The DBL-1/TGF-β signaling pathway tailors behavioral and molecular host responses to a variety of bacteria in Caenorhabditis elegans
Figures
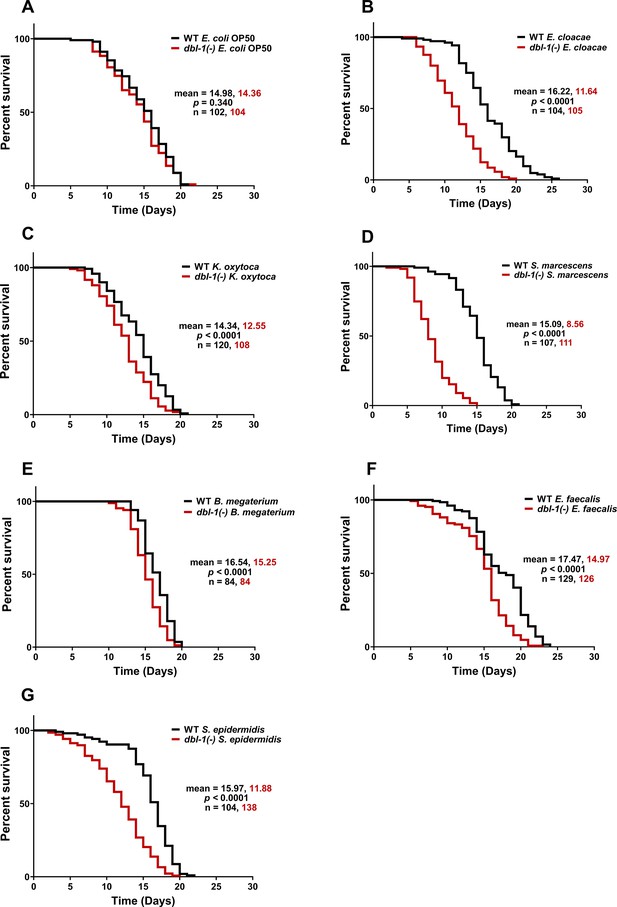
Loss of DBL-1 decreases lifespan of animals exposed to test Gram-negative and Gram-positive bacteria.
Wild-type and dbl-1(-) animals were scored for survival over time from the L4 stage (t = 0 hr) on the following bacteria: (A) E. coli OP50 (control), (B) E. cloacae, (C) K. oxytoca, (D) S. marcescens, (E) B. megaterium, (F) E. faecalis, and (G) S. epidermidis. Survival fraction was calculated by the Kaplan–Meier method. p-Values were calculated using log-rank test and p<0.01 compared to wild-type animals exposed to the same bacteria was considered significant. One representative trial is presented.
-
Figure 1—source data 1
Related to Figure 1.
- https://cdn.elifesciences.org/articles/75831/elife-75831-fig1-data1-v2.xlsx
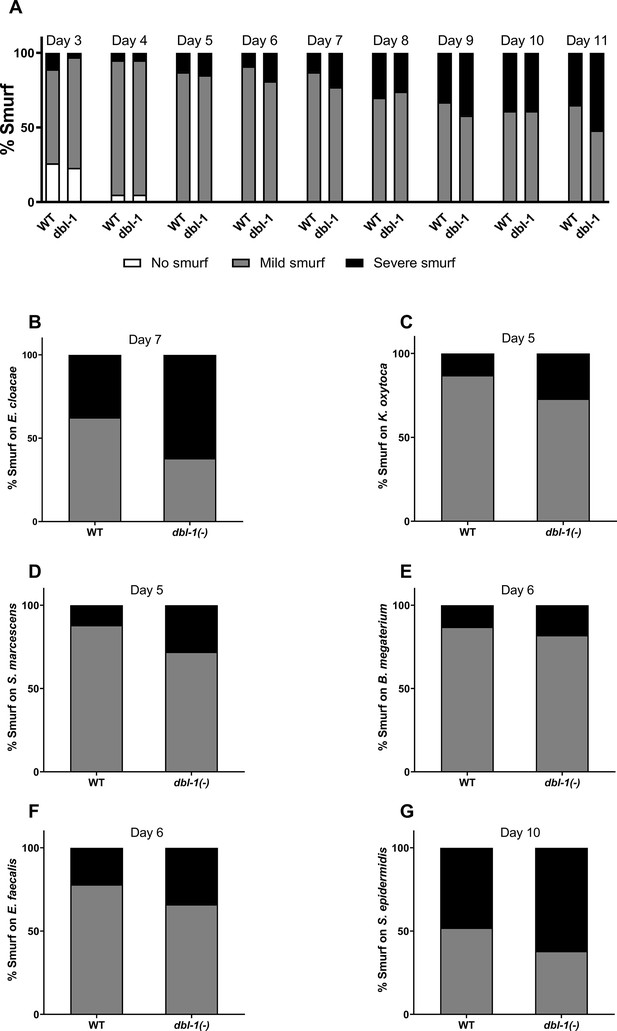
Loss of DBL-1 does not affect intestinal integrity upon exposure to specific bacteria.
