Combining genotypes and T cell receptor distributions to infer genetic loci determining V(D)J recombination probabilities
Figures
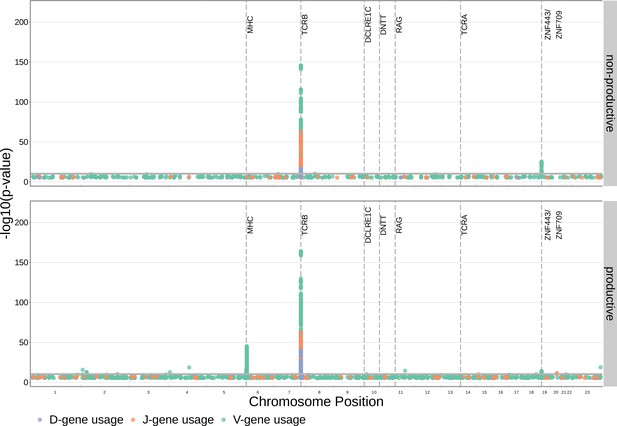
Many strong associations are present between V-, D-, and J-gene usage frequency and various SNPs genome-wide for both productive and non-productive TCRs.
The most significant SNP associations for the frequency of each of the 60 V-genes, 2 D-genes, and 14 J-genes are located within the TCRB and MHC loci. Associations are colored by gene-type instead of by gene identity for simplicity. Only SNP associations whose are shown here. The gray horizontal line corresponds to a Bonferroni-corrected p-value significance threshold of .
-
Figure 1—source data 1
There are 9152 significant associations between the frequency of various V-, D-, and J-genes and the genotype of SNPs genome-wide.
The model type and Bonferroni-corrected p-value significance threshold used to identify these significant associations are described in Table 2.
- https://cdn.elifesciences.org/articles/73475/elife-73475-fig1-data1-v1.txt
-
Figure 1—source data 2
Genomic inflation factor values are less than 1.03 for all paired gene-frequency, productivity GWAS analyses.
This suggests that we have properly controlled for population-substructure-related biases in all of the gene usage analyses. Genomic inflation factor values were calculated as described in Materials and methods.
- https://cdn.elifesciences.org/articles/73475/elife-73475-fig1-data2-v1.txt
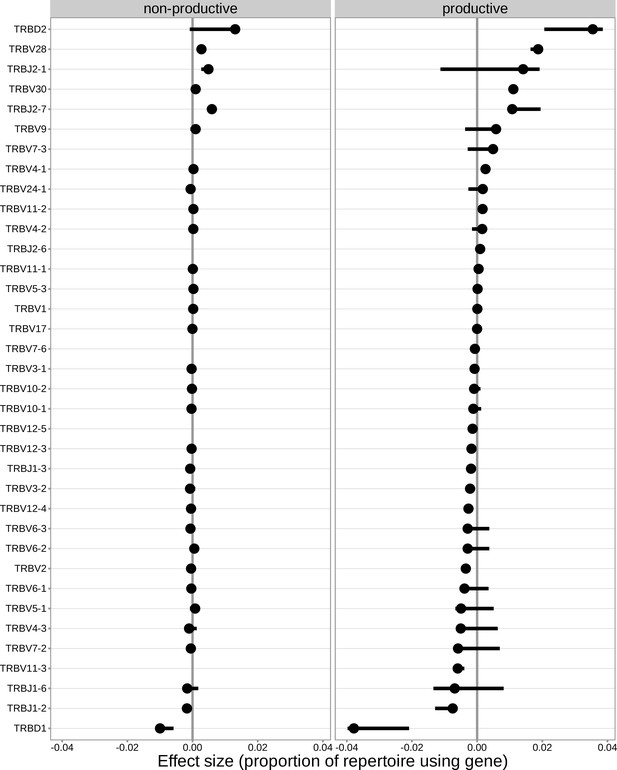
For the significantly associated TCRB locus SNPs, the median association effect magnitude was largest for the expression of TRBD1 followed by the expression of TRBD2 and the expression of TRBV28 all in productive TCRs.
The median association effect magnitude for each gene is shown by each point and the interquartile range of the association effect sizes for each gene is given by each black horizontal line.
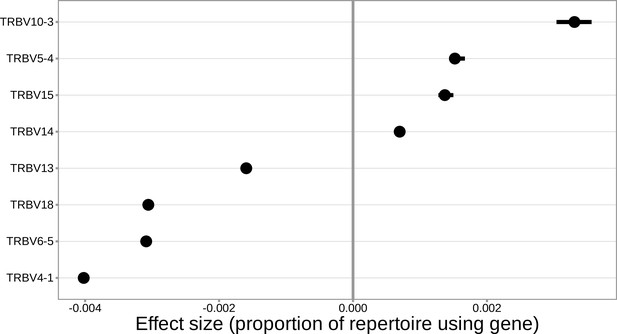
For the significantly associated MHC locus SNPs, the median association effect magnitude was largest for the expression of TRBV4-1 followed by the expression of TRBV10-3.
The median association effect magnitude for each gene is shown by each point and the interquartile range of the association effect sizes for each gene is given by each black horizontal line.
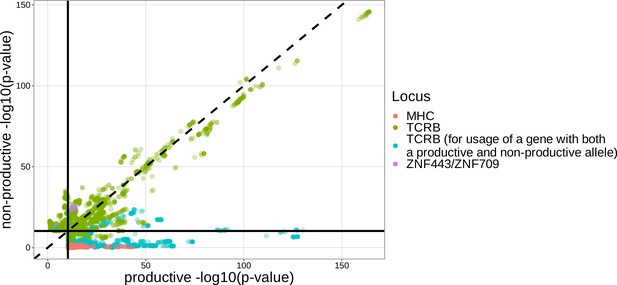
The majority of significantly associated TCRB locus SNPs had similar gene usage association P-values between non-productive and productive TCRs, but significantly associated MHC locus SNPs were only significant for gene usage of productive TCRs.
Notably, the majority of TCRB locus SNPs which were significant for productive TCRs and not significant for non-productive TCRs occurred for the usage of genes which have both productive and non-productive alleles (Dean et al., 2015). Only SNP associations which were significant for either productive TCRs, non-productive TCRs, or both are shown here. There were 15 significant associations which were not located within the MHC, TCRB or ZNF443/ZNF709 loci and are not shown here. The solid black horizontal and vertical lines correspond to the genome-wide Bonferroni-corrected P-value significance threshold of . The dashed black line represents the non-productive -log10(P-value) equals productive -log10(P-value) line.
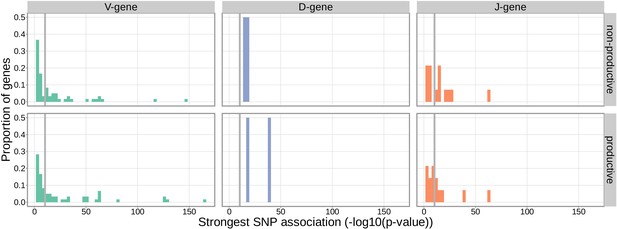
Gene-usage frequency of many V-gene, D-gene, and J-gene segments is significantly associated with variation in the TCRB locus.
