An expanded toolkit for Drosophila gene tagging using synthesized homology donor constructs for CRISPR-mediated homologous recombination
Figures
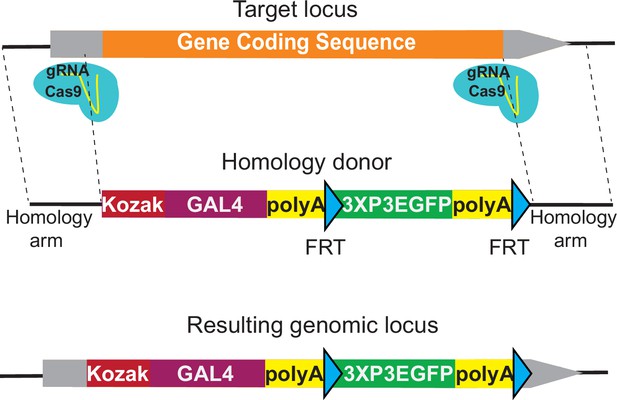
KozakGAL4 strategy can be used to generate GAL4 gene trap alleles.
Schematics of the KozakGAL4 targeting. Gray boxes, UTRs; orange box, gene-coding region.
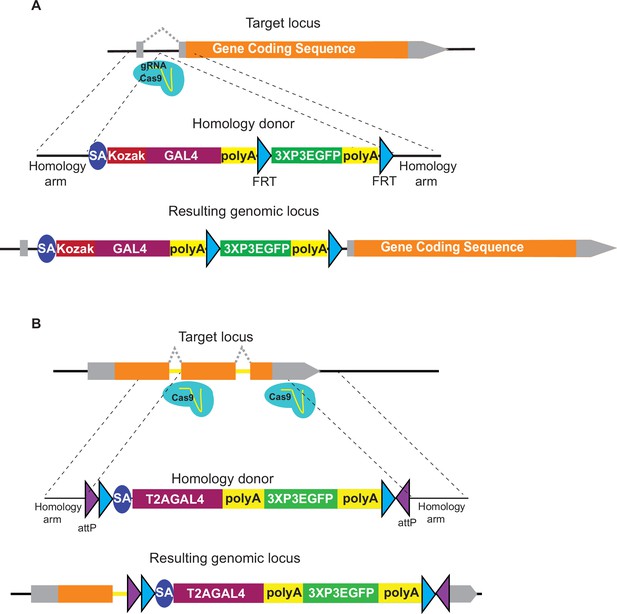
Alternative strategies to generate gene trap alleles in genes without suitable introns.
For genes that cannot be targeted by artificial exon strategies and where suitable sgRNAs could not be found in the 5′ UTR, an artificial exon with SA_KozakGAL4 can be inserted in an intron in the 5′ UTR (A) or in a short intron by deleting the exons following the intron (B).
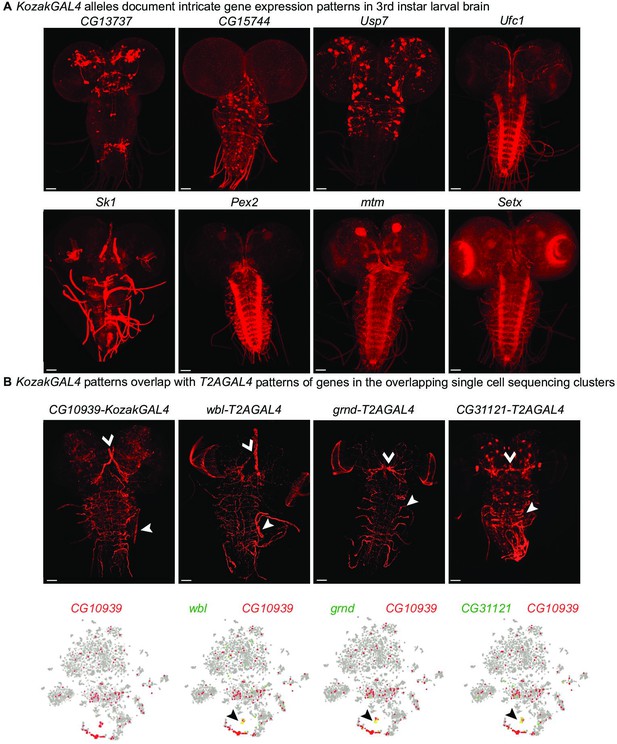
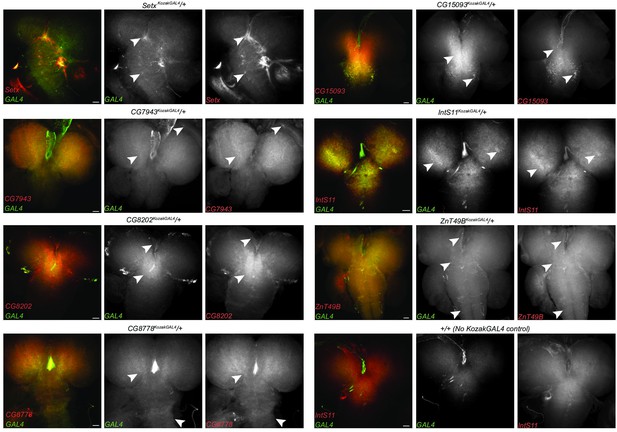
KozakGAL4 alleles document intricate gene expression patterns in third instar larval brains.
