The peroxisomal exportomer directly inhibits phosphoactivation of the pexophagy receptor Atg36 to suppress pexophagy in yeast
Figures
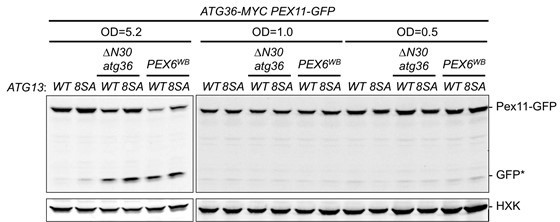
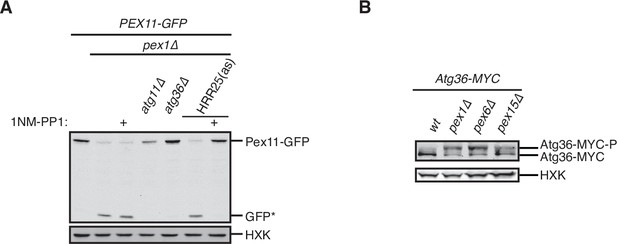
Exportomer mutations enhance Atg36 phosphoactivation by Hrr25.
(A) The indicated extracts were subjected to immunoprecipitation (IP) with anti-MYC magnetic beads. After extensive washing, bound material was treated with lambda phosphatase in the presence or absence of phosphatase (phos.) inhibitors, and then resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) followed by immunoblotting with anti-MYC antibodies. In previous studies, two Atg36 species were detected in the absence of nutrient stress or exportomer mutations (Meguro et al., 2020; Nuttall et al., 2014); the lower-mobility species might correspond to a basally phosphorylated form of Atg36. In our experiments with MYC-tagged Atg36, we were unable to detect more than one major Atg36 species in the absence of oleate stress or exportomer mutations. The higher- and lower-mobility species we detect under oleate stress or in the absence of exportomer function might correspond to basally phosphorylated and hyperphosphorylated species of Atg36, respectively. (B) Extracts derived from cells with indicated genotypes were resolved by SDS–PAGE followed by immunoblotting with anti-MYC antibodies. pex3-177 indicates mutant version of Pex3 unable to bind Atg36 (Motley et al., 2012). (C) Extracts were prepared from cells with indicated genotypes that were treated for 3 hr with either 500 µM auxin or vehicle (DMSO) and resolved by SDS–PAGE, followed by immunoblotting with anti-MYC, anti-V5, and anti-HXK antibodies. (D) Cells expressing Pex11-GFP were grown in oleate medium for 22 hr and then transferred to nitrogen starvation medium. Extracts derived from cells with indicated genotypes were resolved by SDS–PAGE followed by immunoblotting with anti-MYC, anti-GFP, and anti-HXK antibodies. wt, wild-type allele of PEX6; ∆, genomic deletion of PEX6; WB, PEX6 D2 Walker B motif mutant allele (E832A) at the endogenous PEX6 locus. GFP*, GFP fragments produced upon vacuolar degradation of Pex11-GFP. HXK, hexokinase. Data points represent the mean values determined from three independent experiments. Error bars represent standard error.
-
Figure 1—source data 1
Raw immunoblotting data related to Figure 1.
- https://cdn.elifesciences.org/articles/74531/elife-74531-fig1-data1-v1.pdf
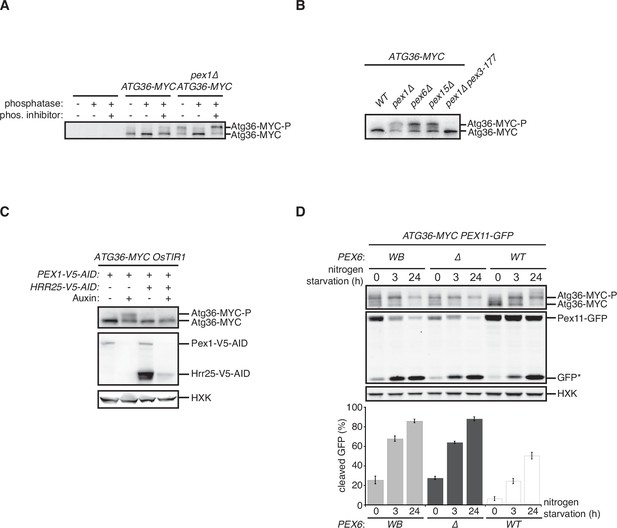
The exportomer inhibits pexophagy in yeast.