Wild-type and dbl-1(-) populations at the L4 stage were exposed to (A) E. coli OP50 (control), (B) E. cloacae, (C) K. oxytoca, (D) S. marcescens, (E) B. megaterium, (F) E. faecalis, or (G) S. epidermidis. Intestinal barrier function was assessed using erioglaucine disodium salt (A) over time or (B–G) when dbl-1(-) populations neared their half lifespan. The leakiness of the intestine was assessed and scored as ‘1’ for no leakage/no Smurf, ‘2’ for mild leakage/mild Smurf, and ‘3’ for severe leakage/severe Smurf phenotypes. The fraction of animals exhibiting each phenotype was calculated. One representative trial of at least three is presented. The intestinal barrier function phenotypes were statistically analyzed by the chi-square test. n ≥ 10 animals per condition at each time point.
-
Figure 1—figure supplement 1—source data 1
Related to Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/75831/elife-75831-fig1-figsupp1-data1-v2.xlsx
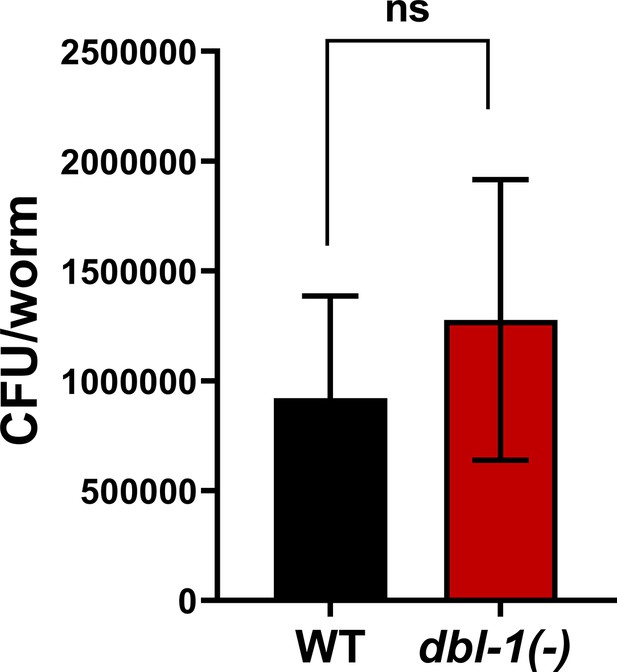
Loss of DBL-1 does not affect bacterial colonization upon exposure to S. marcescens.
Number of colony-forming units (CFU) per worm was compared between wild-type and dbl-1(-) animals exposed to S. marcescens when dbl-1(-) populations neared their half lifespan. n = 5 animals in quadruplicates. Error bars represent standard error mean. Comparison was done using an unpaired t-test, p=0.67.
-
Figure 1—figure supplement 2—source data 1
Related to Figure 1—figure supplement 2.