(A) Examples of third instar larval brain gene expression patterns obtained by crossing KozakGAL4 allele of indicated genes with UAS-CD8mCherry flies. (B) The imaging results of reporter expression generated with KozakGAL4 allele were compared to the expression pattern of genes that are expressed in similar cells by analysis of single-cell sequencing data imaged using T2AGAL4 alleles. Images are taken by crossing the GAL4 alleles with UAS-CD8mCherry. Arrowheads point to the shared expression pattern. Scale bar is 50µm.
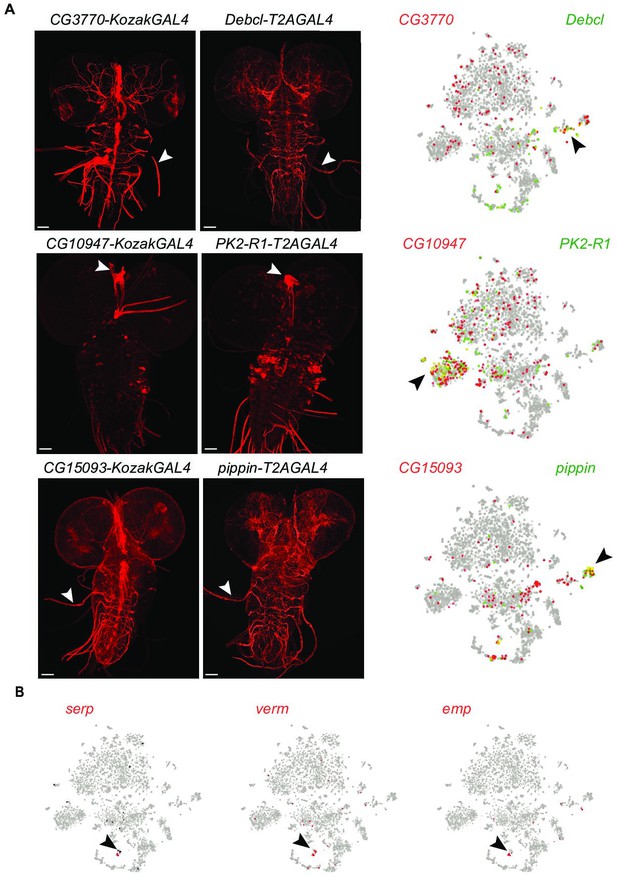
Identification of genes expressed in similar cells to the KozakGAL4 alleles expressed in restricted patterns.
(A) The single-cell sequencing data from Ravenscroft et al., 2020 with the cells expressing the gene targeted by KozakGAL4 marked by red circles and cells expressing the gene targeted with T2AGAL4 allele marked by green circles. The imaging results of reporter expression generated with KozakGAL4 alleles were compared to the expression patterns of genes that are expressed in similar cells by analysis of single-cell sequencing data imaged using T2AGAL4 alleles. Images are taken by crossing the GAL4 alleles with UAS-CD8mCherry. Arrowheads show the regions with the most overlap. (B) Cluster of trachea markers in the scRNA data from Ravenscroft et al., 2020. Scale bar is 50µm.
Comparison of mRNA expression of GAL4 and targeted genes mRNA for KozakGAL4 alleles.
GAL4 mRNA and gene-specific mRNAs are detected simultaneously by two color smiFISH. In each case, there was significant overlap between signals. yw flies are used as control for the specificity of GAL4 probes. Scale bar is 30µm.
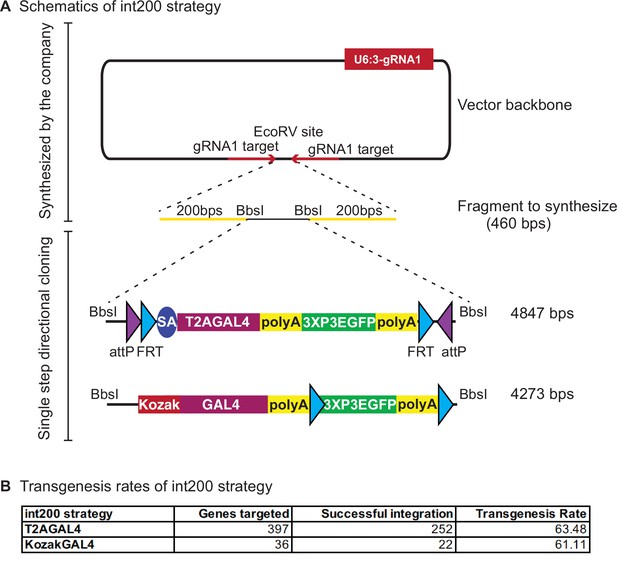
int200 strategy results in similar transgenesis success rates as the long homology arms CRISPR-mediated integration cassettes (CRIMICs).