(A) Cells with indicated genotypes were grown in oleate medium for 22 hr, followed by growth in nitrogen starvation medium for 5 hr in the presence of 100 μM 1NM-PP1, where indicated (+), or vehicle (DMSO), to monitor pexophagy. The corresponding cell extracts were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) followed by immunoblotting with anti-GFP and anti-HXK antibodies. GFP*, GFP fragments produced upon vacuolar degradation of Pex11-GFP. HXK, hexokinase. HRR25(as), ATP analog-sensitive allele of Hrr25. GFP*, GFP fragments produced upon vacuolar degradation of Pex11-GFP. See Materials and methods for additional details. (B) Extracts derived from cells with indicated genotypes were resolved by SDS-PAGE followed by immunoblotting with anti-MYC and anti-HXK antibodies. HXK, hexokinase.
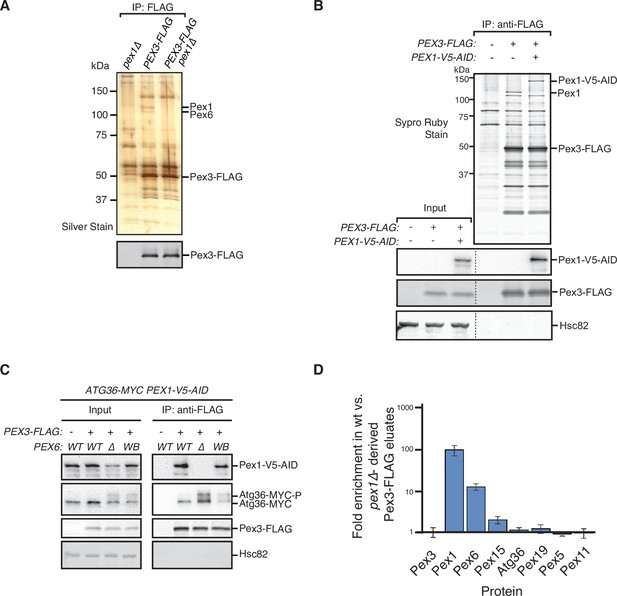
Identification of physical interactions between Pex3 and the exportomer.
(A, B) Detergent-solubilized extracts derived from cells with indicated genotypes were first cleared at 100,000 × g and then immunoprecipitated (IP) with anti-FLAG magnetic beads. (A) Eluates were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) followed by either silver staining or immunoblotting with anti-FLAG antibodies. Selected proteins identified in the Pex3-FLAG eluate by mass spectrometry are indicated based on predicted molecular weight. (B) FLAG peptide eluates and extract (input) samples were resolved by SDS–PAGE followed by either SYPRO Ruby gel staining or immunoblotting with anti-V5, anti-FLAG, and anti-Hsc82 antibodies. Dotted line indicates splicing of identically processed gel-image data from the same gel. Hsc82 was used as sample processing control. (C) The indicated extracts were subjected to immunoprecipitation (IP) with anti-FLAG magnetic beads. FLAG peptide eluates and extract (input) samples were resolved by SDS–PAGE followed by immunoblotting with anti-V5, anti-MYC, anti-FLAG, and anti-Hsc82 antibodies. wt, wild-type allele of PEX6; ∆, genomic deletion of PEX6; WB, PEX6 D2 Walker B motif mutant allele (E832A) at the endogenous PEX6 locus. (D) Quantification of protein abundance in Pex3-FLAG eluates for selected proteins identified by mass spectrometry. Plotted is the mean ratio of protein abundance in wt- versus pex1Δ-derived Pex3-FLAG eluates. Error bars represent standard deviation.
-
Figure 2—source data 1
Raw staining, immunoblotting, and mass spectrometry data related to Figure 2.
- https://cdn.elifesciences.org/articles/74531/elife-74531-fig2-data1-v1.pdf
-
Figure 2—source data 2
The mass spectrometry data related to Figure 2D.
