Mosaic cis-regulatory evolution drives transcriptional partitioning of HERVH endogenous retrovirus in the human embryo
Figures
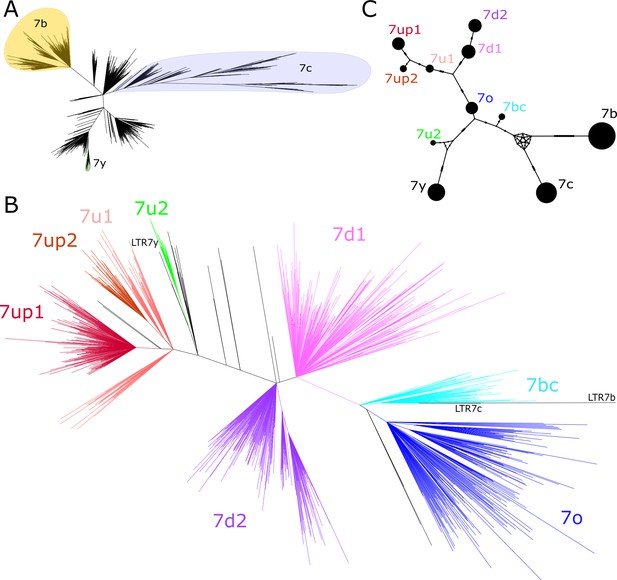
Phylogenetic analysis of LTR7 sequences.
(A) Unrooted phylogeny of all solo and 5’ LTR7 subfamilies from LTR7, 7b, 7c, and 7y. Colors denote clades consisting of previously annotated 7b, 7c, and 7y with >95% concordance. (B) Unrooted phylogeny of all solo and 5’ LTR7 sequences. All nodes with ultrafast bootstraps (UFbootstraps) > 0.95, >10 member insertions, and >1.5 substitutions/100 bp (~6 base pairs) are grouped and colored (see Materials and methods). Previously listed consensus sequences from 7b/c/y were included in the alignment and are shown in black. (C) Median-joining network analysis of all LTR7 and related majority rule consensus sequences. Ticks indicate the number of SNPs at non-gaps between consensus sequences. The size of circles is proportional to the number of members in each subfamily. Only LTR7 insertions that met filtering requirements (see Materials and methods) are included while 7b/c/y counts are from dfam.
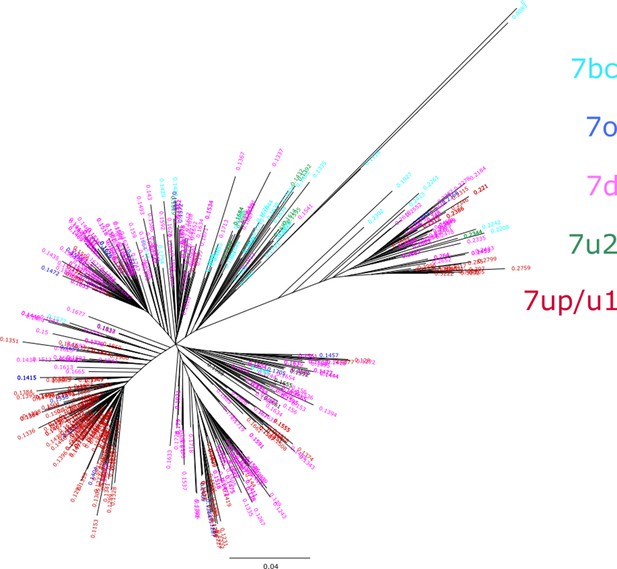
Phylogenetic tree from LTR7 reverse transcriptase (RVT) domains.
The consensus RVT domain was used to extract RT from individual LTR7 insertions. Relative node ages are shown at the end of the terminal branch lengths and are colored corresponding to their subfamily of origin. Only nodes with ultrafast bootstrap support >0.95 are shown. All displayed nodes are considered high confidence.
