All-atom molecular dynamics simulations of Synaptotagmin-SNARE-complexin complexes bridging a vesicle and a flat lipid bilayer
Figures
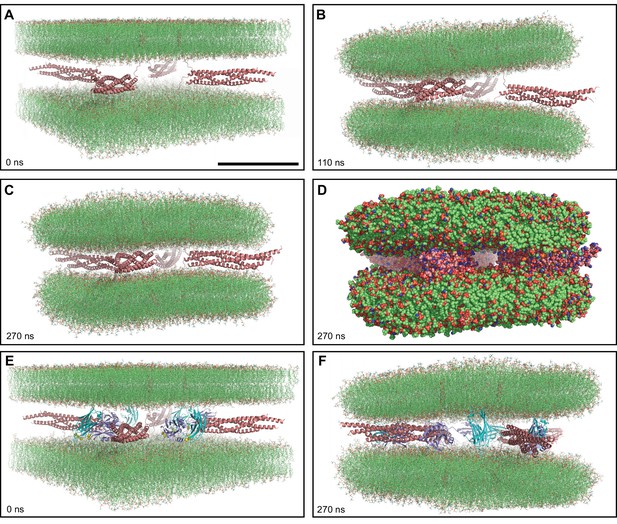
MD simulations of four trans-SNARE complexes bridging two flat bilayers.
(A–C) Initial configuration of the system with SNARE complexes only (A), and snapshots of the MD simulation after 110 and 270 ns (B, C). The SNARE complexes are illustrated by ribbon diagrams in salmon. The lipids are shown as thin stick models. The scale bar in (A) equals 10 nm, which is a little shorter than the length of the SNARE four-helix bundle. (D) Snapshot of the same MD simulation at 270 ns showing all non-solvent atoms as spheres. (E–F) Initial configuration of the system containing four Ca2+-bound Syt1 C2AB molecules in addition to the four trans-SNARE complexes (E) and snapshot of the simulation at 270 ns (F). SNARE complexes are illustrated by ribbon diagrams in salmon and the C2AB molecules are shown as ribbon diagrams with C2A in cyan and C2B in violet. The lipids are shown as thin stick models. The atom color code for the lipids is: carbon lime, oxygen red, nitrogen blue, phosphorous orange. Ca2+ ions are shown as yellow spheres.
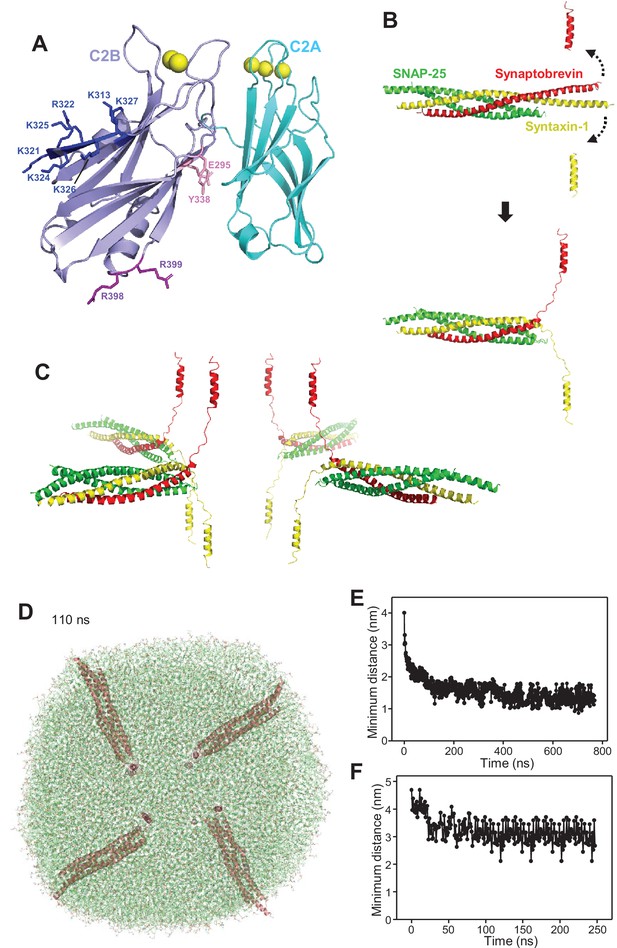
Set up of the system with four trans-SNARE complexes between two flat bilayers.
