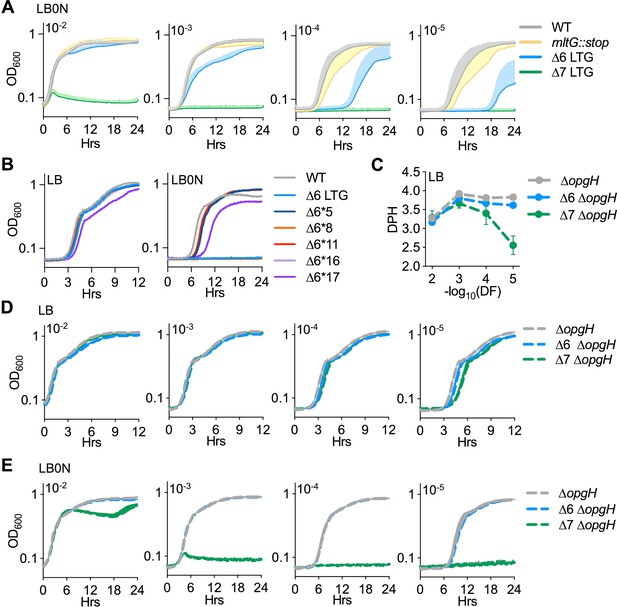
Suppression of growth defects of ∆LTG mutants in low-salt LB.
(A) Mean growth curves in LB0N at 37°C started from 10-fold dilutions of saturated overnight cultures used to generate growth rates in Figure 3A. Error bars, positive standard deviation of the mean, n > 3. Growth plot on the right is reproduced from Figure 3 for comparison. (B) Suppressors of the ∆6 lytic transglycosylase (LTG) LB0N growth defect were isolated on LB and then tested for growth in LB and LB0N from a 10–5 inoculum of saturated overnight (LB) culture. (C) Mass doubling times of ∆opgH mutants calculated from (D) growth curves of ∆opgH mutants in LB at 37°C inoculated with 10-fold serial dilutions of saturated overnight cultures. (E) Mean growth curves in LB0N at 37°C started from 10-fold dilutions of saturated overnight cultures used to generate growth rates in Figure 3D. Error bars, standard deviation of the mean, n = 3.
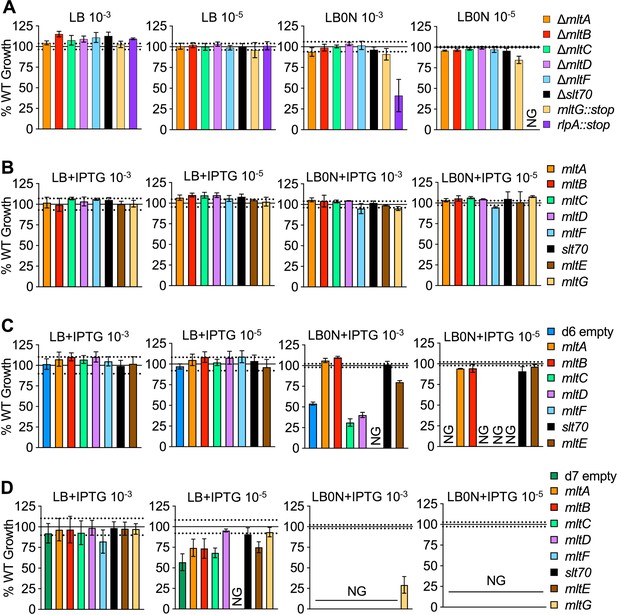
Single LTG contributions to growth in LB and low salt LB.
Mass doubling times of strains diluted 10-3 or 10-5 into LB and LB0N (+ 200μM IPTG when induction was required) normalized to the appropriate WT controls under the same growth conditions. Solid line = WT mean, Dotted lines = +/- standard deviation of WT mean. (A) Single LTG mutant growth normalized to WT growth. (B) WT, (C) ∆6 LTG and (D) ∆7 LTG carrying chromosomal lacZ::Ptac ltg normalized to WT lacZ::Ptac empty. Experiments in panels (C) and (D) were carried out simultaneously and therefore share the same control replicates. Error bars represent standard deviation of 3 biological replicates. NG = No Growth. Raw curves for (A-D) and uninduced data for (B–D) available in Figure 3—source data 1.