(A) Schematics of the int200 strategy. (B) Transgenesis data using int200_T2AGAL4 or int200_KozakGAL4 strategies.
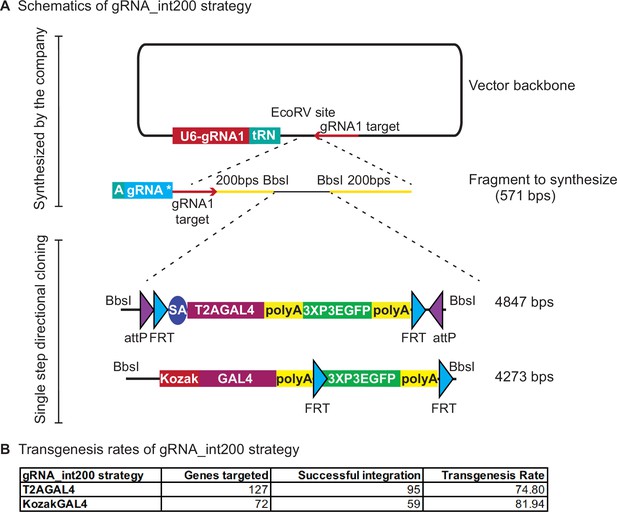
gRNA_int200 strategy increases the transgenesis success rates.
(A) Schematics of the gRNA_int200 strategy. (B) Transgenesis data using gRNA_int200_T2AGAL4 or 2XgRNA_int200_KozakGAL4 strategies.
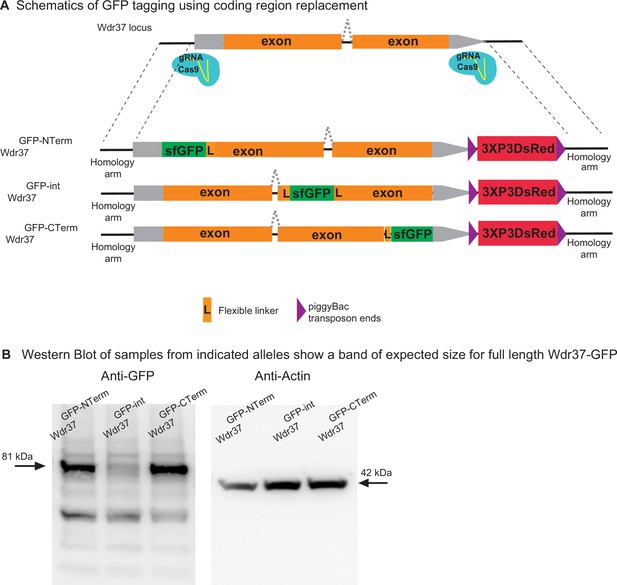
2XgRNA_int200 strategy can be used to tag any gene at any coding region to generate protein trap alleles.
(A) Schematics of the targeting constructs to integrate sfGFP protein tag at an N-terminal, internal, or C-terminal location in Wdr37 gene locus. (B) Western blot analysis from adult flies show full-length protein in all protein trap alleles with the arrow indicating the 81 kDa band that is the length predicted for the Wdr37 protein fused to sfGFP.
-
Figure 5—source data 1
Source data for the western blots from Figure 5.