(A) Ribbon diagram of the conformation of Syt1 C2AB molecules used for the simulation of four trans-SNARE complexes and four C2AB molecules between two bilayers. The C2A domain is colored in cyan and the C2B domain in violet. Key side chains of the C2B domain mentioned in the text, which form the polybasic face (left) or the primary interface (right) are shown as stick models. The polybasic face side chains are colored in blue, and those from the primary interface are in pink (site I) or purple (site II). Ca2+ ions are shown as yellow spheres. (B) Illustration of the procedure used to general the initial structure of a trans-SNARE complex between two flat bilayers. A ribbon diagram of a crystal structure of the neuronal SNARE complex including the TM regions of synaptobrevin and syntaxin-1 (PDB accession code 3HD7) is shown in the middle on the left, with synaptobrevin in red, syntaxin-1 in yellow and SNAP-25 in green. Ribbon diagrams above and below show the positions designed for the TM regions. A 1 ns MD simulation with restraints to force these positions of the TM regions and additional restraints to keep the heavy atoms of the N-terminal half of the SNARE four-helix bundle (up to the polar layer) at their initial positions led to the structure illustrated by the ribbon diagram on the right. (C) Ribbon diagrams of the four trans-SNARE complexes generated for the system with two flat bilayers. Three copies of the original structure obtained by the 1 ns restrained MD simulation were generated and then rotated and translated to yield this final configuration. (D) Snapshot of the MD simulation of four trans-SNARE complexes between two flat bilayers after 110 ns viewed from the top to illustrate that the flat bilayers acquired an almost circular shape. The SNARE complexes are illustrated by ribbon diagrams in salmon. The lipids are shown as thin stick models. The atom color code for the lipids is: carbon lime, oxygen red, nitrogen blue, phosphorous orange. (E) Minimum distance between atoms of the two flat bilayers in frames taken every 1 ns in the simulation of four trans-SNARE complexes between two bilayers. (F) Minimum distance between atoms of the two flat bilayers in frames taken every 1 ns in the simulation of four trans-SNARE complexes and four C2AB molecules between two bilayers.
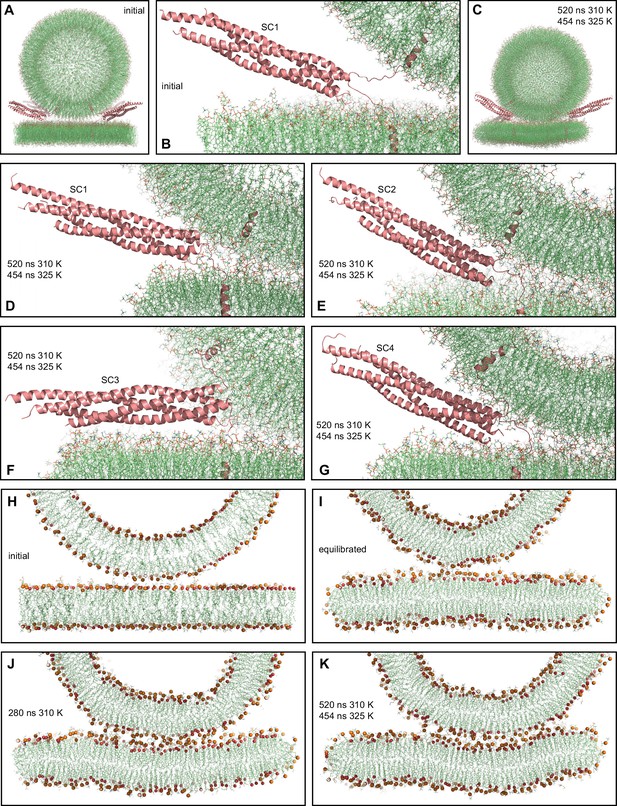
MD simulation of four trans-SNARE complexes bridging a vesicle and a flat bilayer.