Periplasmic glucans do not contribute to division defects in ∆LTG mutants.
(A) ∆opgH mutants in WT, ∆6, and ∆7 backgrounds were grown in LB at 37 °C to OD600 0.3 from 10-3 inoculum of saturated overnight culture, imaged on LB agarose pads, and single rod lengths analyzed in MicrobeJ. Dotted lines denote quartiles. Significance determined by unpaired Mann-Whitney tests, n>70 single rods. (B) T-Coffee alignment of predicted cytoplasmic N-terminal domains from E. coli OpgH (b0149), Y. pseudotuberculosis MdoH (yptb2493), and V. cholerae OpgH (vc1287).
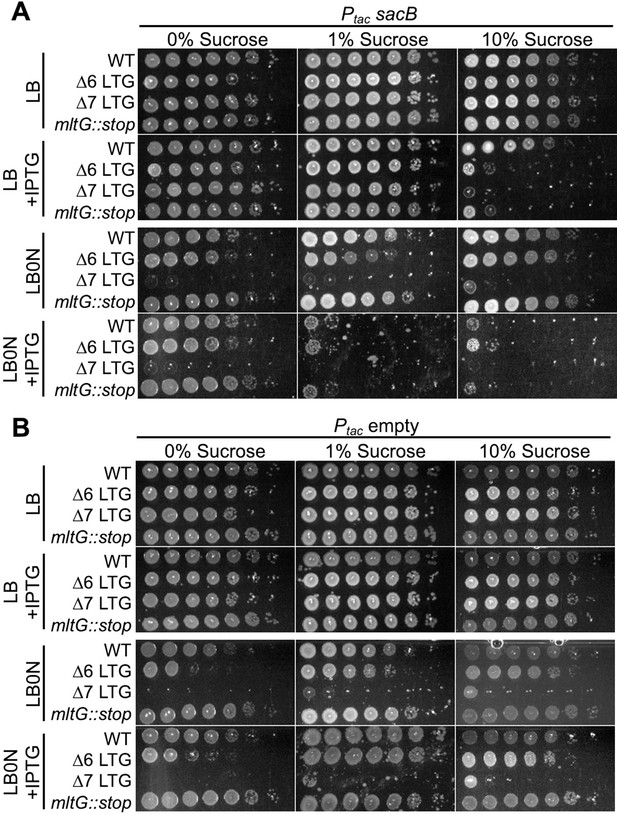
SacB expression is toxic in ∆LTG mutants in a sucrose-dependent manner.
Figure 3—source data 1Saturated overnight cultures harboring (A) IPTG-inducible sacB or (B) empty vector were 10-fold serially diluted (100-10-6) and plated on LB+kan50 and LB0N+kan50 +/- IPTG (200μM) + sucrose (0, 1, or 10% W/V) and incubated at 30 °C for 24 hrs before imaging. Representative of 3 biological replicates.
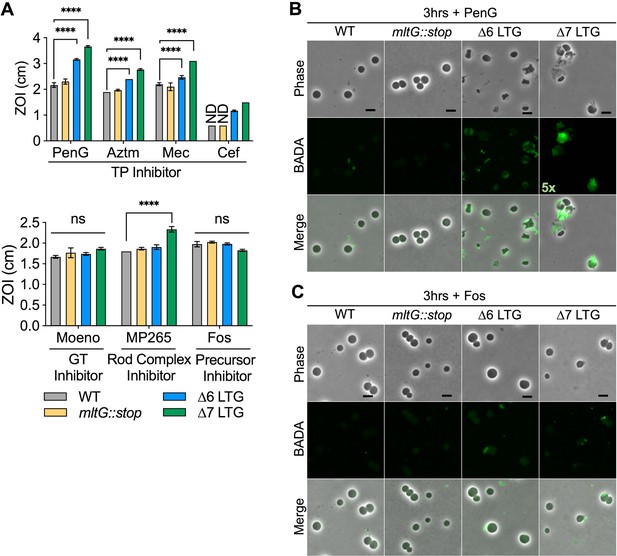
Lytic transglycosylase (LTG) mutants are hypersensitive to antibiotics promoting periplasmic peptidoglycan (PG) accumulation.