- https://cdn.elifesciences.org/articles/75831/elife-75831-fig1-figsupp2-data1-v2.xlsx
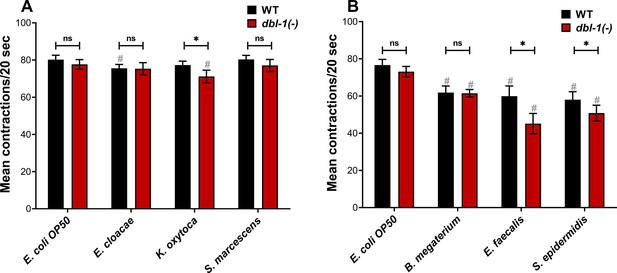
Loss of DBL-1 and exposure to specific bacteria results in decreased pharyngeal pumping.
Wild-type and dbl-1(-) animals at the L4 stage were exposed to the following bacteria: (A, B) E. coli OP50 (control); (A) E. cloacae, K. oxytoca, S. marcescens; (B) B. megaterium, E. faecalis, or S. epidermidis. After 48 hr of exposure, the number of pharyngeal pumps was counted twice per 20 s. The pharyngeal pumps were averaged for each animal. One representative trial is presented. Error bars represent standard deviation. n = 8–12 per condition. *p<0.01, ns, not significant, compared to wild-type animals exposed to the same bacteria, and #p<0.01, respective genotype exposed to test bacteria in comparison to control bacteria by two-way ANOVA using Tukey’s post hoc test.
-
Figure 2—source data 1
Related to Figure 2.
- https://cdn.elifesciences.org/articles/75831/elife-75831-fig2-data1-v2.xlsx
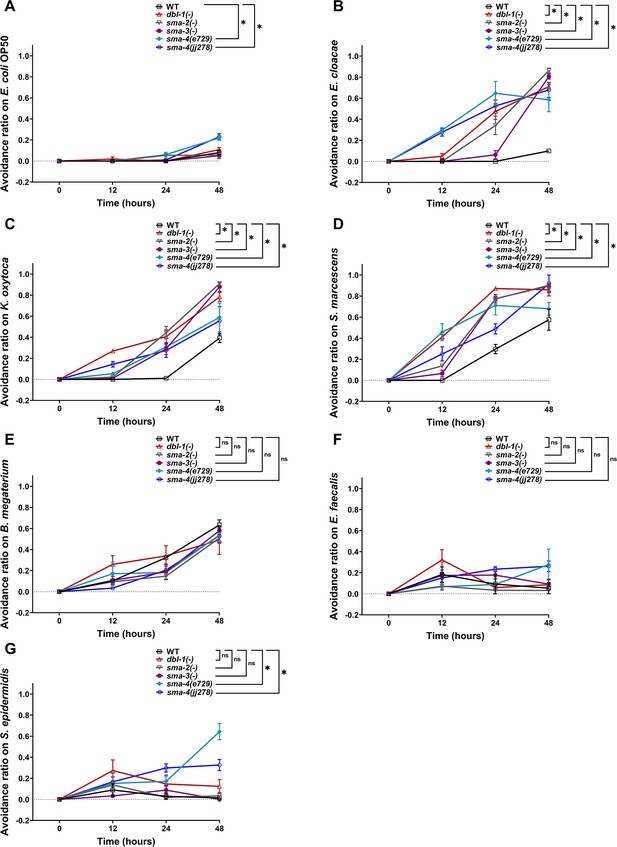
Avoidance to Gram-negative bacteria increases upon loss of canonical DBL-1 signaling over time while avoidance to specific Gram-positive is independent of DBL-1 signaling.
Wild-type, dbl-1(-), sma-2(-), sma-3(-), and two sma-4(-) strains at the L4 stage (t = 0 hr) were exposed to the following bacteria: (A) E. coli OP50 (control), (B) E. cloacae, (C) K. oxytoca, (D) S. marcescens, (E) B. megaterium, (F) E. faecalis, or (G) S. epidermidis. The avoidance ratio was calculated and compared to the wild-type control animals. Each trial used three plates of 30 animals each per condition. One representative trial is presented. Error bars represent standard error mean. *p<0.05, ns, not significant, compared to wild-type animals exposed to the same bacteria by repeated measures ANOVA using Tukey’s post hoc test.