The p-value of the strongest TCRB SNP, gene-usage association for each different V-gene, D-gene, and J-gene segment is given on the X-axis. The proportion of gene segments within each gene type is given on the Y-axis. The gray vertical lines correspond to a whole-genome-level Bonferroni-corrected p-value significance threshold of .
-
Figure 2—source data 1
Top TCRB SNP, gene-usage association p-value for each different V-gene, D-gene, and J-gene.
- https://cdn.elifesciences.org/articles/73475/elife-73475-fig2-data1-v1.txt
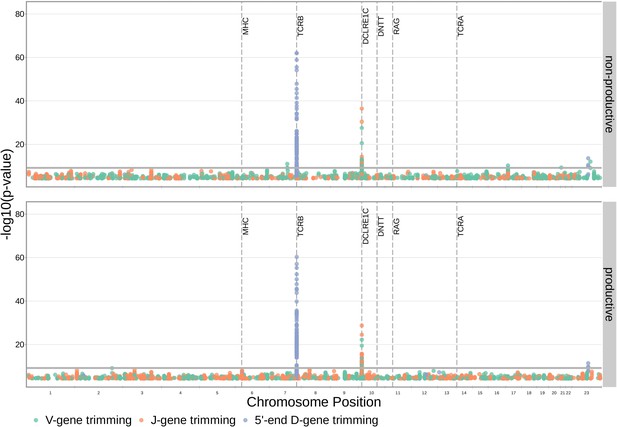
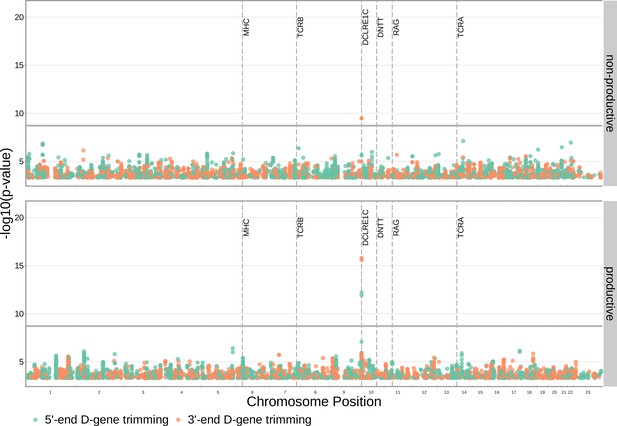
SNP associations for all four trimming types reveal the most significant associations to be located within the TCRB and DCLRE1C loci for 5’ D- gene trimming and J-gene trimming, respectively, when conditioning out effects mediated by gene choice when calculating the strength of association.
Only SNP associations whose are shown here. The gray horizontal line corresponds to a Bonferroni-corrected p-value significance threshold of .
-
Figure 3—source data 1
There are 317 significant SNP associations with the extent of nucleotide trimming for various trimming types.
The model type and Bonferroni-corrected p-value significance threshold used to identify these significant associations are described in Table 2.
- https://cdn.elifesciences.org/articles/73475/elife-73475-fig3-data1-v1.txt
-
Figure 3—source data 2
Genomic inflation factor values are less than 1.03 for all paired nucleotide trimming, productivity GWAS analyses.
This suggests that we have properly controlled for population-substructure-related biases in all the nucleotide trimming analyses. Genomic inflation factor values were calculated as described in Materials and methods.
- https://cdn.elifesciences.org/articles/73475/elife-73475-fig3-data2-v1.txt
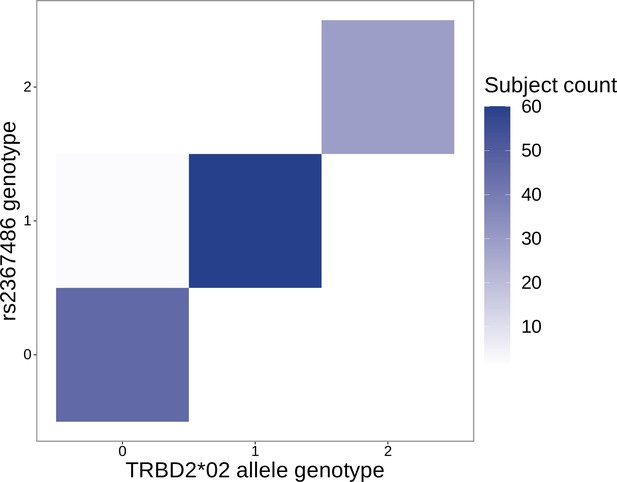
The SNP genotype for the SNP (rs2367486) most significantly associated with 5’ end D-gene trimming within the TCRB locus is also associated with TRBD2*02 allele genotype.
Specifically, SNP genotype and TRBD2*02 allele genotype are significantly correlated ( and ) using a chi-square test of independence. The Y-axis integer genotypes correspond to the number of minor alleles within the rs2367486 SNP genotype. The X-axis integer genotypes correspond to the number of TRBD2*02 alleles within the TRBD2 gene locus genotype.
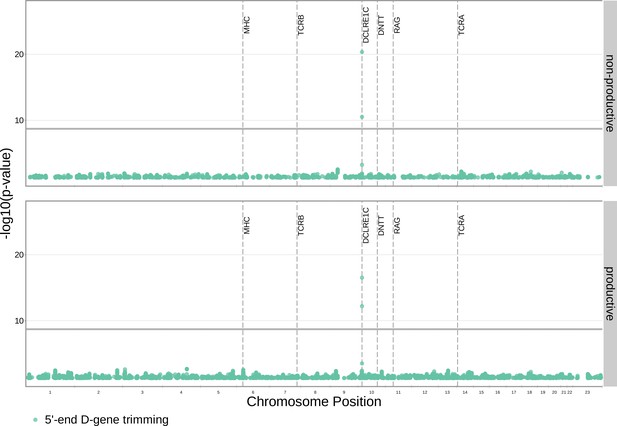
Significant associations are no longer observed between 5’ end D-gene trimming and variation in the TCRB locus after correcting for TRBD2 allele genotype in our model formulation.
Further, four new significant associations are present between 5’ end D-gene trimming and variation in the DCLRE1C locus. Only SNP associations whose are shown here. All genome-wide 3’ end D-gene trimming associations fell above this plotting threshold. The gray horizontal line corresponds to a p-value of (calculated using whole-genome Bonferroni correction, see Materials and methods).
Significant associations are also no longer observed between 5’ end D-gene trimming and variation in the TCRB locus when restricting the analysis to TCRs which contain TRBJ1 genes (and consequently contain TRBD1).