(A) Sensitivity to Penicillin G (PenG), aztreonam (AZTM), mecillinam (Mec), cefsulodin (Cef), moenomycin (Moeno), MP265, and fosfomycin (Fos) measured as zone of inhibition (ZOI) in a disk diffusion assay. ND, no ZOI around disk. Error bars, standard deviation. Significance determined by one-way ANOVA. ns, p>0.05; *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001. Overnight cultures were diluted 1:100 into LB + BADA (100 μM) and grown at 37°C to OD600 0.5 before addition of (B) PenG (100 μg/mL) or (C) Fos (500 μg/mL). Resulting spheroplasts were washed and imaged after 3 hr of antibiotic exposure. Fluorescence was normalized to the same intensity threshold for visual comparison except where indicated (exceptionally bright samples were normalized to a higher-intensity threshold denoted by the multiplier). Representative of two biological replicates. Scale bar = 5 μm.
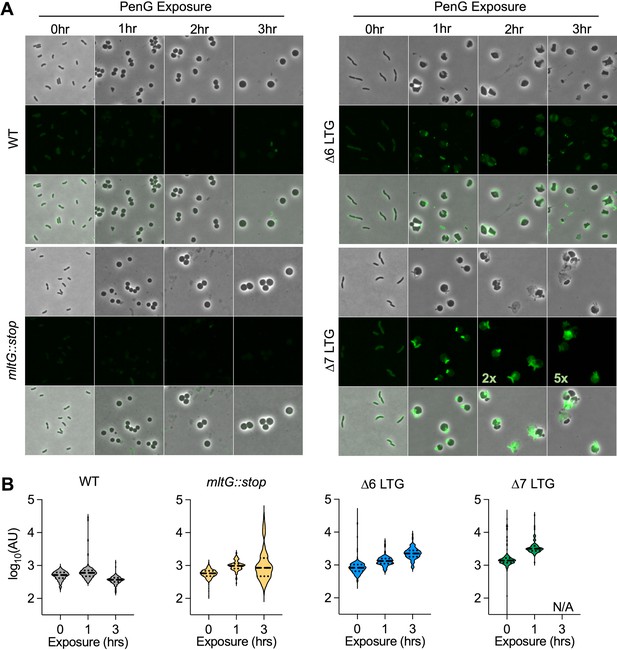
Periplasmic cell wall accumulation in response to Penicillin G (PenG).
Overnight cultures were diluted 1:100 into LB + BADA (100 μM) and grown at 37°C to OD600 0.5 before addition of PenG (100 μg/ml). Resulting spheroplasts were removed periodically, washed, and imaged on LB agarose. (A) Fluorescence was normalized to the same intensity threshold in Figure 4—figure supplement 1 and Figure 4—figure supplement 2 for comparison except where indicated (exceptionally bright samples were normalized to a higher-intensity threshold denoted by the multiplier). Representative of two biological replicates. Scale bar = 5 μm. (B) Mean fluorescence intensity (AU) for >100 cells or spheroplasts was measured in ImageJ. N/A, not applicable due to insufficient intact spheroplasts for measurement.