- https://cdn.elifesciences.org/articles/76077/elife-76077-fig5-data1-v2.zip
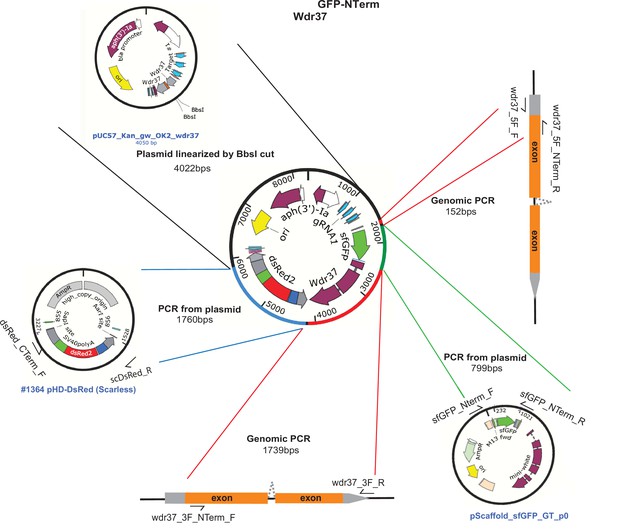
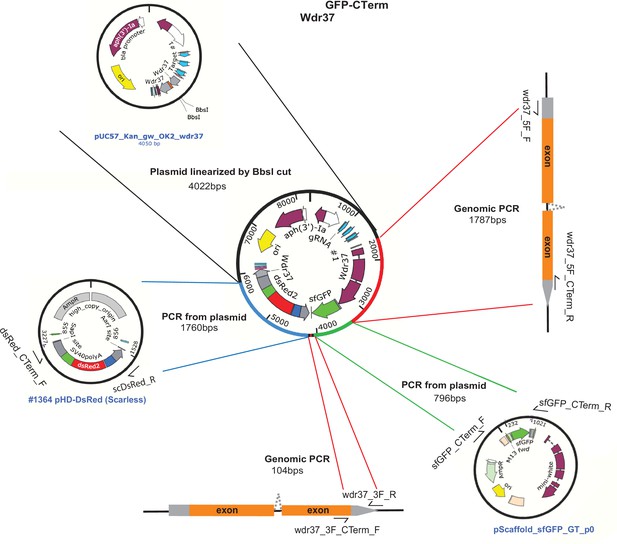
Schematic of Wdr37 GFP-NTerm protein trap allele donor construct.
Schematics of fragments used for NEB-HiFi DNA assembly cloning of Wdr37GFP-NTerm homology donor plasmid.
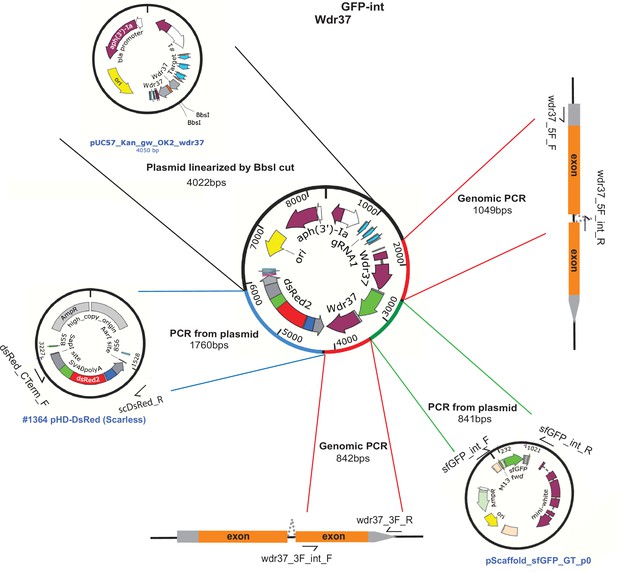
Schematic of Wdr37 GFP-int protein trap allele donor construct.
Schematics of fragments used for NEB-HiFi DNA assembly cloning of Wdr37GFP-int homology donor plasmid.
Additional files
-
Supplementary file 1
Comparison of genes with or without suitable introns for T2AGAL4 strategy.
Comparison of the CDS length of the 5787 protein-coding genes with an intron suitable for the T2AGAL4 strategy with the 8144 genes lacking a suitable intron, because the intron is either absent, <100 nt, or is not present in all annotated transcript isoforms. The left column of the table lists categories of CDS size or the number of alleles annotated in FlyBase (classical and transposon insertions). The number of genes in each category is given for genes with introns that are suitable for the T2AGAL4 strategy (middle column) and for genes lacking a suitable intron (right column). These data for the number of annotated alleles and the CDS size are presented in graphical format below the table.
- https://cdn.elifesciences.org/articles/76077/elife-76077-supp1-v2.xlsx
-
Supplementary file 2
List of the 428 alleles generated in this study.
The genes for which we generated alleles are grouped under headers corresponding to the strategies used to generate the alleles.
- https://cdn.elifesciences.org/articles/76077/elife-76077-supp2-v2.xlsx
-
Supplementary file 3
Detailed design and cloning protocol.
Step-by-step instructions and diagrams depicting construct design and cloning steps. Construct sequences and links to the digital construct sequence files are included in the protocol steps.
- https://cdn.elifesciences.org/articles/76077/elife-76077-supp3-v2.docx
-
Supplementary file 4
Primer sequences.
Sequences of the primers used for PCR verification of alleles and generation of GFP knock-in donor constructs.
- https://cdn.elifesciences.org/articles/76077/elife-76077-supp4-v2.xlsx
-
Supplementary file 5
smiFISH probes.
Sequences of the probes used in smiFISH experiment.
- https://cdn.elifesciences.org/articles/76077/elife-76077-supp5-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/76077/elife-76077-transrepform1-v2.pdf