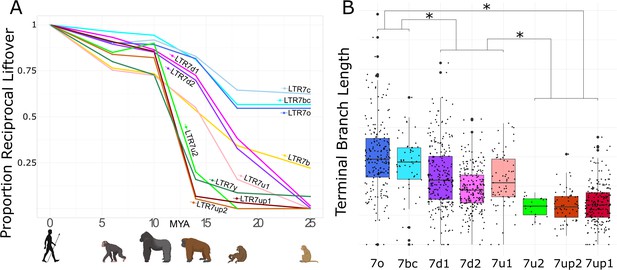
Age analysis of LTR7 subfamilies.
(A) Proportion of a given subfamily that have 1:1 orthologous insertions between human and other primate species. LTR7 subfamilies are from the tree in Figure 1B; 7b/c/y subfamilies are from RepeatMasker annotations. Non-human primates are spaced out on the X axis in accordance with their approximate divergence times to the human lineage. (B) Terminal branch lengths of all LTR7 insertions from Figure 1B. Groups with similar liftover profiles were merged for statistical testing (see Materials and methods). Differences with padj <1e-15 are denoted with * (Wilcoxon rank-sum test with Bonferroni correction).
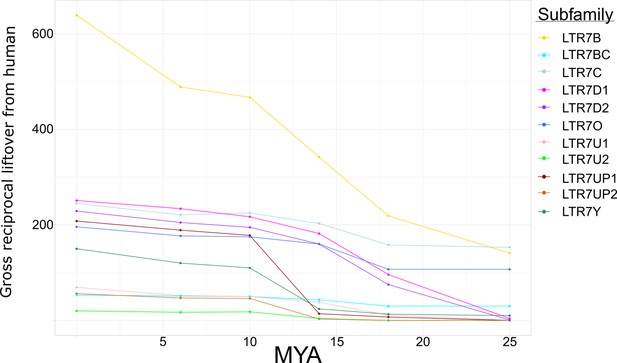
Gross reciprocal liftover from human to non-human primate (NHP).
The absolute number of human endogenous retrovirus type-H (HERVH) long terminal repeats (LTRs) that are in human is listed at MYA 0. Those that reciprocally liftover to chimpanzee, gorilla, orangutan, gibbon, and rhesus macaque are plotted as dots left to right with their relative divergence ages plotted as MYA (bottom). Points are connected with lines, where the slope approximates the rate of propagation of each subfamily between speciation events. This is a sister plot to main text Figure 2A; the only difference is instead of proportion of reciprocal liftover, this is the gross number.
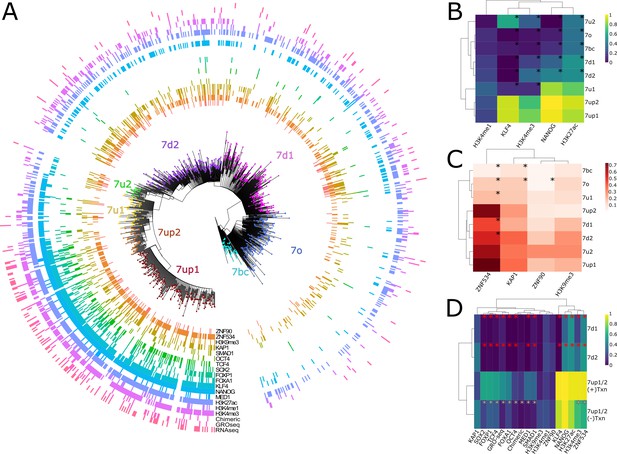
Phyloregulatory analysis of LTR7.