- https://cdn.elifesciences.org/articles/74531/elife-74531-fig2-data2-v1.xlsx
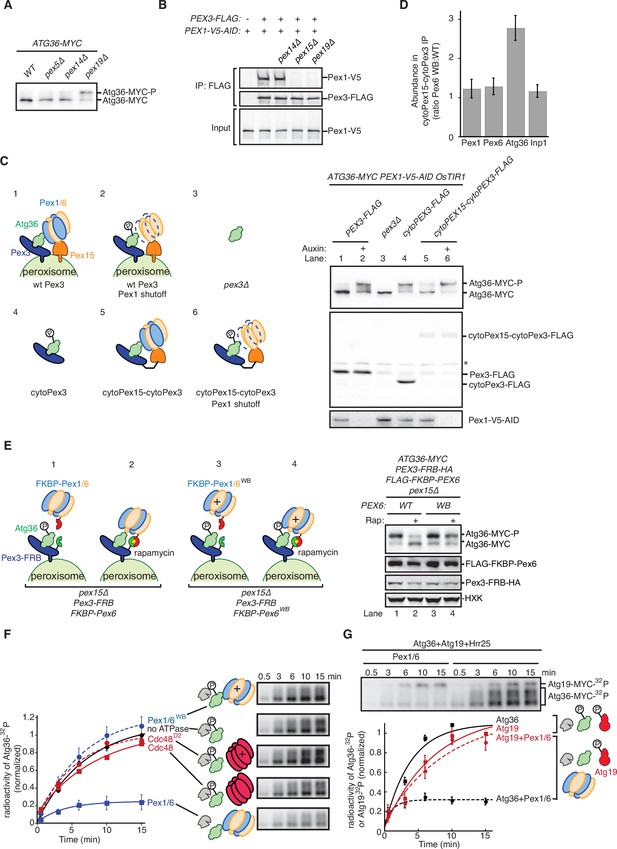
The exportomer inhibits Atg36 both in vivo and in vitro.
(A) Extracts derived from cells with indicated genotypes were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) followed by immunoblotting with anti-MYC. wt, wild-type. (B) The indicated extracts were subjected to immunoprecipitation (IP) with anti-FLAG magnetic beads. FLAG peptide eluates and extract (input) samples were resolved by SDS–PAGE followed by immunoblotting with anti-FLAG and anti-V5 antibodies. (C) Left, schematic showing predicted pattern of Atg36 phosphorylation if exportomer is sufficient to inhibit Atg36. Right, extracts derived from cells with indicated genotypes that were treated for 3 hr with either 500 µM auxin or vehicle (DMSO) were resolved by SDS–PAGE followed by immunoblotting with anti-MYC, anti-FLAG, and anti-V5 antibodies, with lane numbers corresponding to numbered schematics. *, unspecific bands. (D) The abundance ratios of proteins in cytoPex15–cytoPex3-FLAG eluates derived from cells expressing PEX6 D2 Walker B motif mutant allele at the endogenous PEX6 locus (WB) versus eluates from cells expressing PEX6 wild-type (wt). Error bars represent standard error. (E) Left, schematics showing the predicted pattern of Atg36 phosphorylation if bypassing Pex15 by an artificial tethering system (FRB-Rap-FKBP) is sufficient to inhibit Atg36 phosphorylation. Right, extracts derived from cells with indicated genotypes that were treated with 1 µM rapamycin (Rap) or vehicle (90% ethanol, 10% Tween-20) for 3 hr were resolved by SDS–PAGE followed by immunoblotting with anti-MYC, anti-FLAG, anti-HA, and anti-HXK antibodies, corresponding to schematics. WB, PEX6 D2 Walker B motif mutant allele (E832A); HXK, hexokinase; Rap, rapamycin. (F) Purified Atg36 was incubated with 32P-labeled ATP, Hrr25, and indicated AAA-ATPases, and samples at each time point were resolved by SDS–PAGE followed by autoradiography to measure Atg36 phosphorylation. WB, Pex6 D2 Walker B motif mutant (E832A). D2, Cdc48 D2 ring mutant (E588Q). Data points represent the mean values determined from four repeat experiments. Error bars represent standard error. (G) Purified Atg36 and Atg19 were incubated together with 32P-labeled ATP, Hrr25, in the presence or absence of Pex1/6 complexes. Samples at each time point were resolved by SDS–PAGE followed by autoradiography. Data points represent the mean values determined from three repeat experiments. Note that the final time points for Atg19-only and Atg36-only conditions are superimposed. Error bars represent standard error.
-
Figure 3—source data 1
Raw immunoblotting and mass spectrometry data related to Figure 3.
- https://cdn.elifesciences.org/articles/74531/elife-74531-fig3-data1-v1.pdf
-
Figure 3—source data 2
The mass spectrometry data related to Figure 3D.