Additionally, two new associations are present between 5’ end D-gene trimming and variation in the DCLRE1C locus for productive TCRs. Four new associations are present between 3’ end D-gene trimming and variation in the DCLRE1C locus. Only SNP associations whose are shown here. The gray horizontal line corresponds to a P-value of (calculated using whole-genome Bonferroni correction, see Materials and methods).
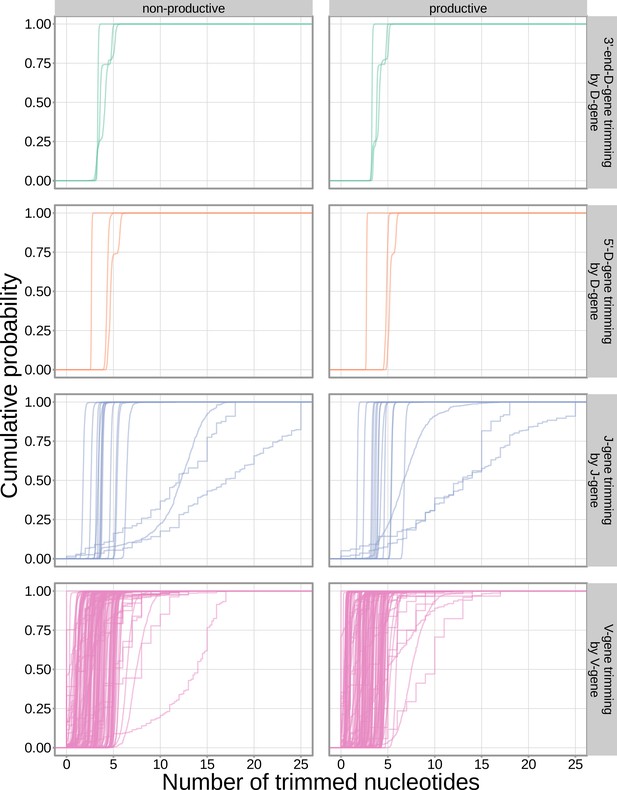
The extent of nucleotide deletion varies by the gene allele identity for all gene types.
An empirical cumulative distribution is drawn for each gene allele type within each indicated gene type (i.e. V-gene, D-gene, J-gene).
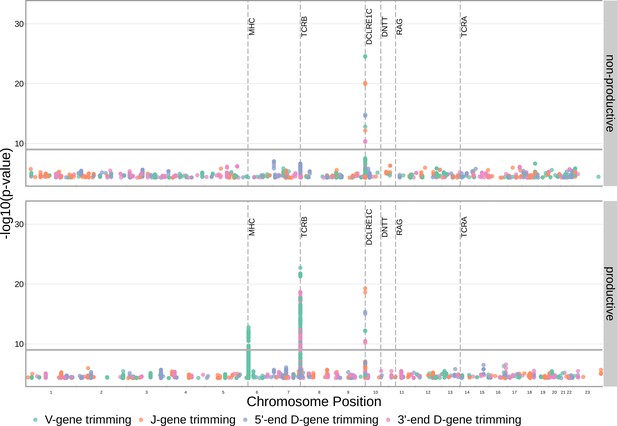
Significant SNP associations are located within the MHC, TCRB, and DCLRE1C loci for all four trimming types when calculating the strength of association without conditioning out effects mediated by gene choice.
Earlier findings relating variations in MHC and TCRB to gene usage changes, however, indicate that many of these associations are likely artefactual. Only SNP associations whose are shown here. The gray horizontal line corresponds to a p-value of .
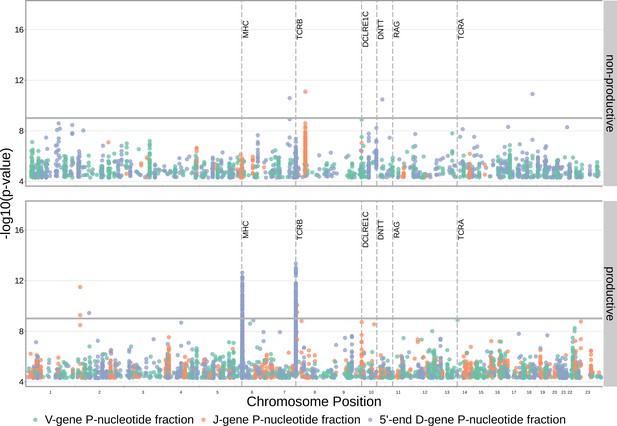
SNP associations for all fractions of non-gene-trimmed TCRs containing P-nucleotides are not significant within the DCLRE1C locus.
However, significant associations are present within the TCRB and MHC loci for the fraction of non-D-gene-trimmed, productive TCRs containing 5’ end D-gene P-nucleotides. Only SNP associations whose are shown here. The gray horizontal line corresponds to a p-value of (calculated using whole-genome Bonferroni correction, see Materials and methods).
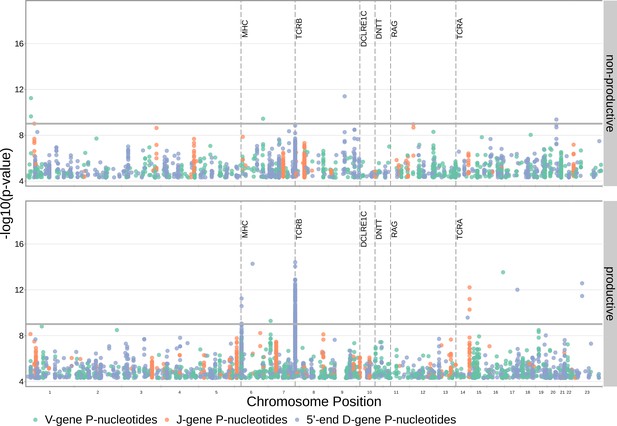
SNP associations for the number of P-nucleotides are not significant within the DCLRE1C locus.
However, significant associations are present within the TCRB and MHC loci. Only SNP associations whose are shown here. The gray horizontal line corresponds to a Bonferroni-corrected whole-genome p-value significance threshold of (see Materials and methods).
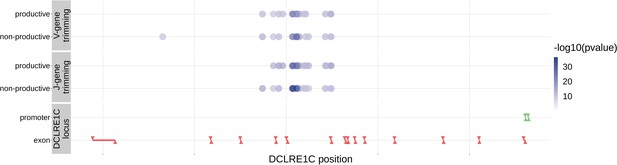
Within the DCLRE1C locus, 93.8% of these significantly associated SNPs were located within introns.
Additionally, many of these significant SNP associations overlapped between trimming types. Downward arrows represent promoter/exon starting positions and upward arrows represent promoter/exon ending positions.
-
Figure 4—source data 1
DCLRE1C locus SNP association p-values and locus positions.
- https://cdn.elifesciences.org/articles/73475/elife-73475-fig4-data1-v1.txt
-
Figure 4—source data 2
There are two independent SNP signals within the DCLRE1C locus for J-gene trimming of non-productive TCRs.
A conditional analysis was performed (as described in Materials and methods) to identify these independent signals.