(A) '“Phyloregulatory’ map of LTR7. The phylogenetic analysis to derive the circular tree is the same as for the tree in Figure 1B but rooted on the 7b consensus. Subfamilies defined in Figure 1 are denoted with dotted colored tips. Positive regulatory calls for each insertion are shown as tick marks of different colors and no tick mark indicates a negative call. All marks are derived from embryonic stem cell (ESC) except for ZNF90 and ZNF534, which are derived from ChIP-exo data after overexpression of these factors in HEK293 cells (see Materials and methods). (B) Heatmap of major activation and repression profiles. Proportions indicate the proportion of each group positive for a given characteristic. Trees group LTR7 subfamilies on regulatory signature, not sequence similarity. Asterisks denote statistical differences between given group and 7up1 (padj < 0.05 Wilcoxon rank-sum with Bonferroni correction). (C) Heatmap done in similar fashion to B but for repression marks. (D) Heatmap of transcribed (>2 fpkm) and untranscribed 7up1/2 (<2 fpkm) and all 7d1/2. Red asterisks denote statistical differences between 7d1/2 and 7up1 (padj < 0.05 chi-square Bonferroni correction). White asterisks denote differences between transcribed and untranscribed LTR7up.

Heatmap and aggregate signal of two replicates of whole-genome bisulfite sequencing (WGBS), GRO-seq (plus strand), H3K9me3, and SOX2 in H1 cells.
Visible window is 500 bp upstream of the start of 5’ (or solo) long terminal repeats (LTR) and extends to 8 kb after this point (~3 kb past 3’ LTR or ~7.5 kb past solo into genomic DNA). Loci are sorted on GRO-seq data in the visible window.
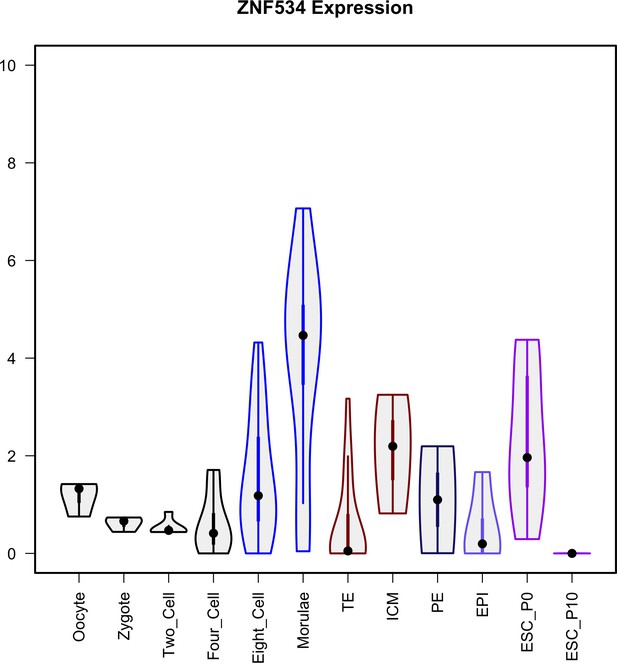
Violin plots visualize the density and distribution of ZNF534 in early embryogenesis and ESCs at passages 0 and 10.
Y axis is (log TPM [transcripts per million]).
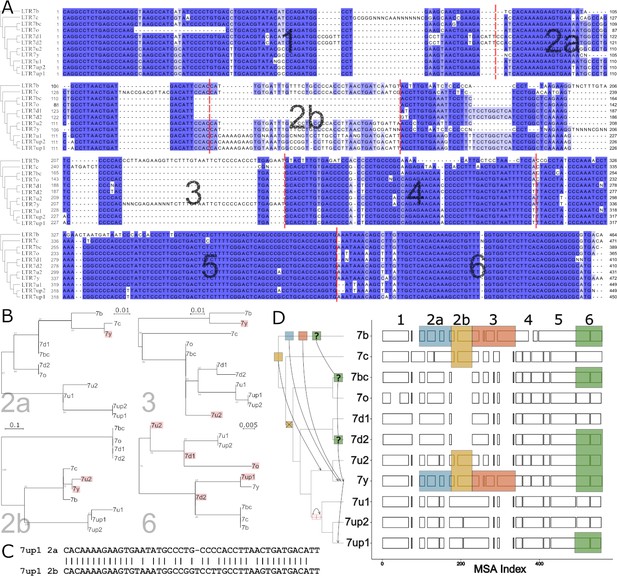
Modular block evolution of LTR7 subfamilies.