- https://cdn.elifesciences.org/articles/74531/elife-74531-fig3-data2-v1.xlsx
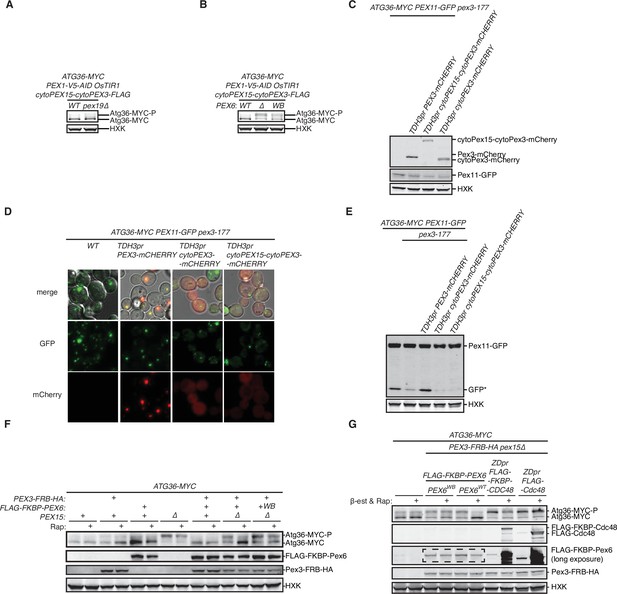
The exportomer inhibits Atg36 phosphorylation in vivo.
(A, B) Extracts derived from cells with indicated genotypes were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) followed by immunoblotting with anti-MYC and anti-HXK antibodies. (A) wt, wild-type; pex19Δ, genomic deletion of PEX19. (B) wt, wild-type allele of PEX6; Δ, genomic deletion of PEX6; WB, PEX6 D2 Walker B motif mutant allele (E832A) at the endogenous PEX6 locus. (C) Extracts derived from cells with indicated genotypes were resolved by SDS–PAGE followed by immunoblotting with anti-mCherry, anti-GFP, and anti-HXK antibodies. A second copy of mCherry-labeled full-length Pex3, cytosolic fragment of Pex3 (cytoPex3), and the chimeric cytoPex15–cytoPex3 were expressed under TDH3 promoter at the trp1 locus. The levels of cytoPex3-mCherry and cytoPex15–cytoPex3-mCherry were lower than Pex3-mCherry, suggesting the cytosolic chimeras are less stable than the wild-type Pex3. (D) Cells with indicated genotypes were imaged by confocal microscopy (594 nm channel for mCherry and 488 nm channel for GFP). mCherry-labeled full-length Pex3 formed puncta that colocalized with Pex11-GFP. mCherry-labeled cytoPex3 and cytoPex15–cytoPex3 showed diffused mCherry signal in the cytoplasm. (E) Cells with indicated genotypes were grown in oleate medium for 22 hr, followed by growth in nitrogen starvation medium for 6 hr to monitor pexophagy. The corresponding cell extracts were resolved by SDS–PAGE followed by immunoblotting with anti-GFP and anti-HXK antibodies. GFP*, GFP fragments produced upon vacuolar degradation of Pex11-GFP. HXK, hexokinase. (F) Extracts derived from cells with indicated genotypes that were treated with 1 μM rapamycin (Rap) or vehicle (90% ethanol, 10% Tween-20) for 3 hr were resolved by SDS–PAGE followed by immunoblotting with anti-MYC, anti-FLAG, anti-HA, and anti-HXK antibodies. WB, PEX6 D2 Walker B motif mutant allele (E832A); HXK, hexokinase. (G) Extracts derived from cells with indicated genotypes that were treated with 1 μM rapamycin (Rap) and 1 μM β-estradiol (β-est), or vehicle (90% ethanol, 10% Tween-20) for 3 hr were resolved by SDS–PAGE followed by immunoblotting with anti-MYC, anti-FLAG, anti-HA, and anti-HXK antibodies. HXK, hexokinase. Box with dotted line on long exposure FLAG immunoblot indicates FLAG-FKBP-Pex6 bands only detectable upon long exposure.