-
Figure 3—source data 1
Related to Figure 3.
- https://cdn.elifesciences.org/articles/75831/elife-75831-fig3-data1-v2.xlsx
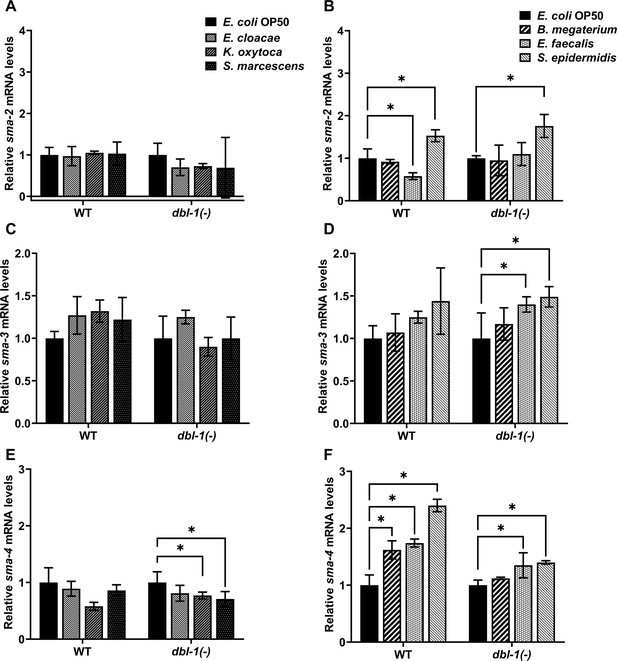
Smad transcription factors gene expression is altered by specific bacteria.
Wild-type and dbl-1(-) animals at the L4 stage were exposed to E. coli OP50 (control), E. cloacae, K. oxytoca, S. marcescens, B. megaterium, E. faecalis, or S. epidermidis for 48 hr. Relative mRNA expression levels of (A, B) sma-2, (C, D) sma-3, and (E, F) sma-4 were quantitated by real-time PCR. Experiments were performed in three technical replicates and in three independent biological trials. One representative trial is presented. Error bars represent standard deviation. *p<0.05, mRNA expression level in respective genotype exposed to test bacteria compared to control E. coli OP50, by one-way ANOVA with Dunnett’s multiple-comparisons test.
-
Figure 4—source data 1
Related to Figure 4.
- https://cdn.elifesciences.org/articles/75831/elife-75831-fig4-data1-v2.xlsx
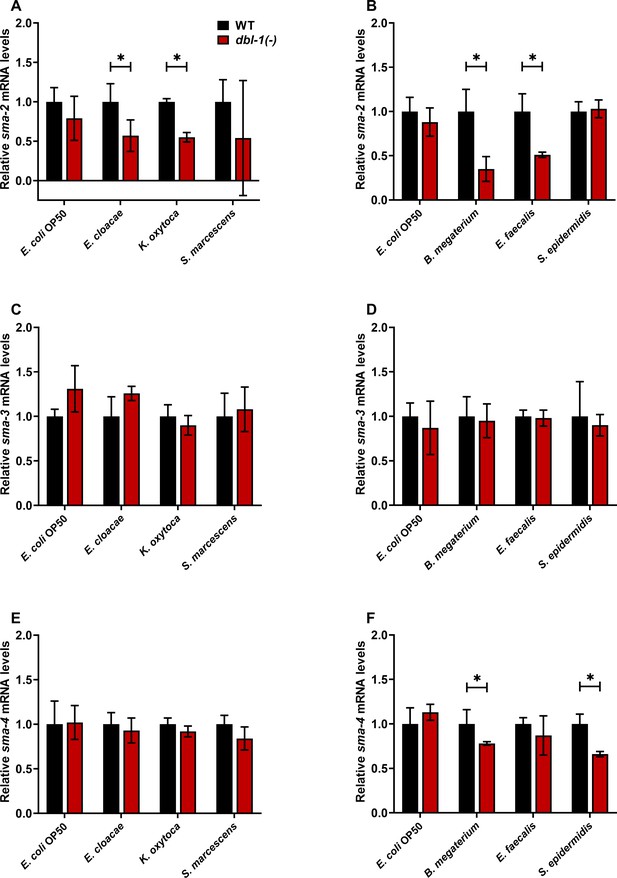
Smad transcription factor gene expression is altered by specific bacteria.