(A) Overall view of the initial system. (B) Close-up view of one of the trans-SNARE complexes in the initial system. (C) Snapshot of the system after a 520 ns MD simulation at 310 K and a 454 ns simulation at 325 K. (D–G) Close-up views of the four trans-SNARE complexes (named SC1-SC4) after the 520 ns MD simulation at 310 K and the 454 ns simulation at 325 K. In (A–G), the SNARE complexes are illustrated by ribbon diagrams in salmon. The lipids are shown as thin stick models (carbon lime, oxygen red, nitrogen blue, phosphorous orange). (H–K) Thin slices of the system in its initial configuration (H), after the equilibration steps (I), after 280 ns at 310 K (J) and after 520 ns at 310 K and 454 ns at 325 K (K). In (H–K) Phosphorous atoms of phospholipids and the oxygen atoms of cholesterol molecules are shown as spheres to illustrate the approximate locations of lipid head groups.
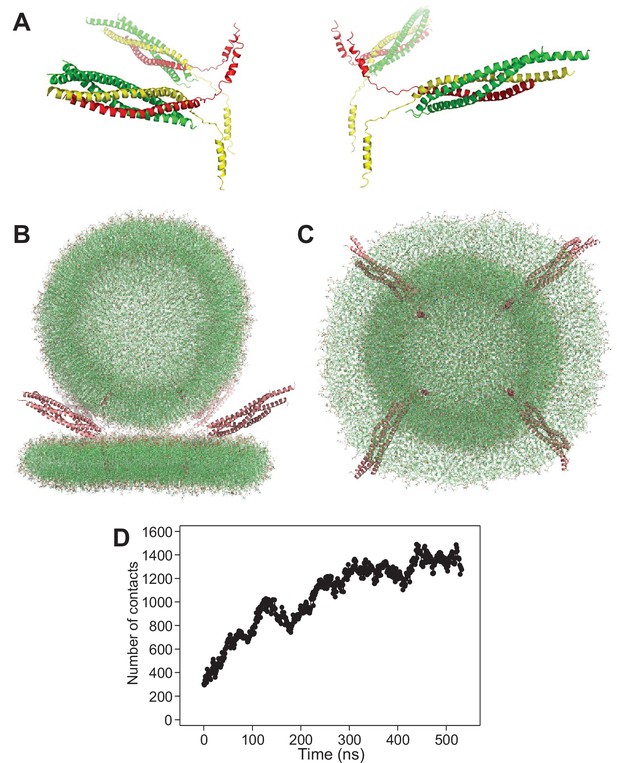
Set up of the system with four trans-SNARE complexes bridging a vesicle and a flat bilayer.
(A) Ribbon diagrams of the four trans-SNARE complexes generated for the initial system with one vesicle and a flat bilayer, with synaptobrevin in red, syntaxin-1 in yellow and SNAP-25 in green. (B,C) The system of four trans-SNARE complexes bridging a vesicle and a flat bilayer after the equilibration steps viewed from the side (B) or from the bottom (C). The SNARE complexes are illustrated by ribbon diagrams in salmon. The lipids are shown as thin stick models (carbon lime, oxygen red, nitrogen blue, phosphorous orange). (D) Number of contacts in frames taken at 1 ns steps in the simulation of four trans-SNARE complexes bridging a vesicle and a flat bilayer. The number of contacts was defined as the number of distances between oxygen atoms of the vesicle and oxygen atoms of the flat bilayer that were smaller than 1 nm.