(A) A multiple sequence alignment of LTR7 subfamily consensus sequences. The phylogenetic topology from Figure 1 is shown on the left. The MSA is broken down into sequence blocks (red lines) with differential patterns of relationships. (B) Parsimony trees from A sequence blocks. Subfamilies whose blocks do not match the overall phylogeny are highlighted in red. Bootstrap values > 0 are shown. (C) Blastn alignment of LTR7up1 block 2a and 2b. (D) A multiple sequence alignment of majority rule consensus sequences from each LTR7 subfamily detailing shared structure. Blocks show aligned sequence; gaps represent absent sequence. Colored sections identify putative phylogeny-breaking events. Recombination events whose directionality can be inferred (via aging) are shown with blocks and arrows on the cladogram. Recombination events with multiple possible routes are denoted with ‘?’. The deletion of 2b is denoted on the cladogram with a red ‘X’; the duplication of 2a is denoted with two red rectangles.
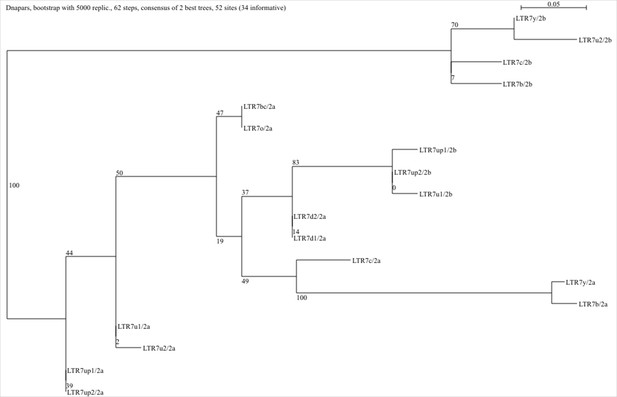
Parsimony tree of all consensus human endogenous retrovirus type-H (HERVH) long terminal repeats (LTR) blocks 2a and 2b.
Sequence blocks 2a and 2b for all subfamilies were aligned using MUSCLE and the output was used to generate a parsimony tree in SEAVIEW (see Materials and methods).
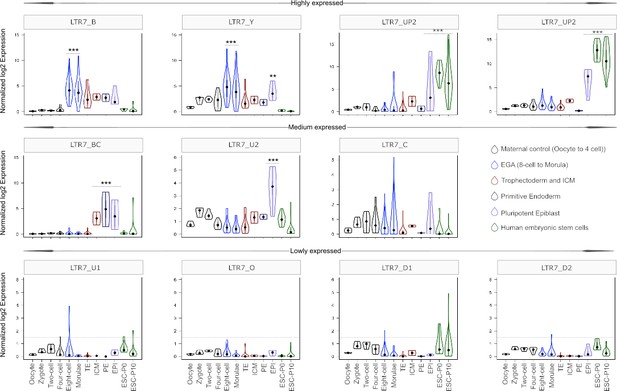
Expression profile of LTR7 subfamilies in human preimplantation embryonic lineages and embryonic stem cells (ESCs).
The solid dots and lines encompassing the violins represent the median and quartiles of single cellular RNA expression. The color scheme is based on embryonic stages, defined as maternal control of early embryos (oocytes, zygote, 2 cell and 4 cell stage), EGA (8 cell and morula), inner cell mass (ICM), trophectoderm (TE), epiblast (EPI), and primitive endoderm (PE) from the blastocyst, and ESCs at passages 0 and 10.
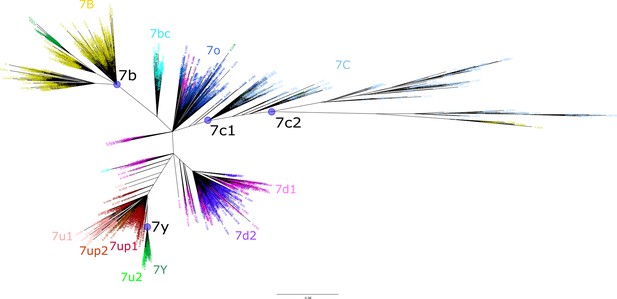
Majority rule consensus sequences used for remasking of human genome.