- https://cdn.elifesciences.org/articles/73475/elife-73475-fig4-data2-v1.txt
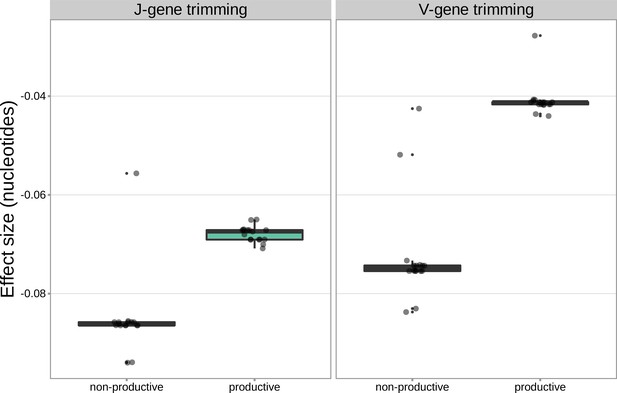
For the significantly associated DCLRE1C locus SNPs, the magnitudes of the effects were greater for non-productive TCRs compared to productive TCRs for both V-gene trimming and J-gene trimming.
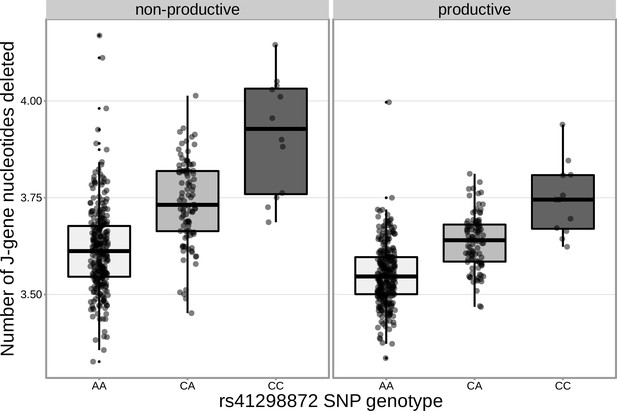
The extent of J-gene trimming changes as a function of SNP genotype for the SNP (rs41298872) most significantly associated with J-gene trimming within the DCLRE1C locus.
Only TCRs containing TRBJ1-1*01 (the most frequently used TRBJ1 gene across subjects) were included when calculating the average number of J-gene nucleotides deleted for each subject.
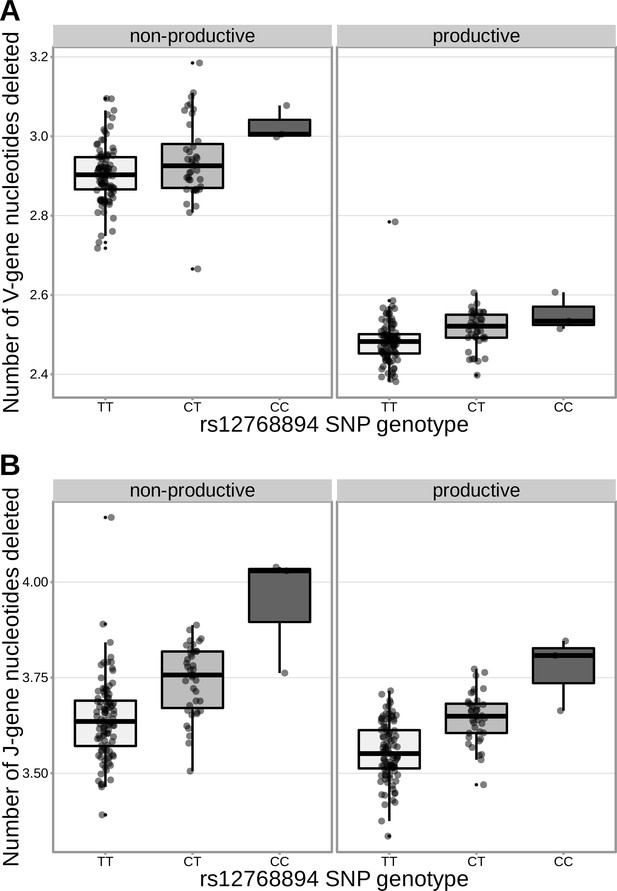
The extent of V- and J-gene trimming of productive and non-productive TCRβ chains changes as a function of SNP genotype within the discovery cohort for a non-synonymous DCLRE1C SNP (rs12768894, c.728A>G).
Only TCRs containing TRBJ1-1*01 (the most frequently used TRBJ1 gene across subjects) were included when calculating the average number of J-gene nucleotides deleted for each subject. Only TCRs containing TRBV5-1*01 (the most frequently used TRBV gene across subjects) were included when calculating the average number of V-gene nucleotides deleted for each subject.
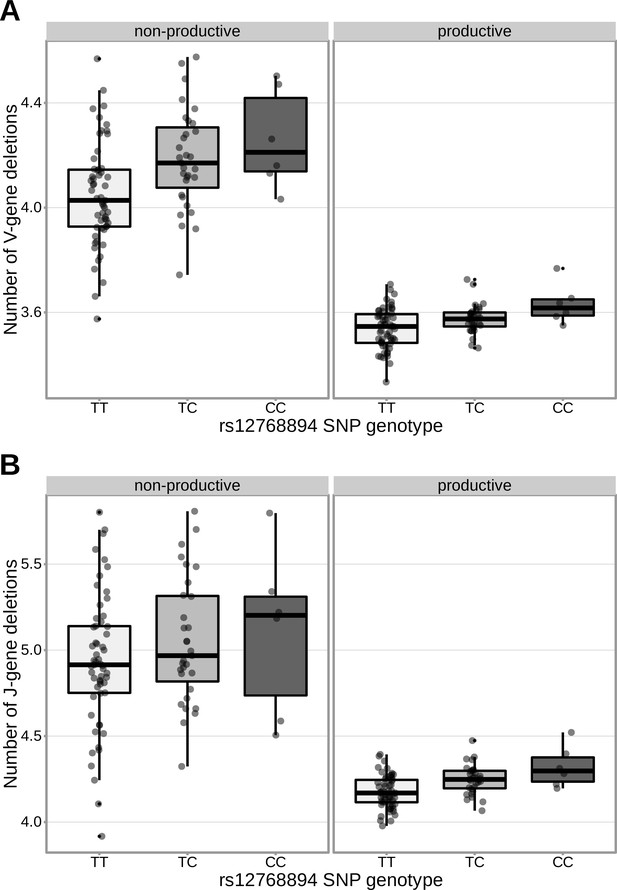
The extent of V-gene trimming.
(A) of productive and non-productive TCRβ chains and J-gene trimming (B) of productive TCRβ chains changes as a function of SNP genotype within the validation cohort for a non-synonymous DCLRE1C SNP (rs12768894, c.728A > G). The average number of nucleotides deleted was calculated across all TCRβ chains for each subject, regardless of gene-usage.