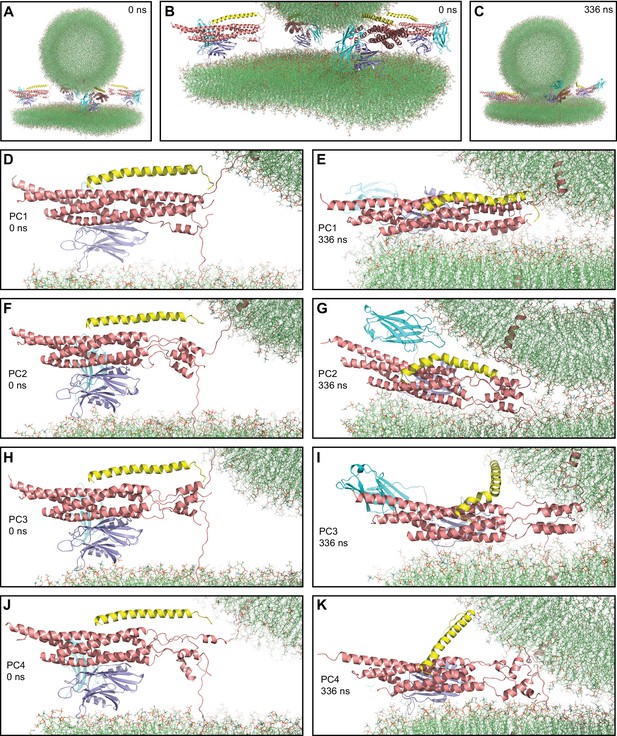
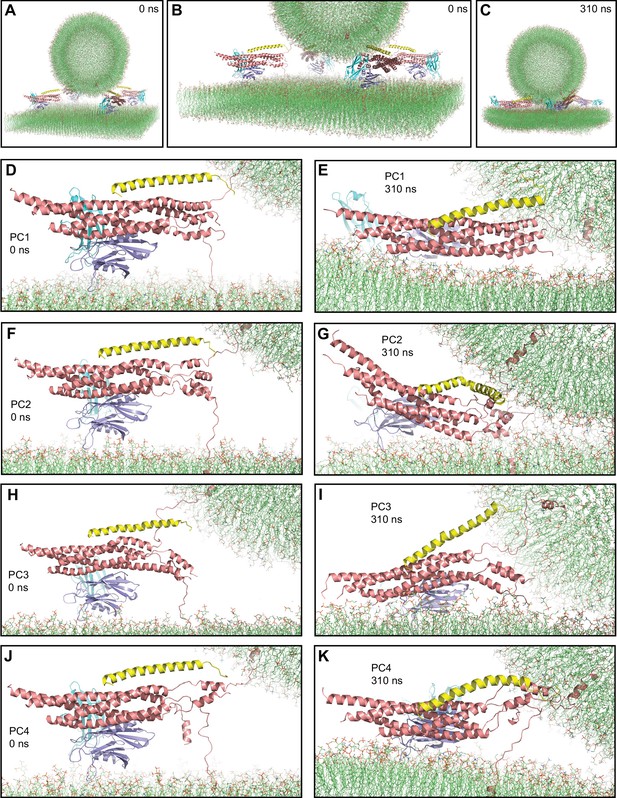
First MD simulation of primed complexes bridging a vesicle and a flat bilayer.
(A) Overall view of the initial system after equilibration. (B) Close-up view of the four primed complexes in the initial system after equilibration. (C) Snapshot of the system after a 336 ns MD simulation. (D–K) Close-up views of the individual primed complexes (named PC1-PC4) in the initial configuration (D,F,H,J) and after the 336 ns MD simulation (E,G,I,K). In all panels, the primed complexes are illustrated by ribbon diagrams, with the SNAREs in salmon, Cpx1(27-72) in yellow and the Syt1 C2AB fragment in cyan (C2A domain) and violet (C2B domain). The lipids are shown as thin stick models (carbon lime, oxygen red, nitrogen blue, phosphorous orange).