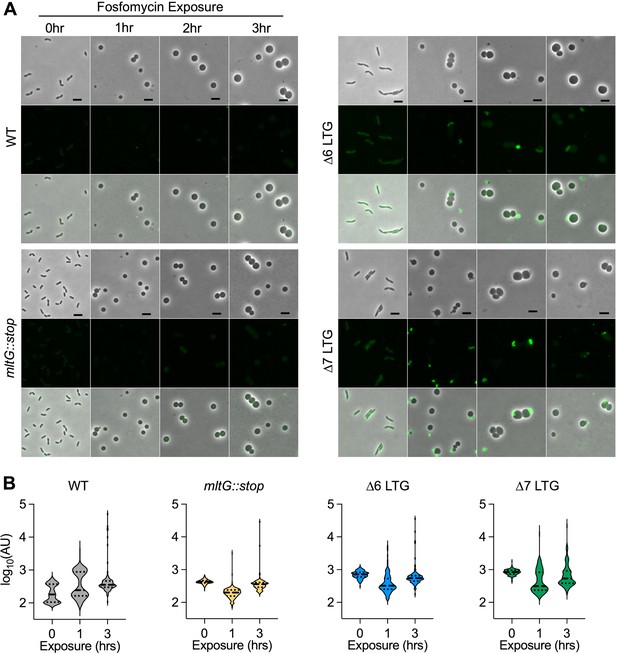
Lack of periplasmic cell wall accumulation in response to fosfomycin.
Overnight cultures were diluted 1:100 into LB + BADA (100 μM) and grown at 37°C to OD600 0.5 before addition of fosfomycin (500 μg/mL). Resulting spheroplasts were removed periodically, washed, and imaged on LB agarose. (A) Fluorescence was normalized to the same intensity threshold in Figure 4—figure supplement 1 and Figure 4—figure supplement 2 for comparison. Representative of two biological replicates. Scale bar = 5 μm. (B) Mean fluorescence intensity (AU) for >100 cells or spheroplasts was measured in ImageJ.
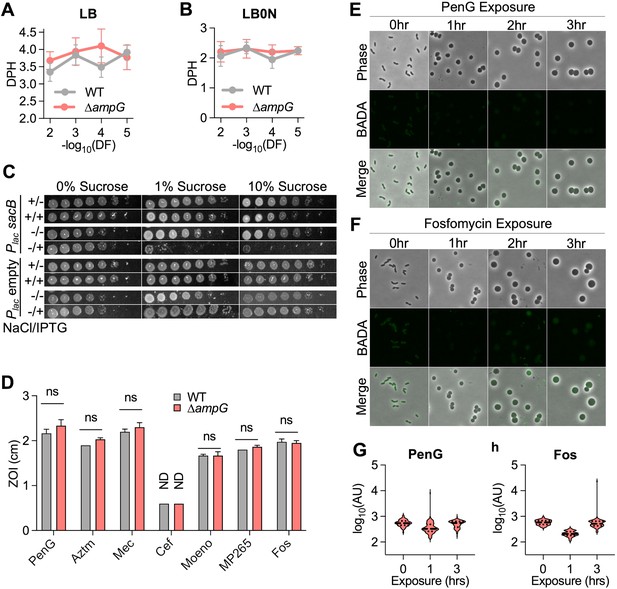
Defects of lytic transglycosylase (LTG)-deficient mutants are independent of peptidoglycan (PG) recycling.