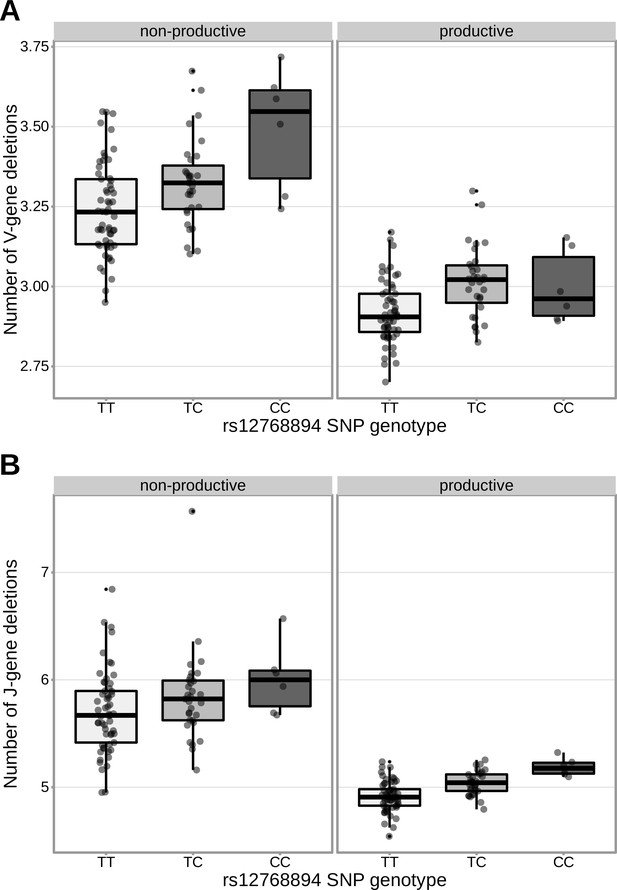
The extent of V- (A) and J-gene (B) trimming of productive and non-productive TCRα chains changes as a function of SNP genotype within the validation cohort for a non-synonymous DCLRE1C SNP (rs12768894, c728A>G).
The average number of nucleotides deleted was calculated across all TCRα chains for each subject, regardless of gene-usage.
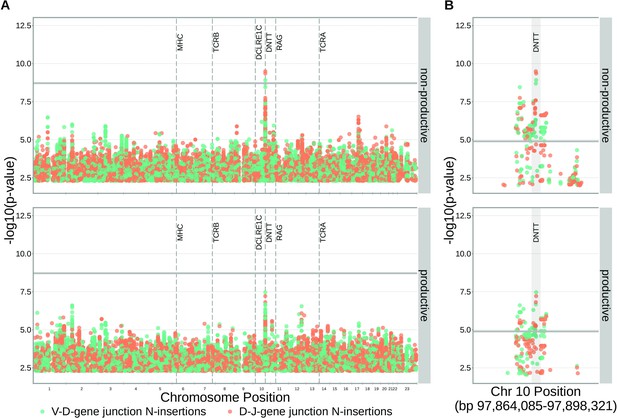
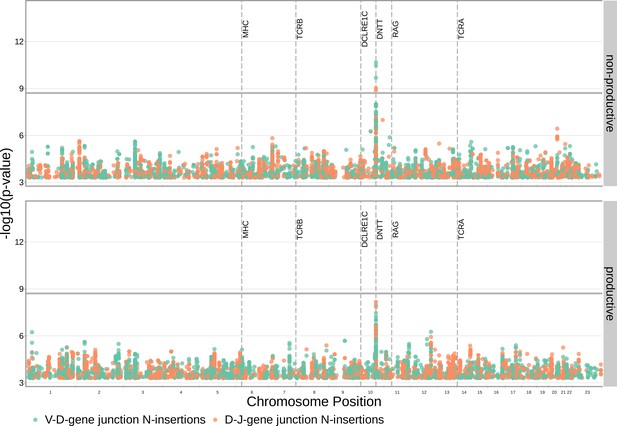
SNPs within the DNTT locus are associated with the extent of N-insertion.
(A) There are three associations for SNPs within the DNTT locus which are significant when considered in the whole-genome context. The gray horizontal line corresponds to a whole-genome Bonferroni-corrected P-value significance threshold of . (B) Using a DNTT gene-level significance threshold, many more SNPs within the extended DNTT locus have significant associations for both N-insertion types. Here, the gray horizontal line corresponds to a gene-level Bonferroni-corrected P-value significance threshold of (calculated using gene-level Bonferroni correction for the 977 SNPs within 200 kb of the DNTT locus, see Materials and methods). For both (A) and (B), only SNP associations whose are shown.
-
Figure 5—source data 1
There are three significant associations between SNPs genome-wide and the number of nucleotide N-insertions.
The model type and Bonferroni-corrected p-value significance threshold used to identify these significant associations are described in Table 2.
- https://cdn.elifesciences.org/articles/73475/elife-73475-fig5-data1-v1.txt
-
Figure 5—source data 2
There are 232 significant associations between SNPs genome-wide and the number of nucleotide N-insertions when restricting the analysis to the extended DNTT locus.
- https://cdn.elifesciences.org/articles/73475/elife-73475-fig5-data2-v1.txt
-
Figure 5—source data 3
Genomic inflation factor values are less than 1.03 for all paired N-insertion, productivity GWAS analyses.
This suggests that we have properly controlled for population-substructure-related biases in all of the N-insertion analyses. Genomic inflation factor values were calculated as described in Materials and methods.
- https://cdn.elifesciences.org/articles/73475/elife-73475-fig5-data3-v1.txt
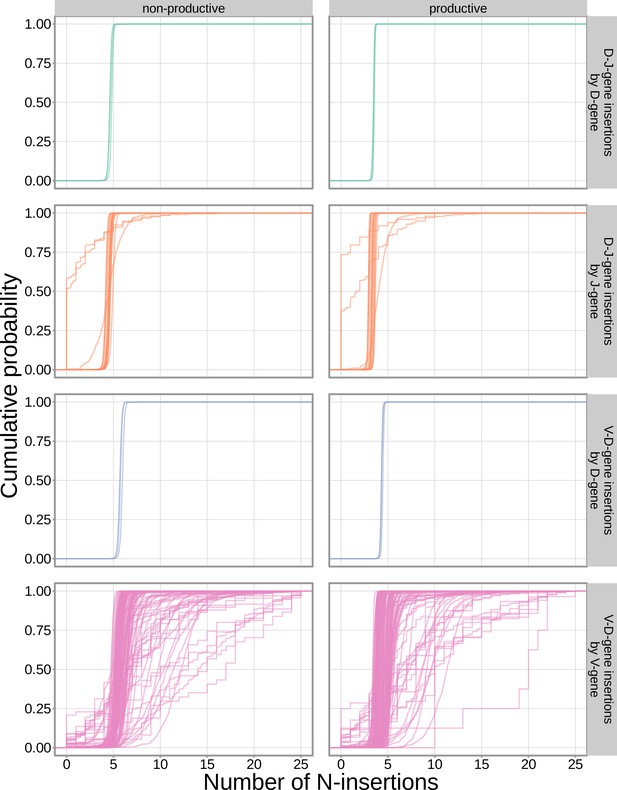
The extent of N-insertion does not vary substantially by the gene allele identity for any gene type.