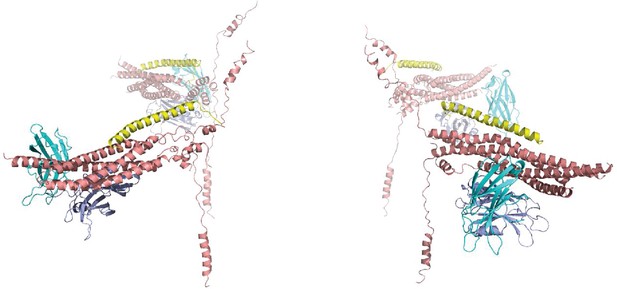
Ribbon diagrams of the four primed complexes generated for the first primed system with one vesicle and a flat bilayer.
The SNAREs are in salmon, Cpx1(27-72) in yellow and the Syt1 C2AB fragment in cyan (C2A domain) and violet (C2B domain).
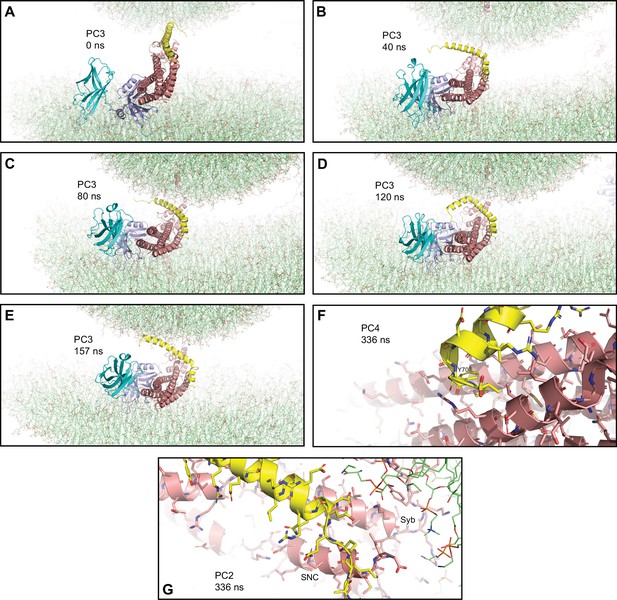
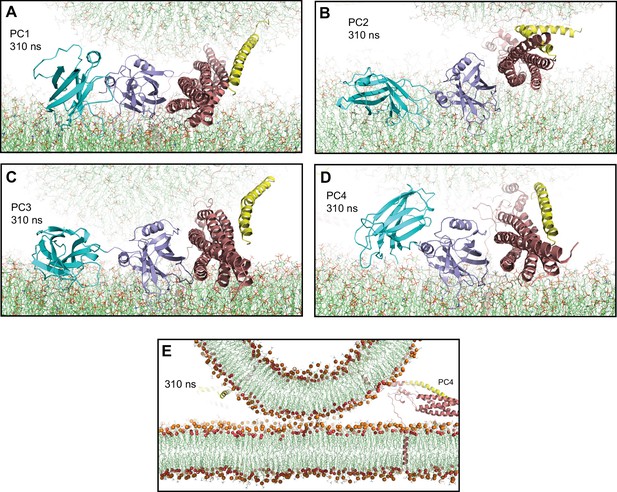
Additional views of the primed complexes bridging a vesicle and a flat bilayer.
(A–E) Close-up views of PC3 in the first MD simulation of primed complexes bridging a vesicle and a flat bilayer in the initial configuration (A) and after 40, 80, 120, and 157 ns (B–E), respectively. PC3 is illustrated by ribbon diagrams, with the SNAREs in salmon, Cpx1(27-72) in yellow and the Syt1 C2AB fragment in cyan (C2A domain) and violet (C2B domain). The lipids are shown as thin stick models (carbon lime, oxygen red, nitrogen blue, phosphorous orange). (F) Close-up view of the region where Cpx1(27-72) remains bound to the SNARE complex in PC4 after 336 ns. The position of the Y70 side chain of Cpx1(27-72), which binds at a hydrophobic pocket of the SNARE complex, is indicated. (G) Close-up view of the region where the Cpx1(27-72) accessory helix interacts with the SNARE motif of synaptobrevin (labeled Syb) and the C-terminal SNARE motif of SNAP-25 (labeled SNC) in PC2 after 336 ns. Cpx1(27-72) and the SNARE complex are illustrated by ribbon diagrams and stick models with oxygen atoms in red, nitrogen atoms in blue, sulfur atoms in light orange and carbon atoms in yellow [for Cpx1(27-72)] or salmon (for the SNAREs).