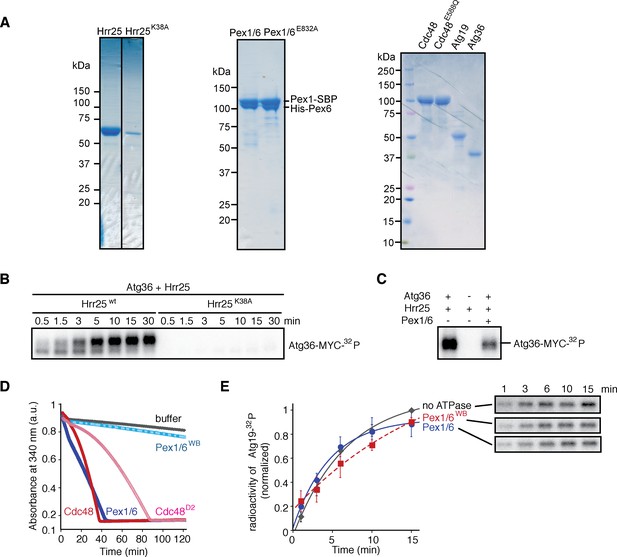
The exportomer inhibits Atg36 phosphorylation in vitro.
(A) Purified proteins used in in vitro kinase assays were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) followed by Coomassie blue staining. K38A, kinase-dead mutant of Hrr25. E832A, Pex1/6 bearing D2 Walker B motif mutant (E832A) in Pex6. D2, Cdc48 D2 ring mutant (E588Q). (B) Purified Atg36 was incubated with 32P-labeled ATP and Hrr25 or Hrr25K38A in vitro, for indicated incubation time. Samples were resolved by SDS–PAGE followed by autoradiography. (C) Purified Atg36 was incubated with 32P-labeled ATP, Hrr25 and Pex1/6 for 15 min. Hrr25 only sample was used as the control. Samples were resolved by SDS–PAGE followed by autoradiography. (D) ATPase activities of AAA+ used in Figure 3F were monitored by measuring the absorbance at 340 nm, using an assay in which regeneration of hydrolyzed ATP was coupled to the oxidation of NADH (see Materials and methods for details). (E) Purified Atg19 was incubated with 32P-labeled ATP, Hrr25, and indicated Pex1/6 complexes. WB, Pex6 D2 Walker B motif mutant (E832A). Samples at each time point were resolved by SDS–PAGE followed by autoradiography. Data points represent the mean values determined from three repeat experiments. Error bars represent standard error.
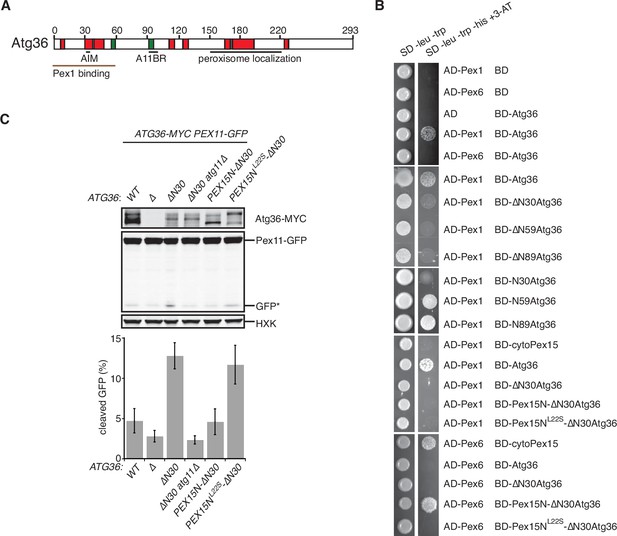
The exportomer inhibits pexophagy by binding to Atg36.
(A) Schematic diagram of Atg36 with predicted secondary structural features indicated (PSIPRED 4.0) (Jones, 1999). Red region, predicted α-helix; green region, predicted β-sheet. AIM, Atg8-family-interacting motif, amino acids 33–36. A11BR, Atg11-binding region, amino acids 94–102. (B) Interaction between exportomer and Atg36 mutants in yeast-two-hybrid assay. Pex1 or Pex6 was fused with the Gal4 activation domain (AD) and Atg36 mutants were fused with Gal4 DNA-binding domain (BD). Cells expressing both AD-exportomer and BD-Atg36 mutants were grown on synthetic defined (SD) media plates as indicated. (C) Cells with indicated genotypes were grown in oleate medium for 22 hr. The extracts were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) followed by immunoblotting with anti-MYC, anti-GFP, and anti-HXK antibodies. GFP*, GFP fragments produced upon vacuolar degradation of Pex11-GFP. HXK, hexokinase. Data points represent the mean values determined from three independent experiments. Error bars represent standard error.
-
Figure 4—source data 1
Raw immunoblotting data related to Figure 4C.