Wild-type and dbl-1(-) animals at the L4 stage were exposed to control or test bacteria for 48 hr. Relative mRNA expression levels of (A, B) sma-2, (C, D) sma-3, and (E, F) sma-4 in wild-type and dbl-1(-) animals exposed to (A–F) E. coli OP50, (A, C, E) E. cloacae, K. oxytoca, S. marcescens, (B, D, F) B. megaterium, E. faecalis, and S. epidermidis were quantitated by real-time PCR. Experiments were performed in triplicate and in three independent trials. One representative trial is presented. Error bars represent standard deviation. *p<0.05, mRNA expression level in dbl-1(-) population compared to wild-type population exposed to the same bacteria by unpaired t-test.
-
Figure 4—figure supplement 1—source data 1
Related to Figure 4—figure supplement 1.
- https://cdn.elifesciences.org/articles/75831/elife-75831-fig4-figsupp1-data1-v2.xlsx
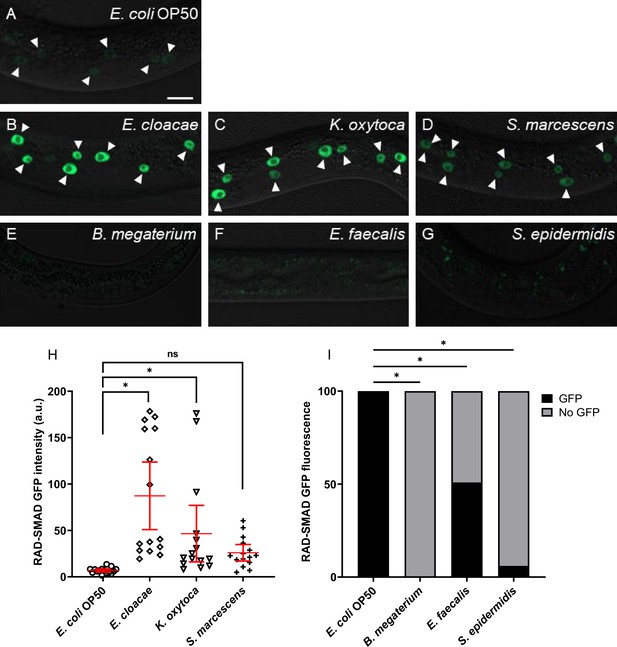
DBL-1 signaling is activated upon exposure to Gram-negative bacteria but is repressed in response to Gram-positive bacteria.
Representative images of L2-stage wild-type animals expressing the RAD-SMAD reporter exposed to (A) E. coli OP50 (control), (B) E. cloacae, (C) K. oxytoca, (D) S. marcescens, (E) B. megaterium, (F) E. faecalis, or (G) S. epidermidis. Imaging conditions were consistent in (A–G). Arrowheads indicate visibly fluorescent hypodermal nuclei. Scale bar, 10 µm. Fluorescent intensities of wild-type animals expressing the RAD-SMAD reporter and fed on control E. coli OP50 or test Gram-negative bacteria are compared in (H). Mean RAD-SMAD fluorescence intensity of five hypodermal nuclei per animal was determined and compared. n = 10 animals per condition in each trial. Experiments were performed in three independent trials. One representative trial is presented. Error bars represent 95% confidence intervals. (I) Qualitative assessments of fluorescence from wild-type animals expressing the RAD-SMAD reporter on control E. coli OP50 or test Gram-positive bacteria. Percentage of animals showing detectable or no detectable RAD-SMAD GFP fluorescence from three trials is presented (n=at least 10 animals/trial). *p<0.05, mean fluorescence intensity in wild-type background on test bacteria compared to control bacteria by chi-square test.