An empirical cumulative distribution is drawn for each gene allele type within each indicated gene type (i.e. V-gene, D-gene, J-gene).
Significant associations continue to be observed within the DNTT locus for both V-D- and D-J-gene-junction N-insertions when restricting the analysis to TCRs which contain TRBJ1 genes and consequently contain TRBD1.
Only SNP associations whose are shown here. The gray horizontal line corresponds to a Bonferroni-corrected p-value significance threshold of (calculated using whole-genome Bonferroni correction, see Materials and methods).
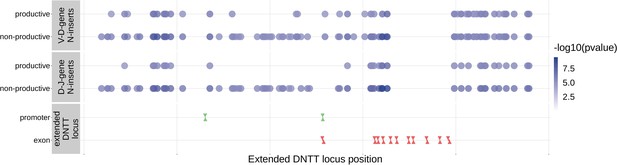
Within the DNTT locus, many of the significant SNP associations overlapped between N-insertion types when using DNTT gene-level Bonferroni-corrected p-value significance threshold of .
Downward arrows represent promoter/exon starting positions and upward arrows represent promoter/exon ending positions.
-
Figure 6—source data 1
DNTT locus SNP association p-values and locus positions.
- https://cdn.elifesciences.org/articles/73475/elife-73475-fig6-data1-v1.txt
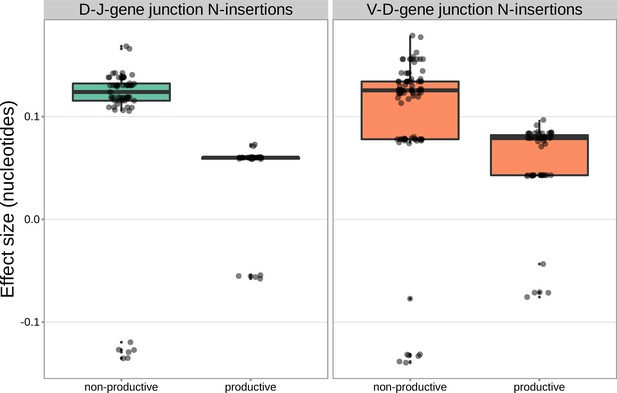
For these significant DNTT locus SNP associations, the magnitudes of the effects were greater for non-productive TCRs compared to productive TCRs for both V-D-gene junction N-insertion and D-J-gene junction N-insertion.
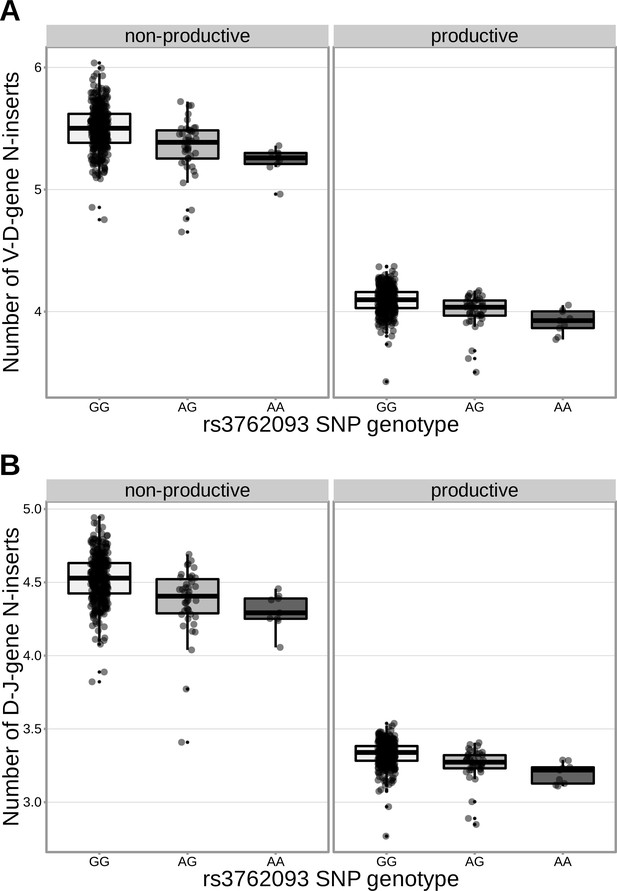
The extent of V-D and D-J N-insertion of productive and non-productive TCRβ chains changes as a function of SNP genotype within the discovery cohort for an intronic DNTT SNP (rs3762093).
The average number of N-insertions was calculated across all TCRβ chains for each subject.
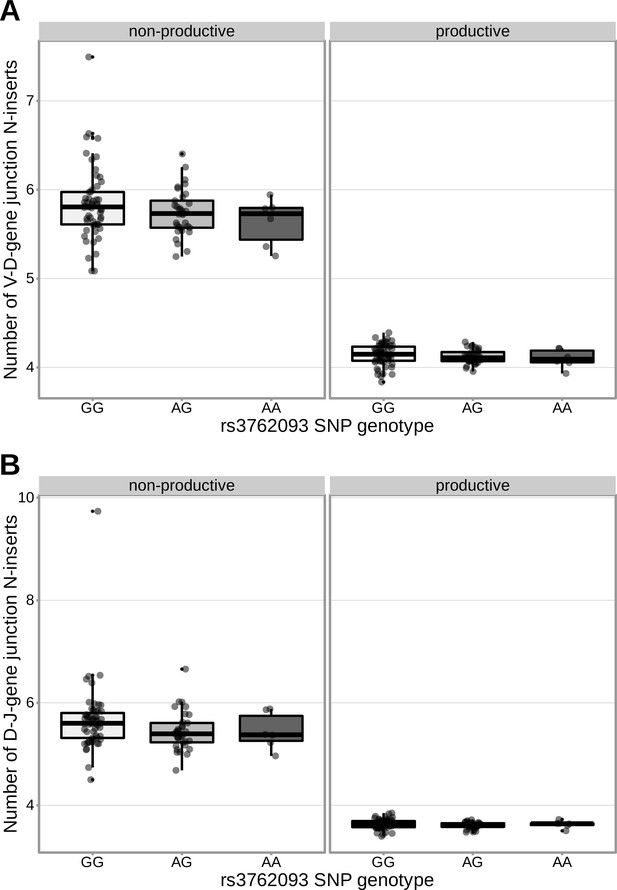
An intronic SNP (rs3762093) within the DNTT gene locus is not strongly associated with the number of V-D (A) or D-J (B) N-inserts within productive or non-productive TCRβ chains in the validation cohort.