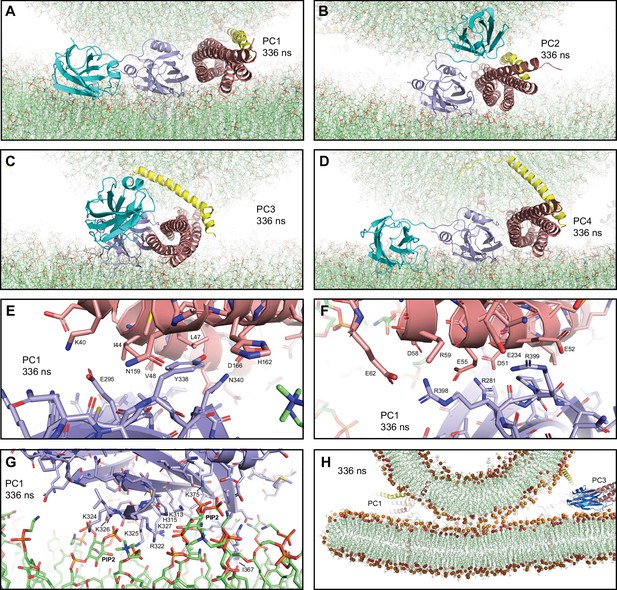
Additional views of the first MD simulation of primed complexes bridging a vesicle and a flat bilayer.
(A–D) Close-up views of the four primed complexes after 336 ns showing how the Syt1 C2B domain binds to the SNARE complex through the primary interface and to the flat bilayer with the polybasic face, which dictates that the Cpx1(27-72) helix is oriented toward the vesicle and bends in different ways and directions to avoid steric clashes. This arrangement forces the SNARE four-helix bundle to be close to the flat bilayer. The primed complexes are illustrated by ribbon diagrams, with the SNAREs in salmon, Cpx1(27-72) in yellow and the Syt1 C2AB fragment in cyan (C2A domain) and violet (C2B domain). The lipids are shown as thin stick models (carbon lime, oxygen red, nitrogen blue, phosphorous orange). (E–F) Two different close-up views of the primary interface between the C2B domain and the SNARE complex in PC1 after 336 ns showing site I of the interface (E) or site II where R398,R399 of the C2B domain are located (F). The C2B domain and the SNARE complex are illustrated by ribbon diagrams and stick models with oxygen atoms in red, nitrogen atoms in blue, sulfur atoms in light orange and carbon atoms in violet [for the C2B domain] or salmon (for the SNAREs). The positions of selected side chains are indicated. (G) Close-up view of the interaction of the C2B domain of PC1 with the flat bilayer after 336 ns. The positions of PIP2 headgroups, basic side chains involved in interactions with the lipids, and the hydrophobic side chain of I367 at the tip of a Ca2+-binding loop that inserts into the bilayer, are indicated. (H) Thin slice of the system showing a point-of-contact interface between the vesicle and the flat bilayer at 336 ns. Phosphorous atoms of phospholipids and the oxygen atoms of cholesterol molecules are shown as spheres to illustrate the approximate locations of lipid head groups. The positions of PC1 and PC3 are indicated.