-
Figure 5—source data 1
Related to Figure 5.
- https://cdn.elifesciences.org/articles/75831/elife-75831-fig5-data1-v2.xlsx
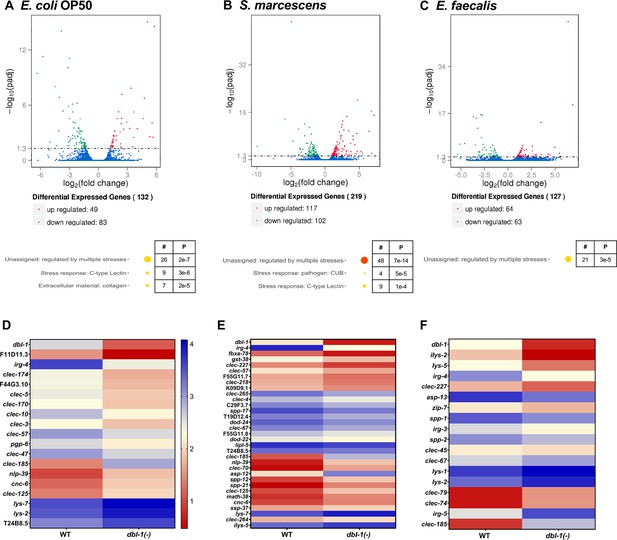
DBL-1 regulates differential gene expression in response to Gram-negative and Gram-positive bacteria.
Wild-type and dbl-1(-) animals were exposed to E. coli OP50 (control), S. marcescens, or E. faecalis for 2 d starting at the L4 stage. RNA-seq analysis using volcano plots shows differential gene expression in animals lacking DBL-1 exposed to (A) E. coli OP50, (B) S. marcescens, and (C) E. faecalis in comparison to wild-type animals exposed to the same bacteria (adjusted p-value<0.01). Genes downregulated in dbl-1(-) animals are represented in green, genes upregulated in dbl-1(-) animals are represented in red, and genes with no change in expression are represented in blue in the volcano plots. Functional categories of differentially expressed genes (WormCat Category 3) are shown. The circle size and color represent relative number of genes in each group and p-value, respectively (orange, p<10–10; yellow, p<0.05; see Figure 6—source data 1 for details). Heatmaps show differential innate immunity gene expression in animals lacking DBL-1 exposed to (D) E. coli OP50, (E) S. marcescens, and (F) E. faecalis in comparison to wild-type animals exposed to the same bacteria. Average log FPKM values from three independent trials are represented.
-
Figure 6—source data 1
Related to Figure 6.
- https://cdn.elifesciences.org/articles/75831/elife-75831-fig6-data1-v2.xlsx
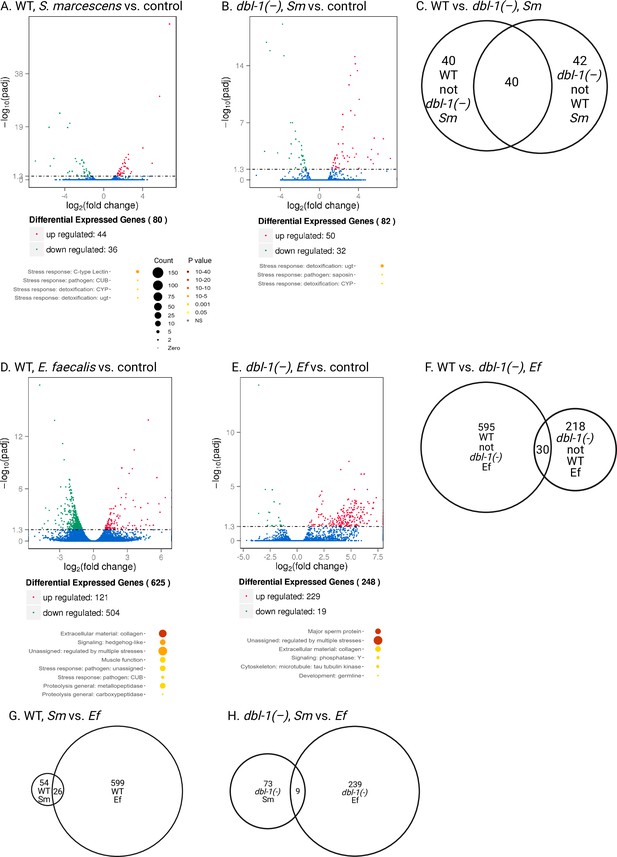
DBL-1-dependent and DBL-1-independent differential gene expression in response to Gram-negative and Gram-positive bacteria.