However, the direction of the effect is the same as the discovery cohort for all N-insertion and productivity types. The average number of N-insertions was calculated across all TCRβ chains for each subject.
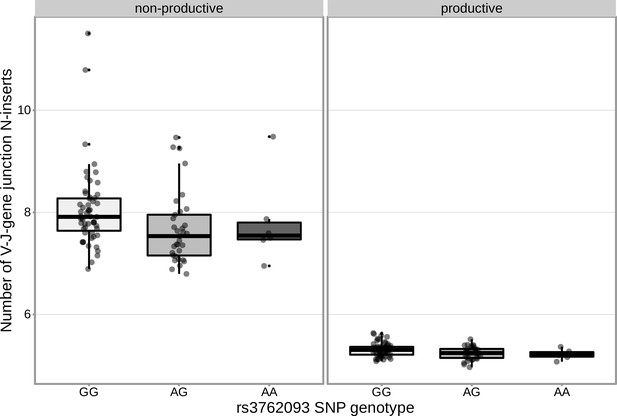
An intronic SNP (rs3762093) within the DNTT gene locus is significantly associated with the number of V-J N-inserts for productive TCRα chains in the validation cohort.
This SNP is not significantly associated with the number of V-J N-inserts for non-productive TCRα chains in the validation cohort. The average number of N-insertions was calculated across all TCRα chains for each subject.
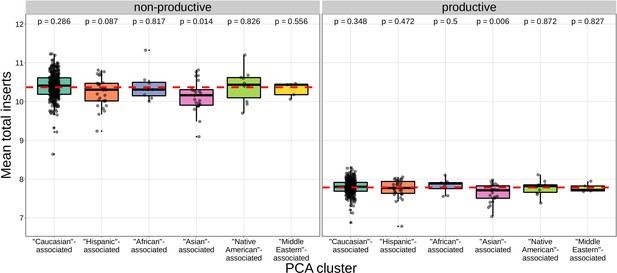
The TCR repertoires for subjects in the ‘Asian’-associated PCA-cluster contain fewer N-insertions for productive TCRs when compared to the population mean computed across all 666 subjects (dashed, red horizontal line).
The p-values from a one-sample t-test (without Bonferroni multiple testing correction) for each PCA cluster compared to the population mean are reported at the top of the plot.
-
Figure 7—source data 1
PCA-cluster and average number of N-insertions by subject.
- https://cdn.elifesciences.org/articles/73475/elife-73475-fig7-data1-v1.txt
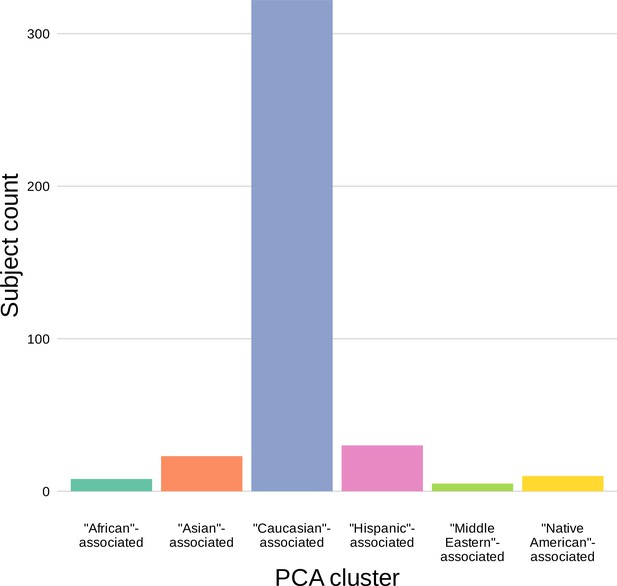
The population mean is dominated by subjects in the ‘Caucasian’-associated PCA-cluster.
Of the 398 subjects in the sample population, 81% are in the ‘Caucasian’-associated PCA-cluster.
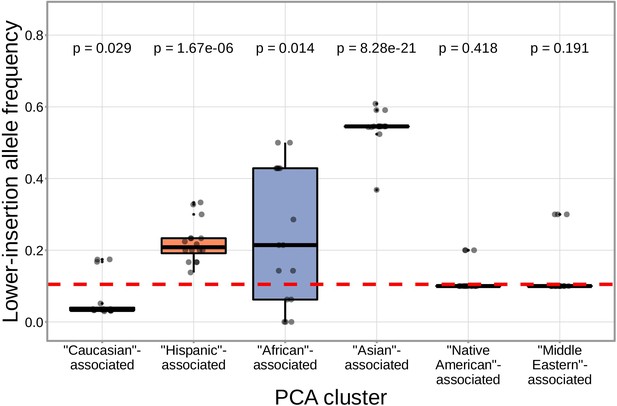
SNPs within the DNTT region that are associated with fewer N-insertions have a higher mean allele frequency within the ‘Asian’-associated PCA-cluster when compared to the population mean allele frequency computed across the 398 discovery cohort subjects (dashed, red horizontal line).
The p-values from a one-sample t-test (without Bonferroni multiple testing correction) for each PCA cluster compared to the population mean are reported at the top of the plot. The population mean is dominated by subjects in the ‘Caucasian’-associated PCA cluster (Figure 7—figure supplement 1).
-
Figure 8—source data 1
Allele frequencies by PCA-cluster for SNPs within the DNTT locus that are associated with fewer N-insertions.
- https://cdn.elifesciences.org/articles/73475/elife-73475-fig8-data1-v1.txt
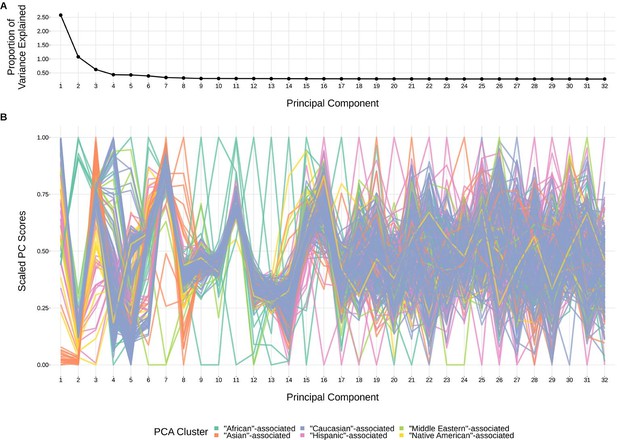
The top principal components calculated from genotype data reflect ancestry structure among samples.
(A) The majority of the ancestry-informative principal component analysis variance is explained by the first eight principal components. (B) The first eight principal components show distinct separation by PCA cluster. Each colored line represents one of the 398 samples. The first 32 principal components are shown on the X-axis and their scaled component values for each subject on the Y-axis.
-
Figure 9—source data 1
Percent variance explained by each principal component.
- https://cdn.elifesciences.org/articles/73475/elife-73475-fig9-data1-v1.txt
-
Figure 9—source data 2
Scaled principal component values by subject.