∆ampG data for this figure was collected during the same experiments in Figures 2A and 3A–F; therefore, WT data are reproduced from those figures and any statistical tests shown here were performed on the complete datasets for those experiments (including WT, ∆ampG, and ∆LTG strains). (A, B) Mass doubling times from growth curves performed in LB or LB0N inoculated with 10-fold serial dilutions (100–10–6) of saturated overnight cultures. Error bars represent standard deviation of the mean, n ≥ 3. (C) Saturated overnight cultures harboring isopropyl-β-D-1-thiolgalactopyranoside (IPTG)-inducible sacB or empty vector were 10-fold serially diluted (10 and plated on LB + kan50) and LB0N + kan50 ± IPTG (200 μM) + sucrose (0, 1, or 10% W/V) and incubated at 30°C for 24 hr before imaging. Representative of three biological replicates. (D) Sensitivity to Penicillin G (PenG), aztreonam (AZTM), mecillinam (Mec), cefsulodin (Cef), moenomycin (Moeno), MP265, and fosfomycin (Fos) measured as zone of inhibition (ZOI) in a disk diffusion assay. ND, no ZOI around disk. Error bars, standard deviation. Significance determined by one-way ANOVA. ns, p>0.05. Overnight cultures were diluted 1:100 into LB + BADA (100 μM) and grown at 37°C to OD600 0.5 before addition of (E) PenG (100 μg/mL) or (F) Fos (500 μg/mL). Samples were washed and imaged after 3 hr of antibiotic exposure. Fluorescence was normalized to the same intensity threshold as Figure 4—figure supplement 1 and Figure 4—figure supplement 2 for comparison. Representative of two biological replicates. Scale bar = 5 μm. (G, H) Mean fluorescence intensity (AU) for >100 BADA-labeled cells or spheroplasts were measured in ImageJ.
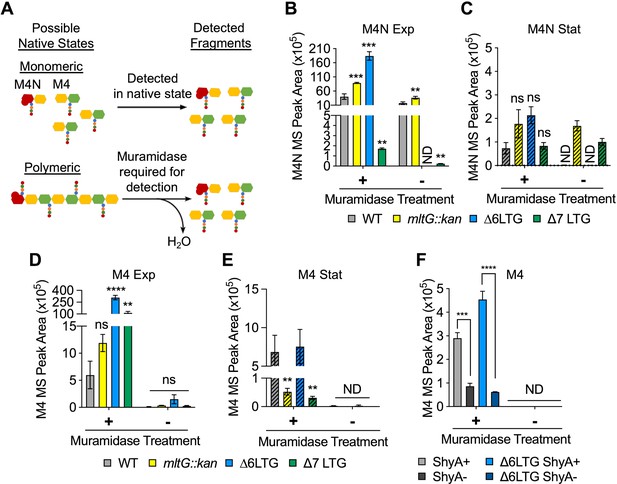
Periplasmic uncrosslinked peptidoglycan (PG) strands accumulate in an endopeptidase-dependent manner during normal growth.
(A) Schema describing muramidase treatment dependence of detection of monomeric or polymeric PG fragments. M4N, anhMurNAc-tetrapeptide; M4, reduced MurNAc-tetrapeptide. (B–E) Overnight cultures of. WT and ∆LTG mutants were diluted 1:100 into LB, grown at 37°C, and harvested at OD600 0.3 (Exp, solid bars) and 1.2 (Stat, striped bars) for soluble PG analysis by LC-MS. MS peak areas for M4N and M4 are shown here, and complete muropeptide profiles can be found in Supplementary file 2. Means compared to WT by unpaired t-tests, n = 3. (F) WT and ∆6 LTG strains harboring a single chromosomal copy of shyA under an isopropyl-β-D-1-thiolgalactopyranoside (IPTG)-inducible promoter were grown from 10–2 inocula for 3 hr (OD600 ~ 1.0) in LB with (ShyA+) or without (ShyA -) 200 μM IPTG at 37°C and harvested for soluble PG analysis by LC-MS. Complete muropeptide profiles can be found in Supplementary file 3. Means compared by unpaired t-test, n = 3. All error bars = standard deviation. ns, p>0.01; **p<0.01; ***p<0.001; ****p<0.0001. ND, not detected in all replicates.