Consensus sequences for LTR7 subfamilies were generated from Figure 1 (main text – see Materials and methods). Here, we show which sequences were included for the LTR7B, LTR7Y, and LTR7C (LTR7C1+ LTR7C2) consensus sequence generations (purple circles, black labels) using the tree from Figure 1A. Previously annotated LTR7B/C/Y and LTR7 subfamily subdivision annotations from Figure 1 (main text) are shown with colored node ages. The LTR7u2 sister clade was not included in the LTR7Y consensus generation.
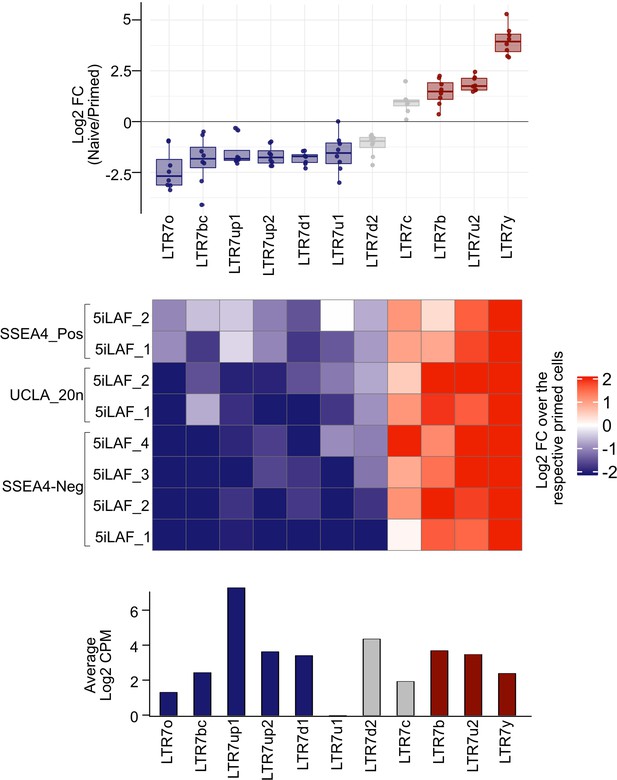
LTR7 subfamily expression in ‘primed’ and ‘naïve’ cell lines.
(Upper panel) Jittered boxplot representing the relative average expression difference (log2 FC) of LTR7 subgroups in naïve and primed cells. Every dot is the log2 FC of one replicate of naïve cells relative to primed cells of the corresponding subfamily. (Middle panel) Heatmap showing the differential expression of distinctive LTR7 subgroups (log2 FC) in individual naïve cells compared with their respective primed cells. (Lower panel) Bar plot showing the average expression across samples of each LTR7 subgroup.
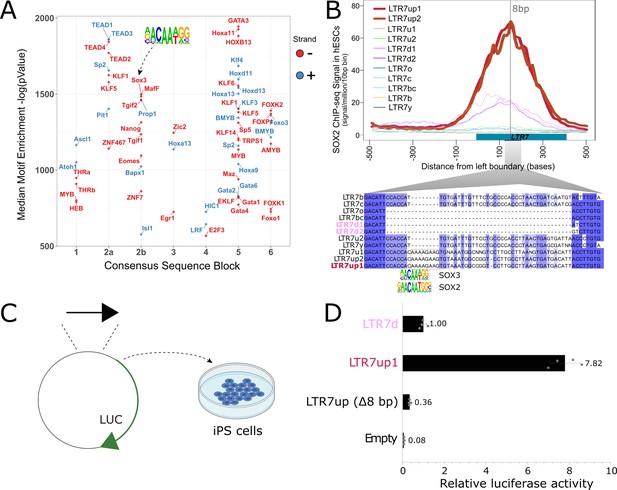
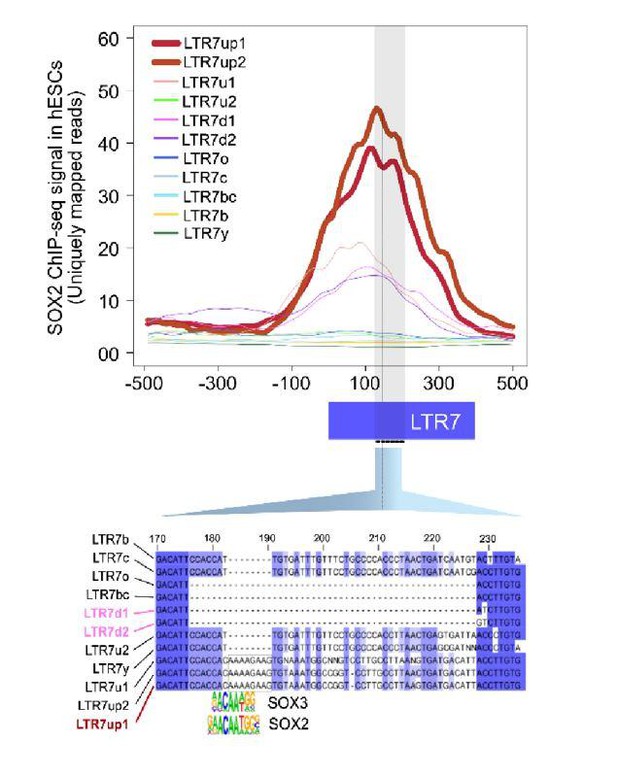
An 8 bp insertion, SOX2/3 binding site necessary for LTR7up transcription.
(A) (log) p-values > 500 for HOMER motifs enriched in 7up1 insertion’s sequence blocks vs. the same blocks from other insertions from other human endogenous retrovirus type-H (HERVH) subfamilies are shown. (B) Line plots show SOX2 ChIP-seq signal at LTR7 subfamily loci in human embryonic stem cells (ESCs). Signal from genomic loci was compiled relative to position 0. The 7up/u1 8 bp insertion position