- https://cdn.elifesciences.org/articles/74531/elife-74531-fig4-data1-v1.pdf
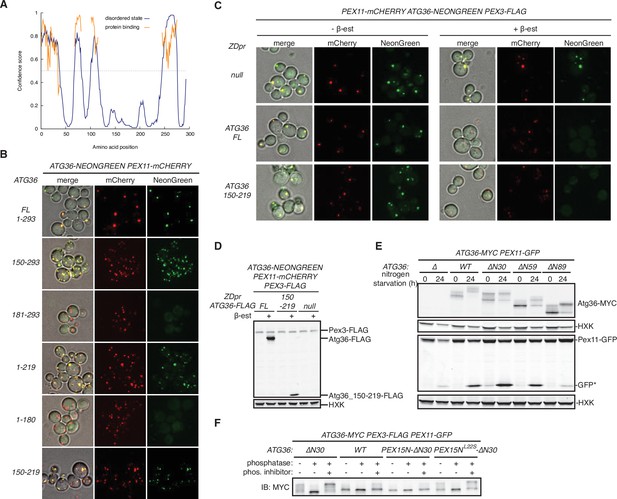
The exportomer inhibits pexophagy by binding to Atg36.
(A) Disordered regions in Atg36 predicted by DISOPRED3 (Jones and Cozzetto, 2015). (B) Cells expressing Pex11-mCherry and NeonGreen-tagged Atg36 mutants were imaged by confocal microscopy (594 nm channel for mCherry and 488 nm channel for NeonGreen). NeonGreen-tagged Atg36 mutants without N-terminal 149 (150–293 mutant) or C-terminal 74 (1–219 mutant) amino acid residues still formed puncta that colocalized with Pex11-mCherry. The truncations that interrupted the fragment between amino acids 150 and 219 (181–293 and 1–180 mutants) abolished this colocalization. Fusion of a fragment containing residues 150–219 of Atg36 is sufficient to target NeonGreen to the peroxisome. Thus, amino acids 150–219 of Atg36 are necessary and sufficient for the peroxisomal localization of Atg36. Cells expressing 150–293 and 1–219 Atg36 mutants exhibit increased numbers of Pex11-mCherry puncta, suggesting the N- and C-terminal domains of Atg36 may be involved in the regulation of peroxisome size, clustering, or abundance. (C) Cells expressing Pex11-mCherry and Atg36-NeonGreen under control of their endogenous promoters, respectively, and a second copy of FLAG-tagged Atg36 (wild-type or amino acids 150–219 of Atg36; no fluorescent protein tag) expressed on a CEN plasmid under control of the ZD promoter were imaged by confocal microscopy. Overexpression of either FLAG-tagged full-length Atg36 or amino acids 150–219 caused reduced colocalization of Atg36-NeonGreen signal with Pex11-mCherry-labeled peroxisomes. Where indicated, cells were treated with 1 μM β-estradiol (β-est) for 3 hr before imaging. (D) Extracts derived from cells imaged in (C) with indicated genotypes were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) followed by immunoblotting with anti-FLAG and anti-HXK antibodies. HXK, hexokinase. (E) Cells expressing Pex11-GFP were grown in oleate medium for 22 hr and then transferred to nitrogen starvation medium. Extracts derived from cells with indicated genotypes were resolved by SDS–PAGE followed by immunoblotting with anti-MYC, anti-HXK, and anti-GFP antibodies. GFP*, GFP fragments produced upon vacuolar degradation of Pex11-GFP. HXK, hexokinase. (F) The indicated extracts were subjected to immunoprecipitation (IP) with anti-MYC magnetic beads. After extensive washing, bound material was treated with lambda phosphatase in the presence or absence of phosphatase (phos.) inhibitors, and then resolved by SDS–PAGE followed by immunoblotting (IB) with anti-MYC antibodies.
Additional files
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/74531/elife-74531-transrepform1-v1.docx
-
Supplementary file 1
Table of yeast strains used in this study.
- https://cdn.elifesciences.org/articles/74531/elife-74531-supp1-v1.docx
-
Source data 1
- https://cdn.elifesciences.org/articles/74531/elife-74531-data1-v1.zip
-
Source data 2
Raw immunoblot and autoradiography data, related to Figures 1—4.
- https://cdn.elifesciences.org/articles/74531/elife-74531-data2-v1.zip
-
Source data 3
Raw immunoblot and autoradiography data, related to figure supplements.
- https://cdn.elifesciences.org/articles/74531/elife-74531-data3-v1.zip