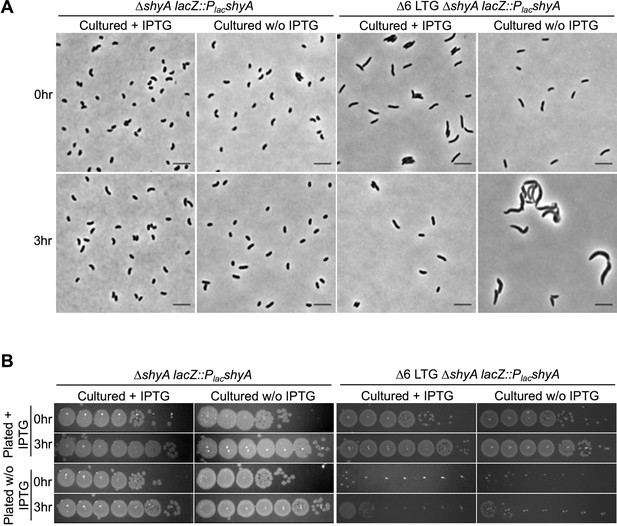
Endopeptidase depletion phenotypes in lytic transglycosylase (LTG)-deficient mutants.
WT and ∆6 LTG strains harboring a single chromosomal copy of shyA under an isopropyl-β-D-1-thiolgalactopyranoside (IPTG)-inducible promoter were grown from 10–2 inocula for 3 hr (OD600 ~1.0) in LB ± IPTG (200 μM) at 37°C. Samples were (A) imaged on LB + agarose pads and (B) spot plated in 10-fold serial dilutions (100–10–6) onto LB ± IPTG (200 μM), then incubated at 30°C for 24 hs before imaging. Representative of three biological replicates.
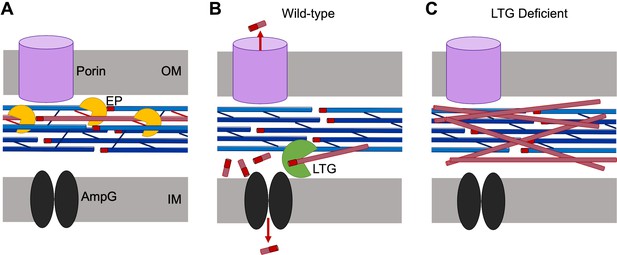
Model for lytic transglycosylase (LTG)-mediated removal of toxic peptidoglycan (PG) debris.
(A) Endopeptidases (EPs, yellow) excise PG strands (red) from the sacculus (blue), permitting sacculus expansion. (B) In wild-type cells, LTGs (green) digest excised, uncrosslinked PG strands into smaller fragments that can be recycled by AmpG (black) or released through porins (violet). (C) In LTG-deficient cells, excised PG debris crowds the periplasm and becomes toxic.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene(Vibrio cholerae) | mltA | UniProtKB | vc_2312 | |
| Gene(V. cholerae) | mltB | UniProtKB | vc_1956 | |
| Gene(V. cholerae) | mltC | UniProtKB | vc_0450 | |
| Gene(V. cholerae) | mltD | UniProtKB | vc_2237 | |
| Gene(V. cholerae) | mltF | UniProtKB | vc_0866 | |
| Gene(V. cholerae) | mltG | UniProtKB | vc_2017 | |
| Gene(V. cholerae) | rlpA | UniProtKB | vc_0948 | |
| Gene(V. cholerae) | slt70 | UniProtKB | vc_0700 | |
| Gene(V. cholerae) | shyA | UniProtKB | vc_a0079 | |
| Gene(V. cholerae) | opgH | UniProtKB | vc_1287 | |
| Gene(V. cholerae) | ampG | UniProtKB | vc_2300 | |
| Gene(V. cholerae) | lacZ | Kegg2 Gene DB | vc_2338 | |
| Gene (Escherichia coli) | EcrlpA | UniProtKB | b0633 | |
| Gene (E. coli) | EcmltE | UniProtKB | b1193 | |
| Gene(Pseudomonas aeruginosa) | ParlpA | UniProtKB | pa4000 | |
| Gene(Bacillus subtilis) | sacB | UniProtKB | bsu34450 | |
| Strain, strain background(V. cholerae) | El Tor N16961; wild-type | PMID:243277 | ||
| Recombinant DNA reagent | ∆mltABCDF ∆slt70; ∆6 LTG | PMID:31286580 | See Supplementary file 4; request from Doerr Lab | |
| Recombinant DNA reagent | ∆mltABCDF ∆slt70 mltG::stop;∆7 LTG | This study | See Supplementary file 4; request from Doerr lab | |
| Recombinant DNA reagent | mltG::stop | This study | See Supplementary file 4; request from Doerr Lab | |
| Recombinant DNA reagent | pCVD442 | PMID:15109831 | ||
| Recombinant DNA reagent | pTOX5 | PMID:31201277 | Addgene# 127450 | |
| Recombinant DNA reagent | pAM224; pGP704-kanR | PMID:2836362 | ||
| Recombinant DNA reagent | pAM299 | PMID:25631756 | ||
| Recombinant DNA reagent | pJL1 | PMID:24348240 | ||