is shown with a dotted line. Region 2b harboring SOX2/3 transcription factor binding site (TFBS) is detailed below. (C) Scheme of DNA fragments cloned into pGL3-basic vector driving luciferase gene expression (LUC) with identified SOX2/3 motifs. Three constructs were analyzed: Entire LTR7up (7up1), 7d1/2 consensus sequence (approximate ancestral sequence for all LTR7d) and LTR7up with eight nucleotides deleted (LTR7up (Δ8bp – AAAAGAAG)) (see panel B). (D) Normalized relative luciferase activity of tested fragments compared to LTR7 down; n = 4 measurements; bars, means across replicates; error bars, standard deviation of the mean, dots, individual replicates.
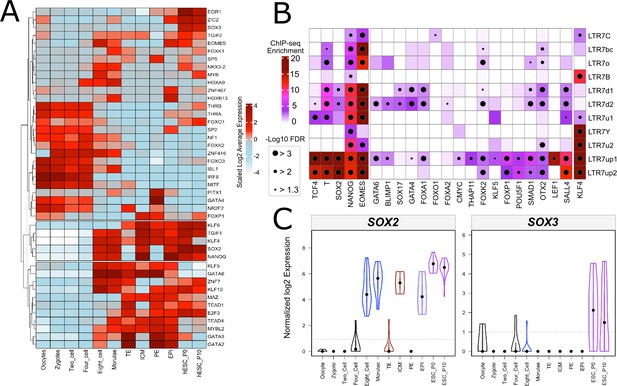
Transcription factor (TF) expression in the preimplantation embryo and TF binding in embryonic stem cells.
(A) Heatmap showing averaged and scaled expression (log of normalized CPM values) of human TFs with enriched motifs in LTR7-up1 subgroup and those are expressed ([log TPM (transcripts per million)] > 1) at least in one cell of early human embryos (A-C). Color scheme is based on z-score distribution, from –4 (light blue) to 4 (dark red). (B) The merged dotplot and heatmap demonstrates overrepresentation analysis of pluripotency TFs binding in distinctive LTR7 subgroups. Color intensity corresponds to log scaled enrichment of the analyzed ChIP-seq peaks within LTR7 subgroups compared to randomized 500 bps bins of human genome. Dots represent the significance (−log10 [FDR]) of ChIP-seq enrichment within a given LTR7 subgroup (see Materials and methods). (C) Violin plots detail the spread of expression of SOX2/3 in the preimplantation embryo. Y axis is log2 TPM.