- https://cdn.elifesciences.org/articles/73475/elife-73475-fig9-data2-v1.txt
Tables
Discovery cohort demographics.
| Count | ||
|---|---|---|
| Sex | Female | 179 |
| Male | 197 | |
| Unknown | 22 | |
| Age (in years) | < 10 | 12 |
| 11–20 | 11 | |
| 21–30 | 48 | |
| 31–40 | 70 | |
| 41–50 | 103 | |
| 51–60 | 70 | |
| > 60 | 22 | |
| Unknown | 62 | |
| Ancestry-informative PCA cluster (see Materials and methods) | “African”-associated | 8 |
| “Asian”-associated | 23 | |
| “Caucasian”-associated | 322 | |
| “Hispanic”-associated | 30 | |
| “Middle Eastern”-associated | 5 | |
| “Native American”-associated | 10 | |
| CMV serostatus | Positive | 171 |
| Negative | 204 | |
| Unknown | 23 | |
| Total | 398 |
-
Table 1—source data 1
Subjects map from the original self-identified ancestry groups to ancestry-informative PCA clusters (see Materials and methods).
- https://cdn.elifesciences.org/articles/73475/elife-73475-table1-data1-v1.txt
We inferred the associations between genome-wide variation and many different TCR repertoire features for productive and non-productive TCR sequences, separately.
For each TCR repertoire feature, we considered the significance of associations using a Bonferroni-corrected threshold established to correct for each TCR feature subtype, the two TCR productivity types, and the total number of SNPs tested (described in detail in Methods).
| Repertoire feature(significance threshold) | Model type | Feature subtype | Productivity | Significant association |
|---|---|---|---|---|
| V(D)J gene usage(5.09 × 10−11) | simple | Each of 60 V-genes | Productive | Yes, for 36 V-genes |
| Non-productive | Yes, for 26 V-genes | |||
| Each of 2 D-genes | Productive | Yes, for both D-genes | ||
| Non-productive | Yes, for both D-genes | |||
| Each of 14 J-genes | Productive | Yes, for 7 J-genes | ||
| Non-productive | Yes, for 8 J-genes | |||
| Amount of nucleotide trimming(9.68 × 10−10) | gene-conditioned | V-gene trimming | Productive | Yes |
| Non-productive | Yes | |||
| 5’ end D-gene trimming | Productive | No | ||
| Non-productive | No | |||
| 3’ end D-gene trimming | Productive | No | ||
| Non-productive | No | |||
| J-gene trimming | Productive | Yes | ||
| Non-productive | Yes | |||
| Number of N-insertions(1.94 × 10−9) | simple | V-D-gene N-insertions | Productive | No |
| Non-productive | Yes | |||
| D-J-gene N-insertions | Productive | No | ||
| Non-productive | Yes |
Validation cohort demographics.
| Count | ||
|---|---|---|
| Sex | Female | 58 |
| Male | 36 | |
| Age (in years) | < 10 | 26 |
| 11–20 | 15 | |
| 21–30 | 13 | |
| 31–40 | 12 | |
| 41–50 | 11 | |
| 51–60 | 9 | |
| > 60 | 8 | |
| Self-reported ethnicity | Hispanic or Latino | 94 |
| CMV serostatus | Positive | 37 |
| Negative | 57 | |
| Total | 94 |
We inferred the associations between SNP genotype and TCR repertoire features for two SNPs overlapping between discovery-cohort and validation-cohort SNP sets.
We considered the significance of the validation cohort associations at a Bonferroni-corrected SNP-level p-value significance threshold of 0.0042 for trimming and 0.0083 for N-insertion (see Materials and methods). Validation cohort p-values are one-tailed. * Discovery-cohort associations were only significant when considered at the DNTT -gene level significance threshold, not at the whole-genome significance threshold.
| SNP | TCR chain | Repertoire feature | Productivity type | Discovery cohort significant association | Validation cohort significant association |
|---|---|---|---|---|---|
| rs12768894 | TCRβ | V-gene trimming | Productive | Yes (2.16 × 10−14) | Yes (7.17 × 10−7) |
| Non-productive | Yes (7.21 × 10−14) | Yes (8.75 × 10−6) | |||
| J-gene trimming | Productive | Yes (1.23 × 10−11) | Yes (5.16 × 10−10) | ||
| Non-productive | Yes (6.62 × 10−12) | No (4.18 × 10−2) | |||
| TCRα | V-gene trimming | Productive | N/A | Yes (2.59 × 10−5) | |
| Non-productive | N/A | Yes (2.68 × 10−7) | |||
| J-gene trimming | Productive | N/A | Yes (6.29 × 10−12) | ||
| Non-productive | N/A | No (9.99 × 10−3) | |||
| rs3762093 | TCRβ | V-D N-insertion | Productive | Yes* (1.37 × 10−6) | No (0.153) |
| Non-productive | Yes* (1.50 × 10−7) | No (0.059) | |||
| D-J N-insertion | Productive | Yes* (9.43 ×10−6) | No (0.137) | ||
| Non-productive | Yes* (1.94 × 10−7) | No (0.006) | |||
| TCRα | V-J N-insertion | Productive | N/A | Yes (0.006) | |
| Non-productive | N/A | No (0.031) |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Software, Algorithm | TCRdist | Dash et al., 2017, Bradley et al., 2017 | Version 0.0.2; Software can be found on GitHub | |
| Software, Algorithm | migec | Shugay et al., 2014 | RRID: SCR_016337 | Version 1.2.9; Software can be found on GitHub |
| Antibody | CD3-PerCP eFluor710 (Mouse monoclonal) | Thermo Fisher Scientific | Cat: 46-0037-42; RRID: AB_1834395 | 0.012 μg per 1 million cells (1:100) |
| Antibody | CD4-BV650 (Mouse monoclonal) | BD Biosciences | Cat: 563875; RRID: AB_2687486 | 2 μl per 1 million cells (1:50) |
| Antibody | CD8-APC Fire750 (Mouse monoclonal) | Biolegend | Cat: 344746; RRID: AB_2572095 | 0.1 μg per 1 million cells (1:100) |
| Antibody | TCRγ/-PE Cy7 (Mouse monoclonal) | Biolegend | Cat: 331222; RRID: AB_2562891 | 1 μg per 1 million cells (1:40) |
| Other | Fc Block | BD Biosciences | Cat: 564220; RRID: AB_2728082 | 2.5 μg per 1 million cells (1:20) |
| Other | Live/Dead Aqua | Tonbo Biosciences | Cat: 13–0870 T100 | 1 μl per 1 million cells (1:100) |
| Commercial assay, kit | Qiagen QIAamp DNA Mini Kit | Qiagen | Cat: 51,306 | |
| Commercial assay, kit | Taqman SNP Genotyping Assay | Thermo Fisher Scientific | Cat: 4351379 | |
| Commercial assay, kit | TaqMan Genotyping Master Mix | Thermo Fisher Scientific | Cat: 4371353 |