| Antibody | Anti-mCherry (rabbit polyclonal) | GeneTex | GTX59788 | (1:5000) |
| Antibody | Goat anti-rabbit IRDye 800CW secondary | LI-COR | 926-32211 | (1:16,000) |
| Antibody | Anti-RpoA (mouse monoclonal) | BioLegend | 663104 | (1:10,000) |
| Antibody | Goat anti-mouse IRDye 800CW secondary | LI-COR | 926-32210 | (1:16,000) |
| Chemical compound, drug | Penicillin G potassium salt | Fisher Scientific | CAS:113-98-4 | |
| Chemical compound, drug | Aztreonam | Fisher Scientific | CAS:78110-38-0 | |
| Chemical compound, drug | Mecillinam | Sigma-Aldrich | CAS:32887-01-7 | |
| Chemical compound, drug | Cefsulodin sodium salt hydrate | TCI Chemicals | CAS:1426397-23-0 | |
| Chemical compound, drug | S-(4-Chlorobenzyl) Isothiouronium chloride (MP265) | Chem-Impex International | CAS:544-47-8 | |
| Chemical compound, drug | Phosphomycin disodium salt (fosfomycin) | Sigma-Aldrich | CAS:26016-99-9 | |
| Software, algorithm | Fiji | PMID:22743772 | ||
| Software, algorithm | MicrobeJ | PMID:27572972 | ||
| Software, algorithm | Oufti | PMID:26538279 | ||
| Other | BADA | PMID:28989665 |
Additional files
-
Supplementary file 1
Complete sacculus muropeptide composition profiles of lytic transglycosylase (LTG)-deficient mutants.
Supports Figure 1—figure supplement 1D. Presents mean relative muropeptide abundance ± standard deviation of the mean for all detectable muropeptide species from muramidase-treated sacculi isolated by ultracentrifugation from LTG-deficient mutant cultures during exponential and stationary growth phases. n = 3.
- https://cdn.elifesciences.org/articles/73178/elife-73178-supp1-v2.xlsx
-
Supplementary file 2
Complete muropeptide detection profiles of soluble peptidoglycan (PG) material from lytic transglycosylase (LTG)-deficient mutants.
Supports Figure 5B–E. Presents mean mass peak area ± standard deviation of the mean for all soluble, detectable muropeptide species from muramidase-treated and untreated lysates of LTG-deficient mutant cultures during exponential and stationary growth phases. n = 3.
- https://cdn.elifesciences.org/articles/73178/elife-73178-supp2-v2.xlsx
-
Supplementary file 3
Complete muropeptide detection profiles of soluble peptidoglycan (PG) material from ShyA-depleted mutants.
Supports Figure 5F. Presents mean mass peak area ± standard deviation of the mean for all soluble, detectable muropeptide species from muramidase-treated and untreated lysates of cultures grown with and without ShyA induction. n = 3.
- https://cdn.elifesciences.org/articles/73178/elife-73178-supp3-v2.xlsx
-
Supplementary file 4
Bacterial strains, plasmids, and primers used in this study.
- https://cdn.elifesciences.org/articles/73178/elife-73178-supp4-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/73178/elife-73178-transrepform1-v2.docx