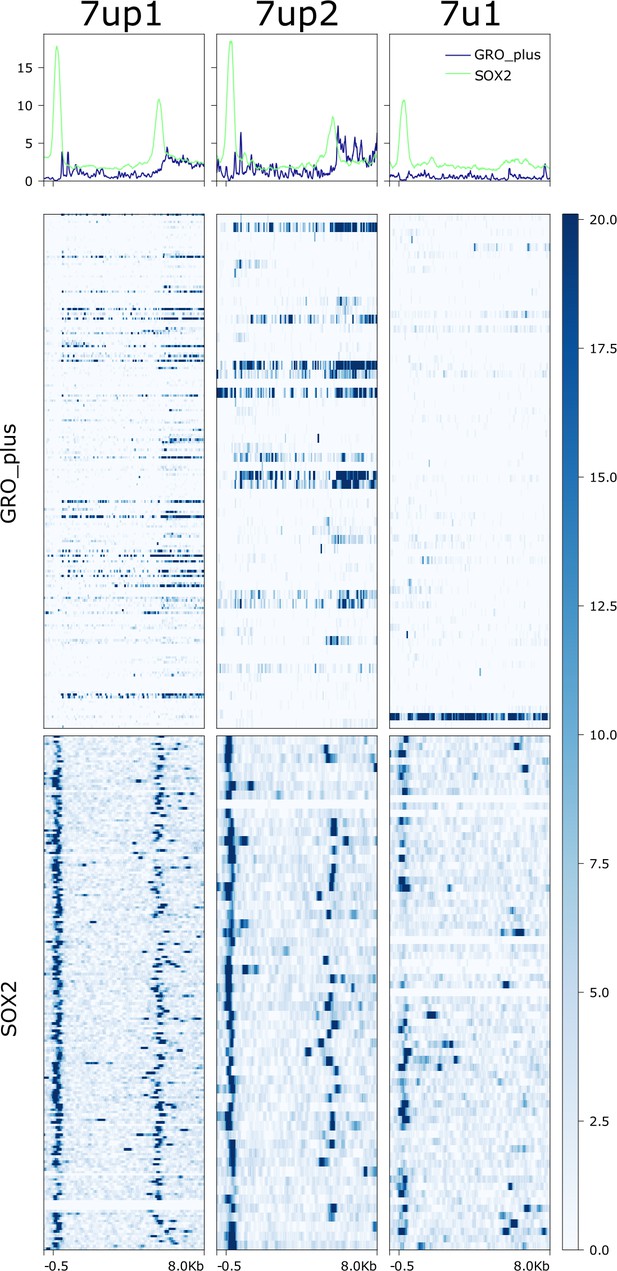
Heatmap showing read pileup from GRO-seq plus strand and SOX2 ChIP-seq on 7up and 7u1 insertions.
Insertions are ranked from highest to lowest in SOX3 motif (HOMER – see Materials and methods) strength. Visible window is -500 bp to +8kb from start of 5' or solo LTR. SOX2 signal intensity over long terminal repeats (LTRs - not entire visible window) with this SOX3 motif and those without was not significantly different (Wilcoxon rank-sum test with Bonferroni correction).