Wild-type and dbl-1(-) animals were exposed to E. coli OP50 (control), S. marcescens (Sm), or E. faecalis (Ef) for 2 d starting at the L4 stage. RNA-seq analysis using volcano plots shows differential gene expression in wild-type animals fed on control bacteria compared to (A) S. marcescens or (D) E. faecalis (adjusted p-value<0.01). Genes downregulated in wild-type animals upon exposure to S. marcescens or E. faecalis are represented in green, whereas the upregulated genes are represented in red, and genes with no change in expression are represented in blue. Functional categories of differentially expressed genes (WormCat Category 3) are shown. Circle size and color represent relative number of genes in each group and p-value, respectively (orange, p<10–10; yellow, p<0.05; see Figure 6—source data 1 for details). (C) Venn diagram indicating overlapping and nonoverlapping differentially expressed genes in wild-type and dbl-1(-) populations exposed to S. marcescens. Volcano plots show differential gene expression in dbl-1(-) animals fed on control bacteria compared to (B) S. marcescens or (E) E. faecalis (adjusted p-value<0.01). Genes downregulated in dbl-1(-) animals upon exposure to S. marcescens or E. faecalis are represented in green, whereas the upregulated genes are represented in red, and genes with no change in expression are represented in blue. Functional categories of differentially expressed genes (WormCat Category 3) are shown. (F) Venn diagram indicating overlapping and non-overlapping differentially expressed genes in wild-type and dbl-1(-) populations exposed to E. faecalis. (G) Venn diagram indicating overlapping and nonoverlapping differentially expressed genes in wild-type populations exposed to S. marcescens and E. faecalis. (H) Venn diagram indicating overlapping and non-overlapping differentially expressed genes in dbl-1(-) populations exposed to S. marcescens and E. faecalis.
-
Figure 6—figure supplement 1—source data 1
Related to Figure 6—figure supplement 1.
- https://cdn.elifesciences.org/articles/75831/elife-75831-fig6-figsupp1-data1-v2.xlsx
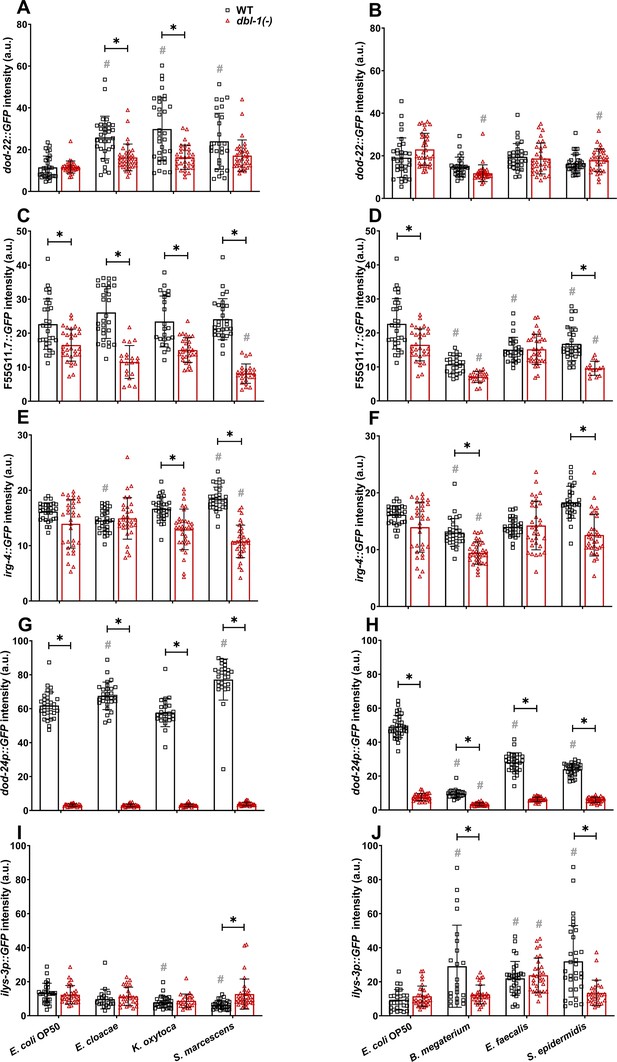
Innate immune reporter activity is regulated by exposure to specific bacteria and by DBL-1 signaling.