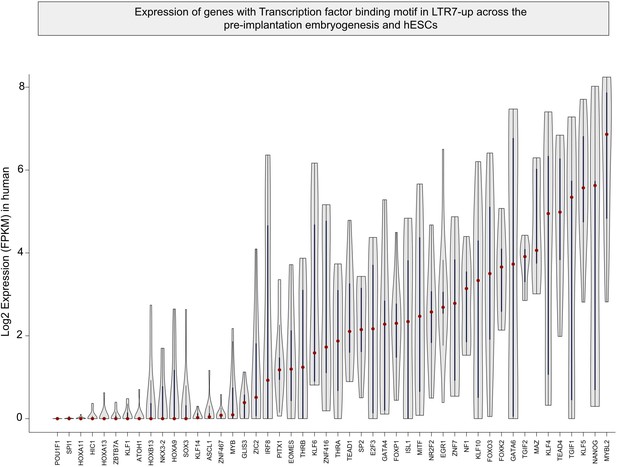
Violin plots detailing the distribution of transcription factor (TF) expression shown in Figure 6—figure supplement 1 in early human embryos and embryonic stem cells.
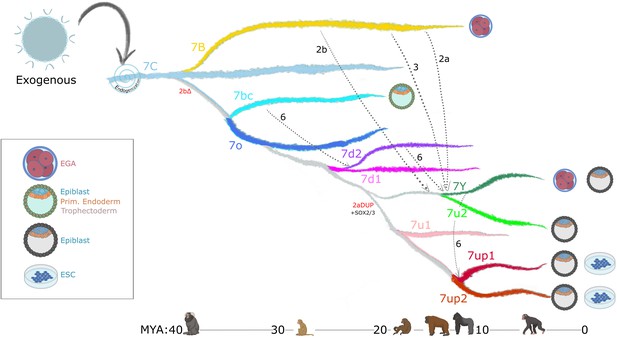
Model of LTR7 subfamily evolution.
Estimated LTR7 subfamily transpositional activity in million years ago (mya) is listed with corresponding approximate primate divergence times (bottom). The positioning and duration of transpositional activity are based on analysis from Figure 3B. The gray connections between subfamilies indicate average tree topology which is driven by overall pairwise sequence similarity. Dashed lines indicate likely recombination events which led to the founding of new subfamilies. Stage-specific expression profiles from Figure 5A are detailed to the right of each corresponding branch.
Additional files
-
Supplementary file 1
List of all LTR7 insertions included in phyloregulatory analysis in hg38 coordinates with accompanying binary regulatory calls.
- https://cdn.elifesciences.org/articles/76257/elife-76257-supp1-v2.xlsx
-
Supplementary file 2
OneCodeToFindThemAll.pl calls for remasked LTR7, LTR7B, LTR7C, and LTR7Y (hg38 coordinates) and quantification of subfamily annotation changes between old and new repeat masking.
- https://cdn.elifesciences.org/articles/76257/elife-76257-supp2-v2.xlsx
-
Supplementary file 3
Complete statistical support for HOMER motif enrichment for LTR7 subfamilies by sequence block.
- https://cdn.elifesciences.org/articles/76257/elife-76257-supp3-v2.txt
-
Supplementary file 4
Most enriched HOMER motifs for each human endogenous retrovirus type-H (HERVH) subfamily by sequence block.
- https://cdn.elifesciences.org/articles/76257/elife-76257-supp4-v2.pdf
-
Supplementary file 5
Alignment of all human endogenous retrovirus type-H (HERVH) long terminal repeats (LTRs) used in study (including 7b/c/y).
- https://cdn.elifesciences.org/articles/76257/elife-76257-supp5-v2.txt
-
Supplementary file 6
Alignment of only LTR7 long terminal repeats (LTRs) used in study.
- https://cdn.elifesciences.org/articles/76257/elife-76257-supp6-v2.txt
-
Supplementary file 7
Alignment of human endogenous retrovirus type-H (HERVH) long terminal repeats (LTR) consensus sequences.
- https://cdn.elifesciences.org/articles/76257/elife-76257-supp7-v2.txt
-
Supplementary file 8
Full statistical support for phyloregulatory analysis.
- https://cdn.elifesciences.org/articles/76257/elife-76257-supp8-v2.xlsx
-
Supplementary file 9
Statistical support for transcribed vs. non-transcribed LTR7up1/2.
- https://cdn.elifesciences.org/articles/76257/elife-76257-supp9-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/76257/elife-76257-transrepform1-v2.docx