Comparison of (A, B) dod-22::GFP, (C, D) F55G11.7::GFP, (E, F) irg-4::GFP, (G, H) dod-24p::GFP, and (I, J) ilys-3p::GFP intensities in adult wild-type and dbl-1(-) animals after a 2-day exposure to the following bacteria; control E. coli OP50, E. cloacae, K. oxytoca, S. marcescens, B. megaterium, E. faecalis, or S. epidermidis. Three independent trials were performed. One representative trial is shown. Error bars represent standard deviation. n ≥ 14 per condition in each trial. **p<0.05 compared to wild-type animals exposed to the same bacteria, and #p<0.05 respective genotype exposed to test bacteria in comparison to control bacteria, by two-way ANOVA using Tukey’s post hoc test.
-
Figure 7—source data 1
Related to Figure 7.
- https://cdn.elifesciences.org/articles/75831/elife-75831-fig7-data1-v2.xlsx
Tables
| gene | Published change onGram-negative bacteria | Published change on Gram-positive bacteria | Published target of IIR pathway | Change with dbl-1(-) compared to wild typeon E. coli OP50 (thiswork) | Change with dbl-1(-) compared to wild typeon S. marcescens (thiswork) | Change with dbl-1(-) compared to wild typeon E. faecalis (our work) | Change with wild-type on S. marcescenscompared to wild typeon E. coli OP50 (ourwork) | Change with wild-type on E. faecalis compared to wild typeon E. coli OP50 (ourwork) |
|---|---|---|---|---|---|---|---|---|
| dod-22 | Y | N | DAF-16 | N | N | Y | Y | Y |
| F55G11.7 | Y | Y | DAF-16, MAPK, and DBL-1 | N | Y | N | Y | N |
| irg-4 | Y | N | DAF-16, MAPK, and DBL-1 | Y | Y | Y | Y | Y |
| dod-24 | Y | N | DAF-16 | N | N | Y | Y | Y |
| ilys-3 | N | Y | ERK MAPK | N | N | N | N | Y |
Additional files
-
Supplementary file 1
Summary of survival assay reported in Figure 1.
- https://cdn.elifesciences.org/articles/75831/elife-75831-supp1-v2.docx
-
Supplementary file 2
Fraction of animals expressing detectable RAD-SMAD reporter fluorescence in dbl-1(-) animals upon exposure to control and test Gram-negative and Gram-positive bacteria.
- https://cdn.elifesciences.org/articles/75831/elife-75831-supp2-v2.xlsx
-
Supplementary file 3
List of strains used and created for this work.
- https://cdn.elifesciences.org/articles/75831/elife-75831-supp3-v2.docx
-
Supplementary file 4
List of primers used for qRT-PCR.
- https://cdn.elifesciences.org/articles/75831/elife-75831-supp4-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/75831/elife-75831-transrepform1-v2.pdf