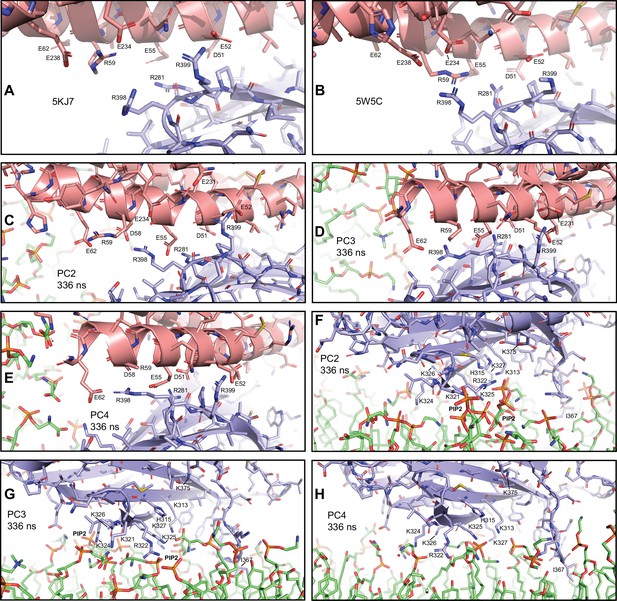
Additional close-up views of primed complexes bridging a vesicle and a flat bilayer.
(A–E) Close-up views of site II of the primary interface between the C2B domain and the SNARE complex in two crystal structures (A), PDB accession number 5KJ7; (B), PDB accession number 5W5C, and in PC2, PC3, and PC4 (C–E), respectively after 336 ns of the first MD simulation of primed complexes bridging a vesicle and a flat bilayer.
The C2B domain and the SNARE complex are illustrated by ribbon diagrams and stick models with oxygen atoms in red, nitrogen atoms in blue, sulfur atoms in light orange and carbon atoms in violet [for the C2B domain] or salmon (for the SNAREs). The positions of selected side chains are indicated. (F–H) Close-up views of the interaction of the C2B domain of PC2, PC3, or PC4 with the flat bilayer after 336 ns (F–H), respectively. The positions of PIP2 headgroups, basic side chains involved in interactions with the lipids, and the hydrophobic side chain of I367 at the tip of a Ca2+-binding loop that inserts into the bilayer, are indicated.
Second MD simulation of primed complexes bridging a vesicle and a flat bilayer.
(A) Overall view of the initial system. (B) Close-up view of the four primed complexes in the initial system. (C) Snapshot of the system after a 310 ns MD simulation. (D–K) Close-up views of the individual primed complexes (named PC1-PC4) in the initial configuration (D,F,H,J) and after the 310 ns MD simulation (E,G,I,K). In all panels, the primed complexes are illustrated by ribbon diagrams, with the SNAREs in salmon, Cpx1(27-72) in yellow and the Syt1 C2AB fragment in cyan (C2A domain) and violet (C2B domain). The lipids are shown as thin stick models (carbon lime, oxygen red, nitrogen blue, phosphorous orange).
Additional views of the second MD simulation of primed complexes bridging a vesicle and a flat bilayer.
(A–D) Close-up views of the four primed complexes after 310 ns showing how the Syt1 C2B domain binds to the SNARE complex through the primary interface and to the flat bilayer with the polybasic face, which dictates that the Cpx1(27-72) helix is oriented toward the vesicle and bends in different ways and directions to avoid steric clashes. This arrangement forces the SNARE four-helix bundle to be close to the flat bilayer. The primed complexes are illustrated by ribbon diagrams, with the SNAREs in salmon, Cpx1(27-72) in yellow and the Syt1 C2AB fragment in cyan (C2A domain) and violet (C2B domain). The lipids are shown as thin stick models (carbon lime, oxygen red, nitrogen blue, phosphorous orange). (E) Thin slice of the system showing a point-of-contact interface between the vesicle and the flat bilayer at 310 ns. Phosphorous atoms of phospholipids and the oxygen atoms of cholesterol molecules are shown as spheres to illustrate the approximate locations of lipid head groups. The positions of PC1 and PC3 are indicated.
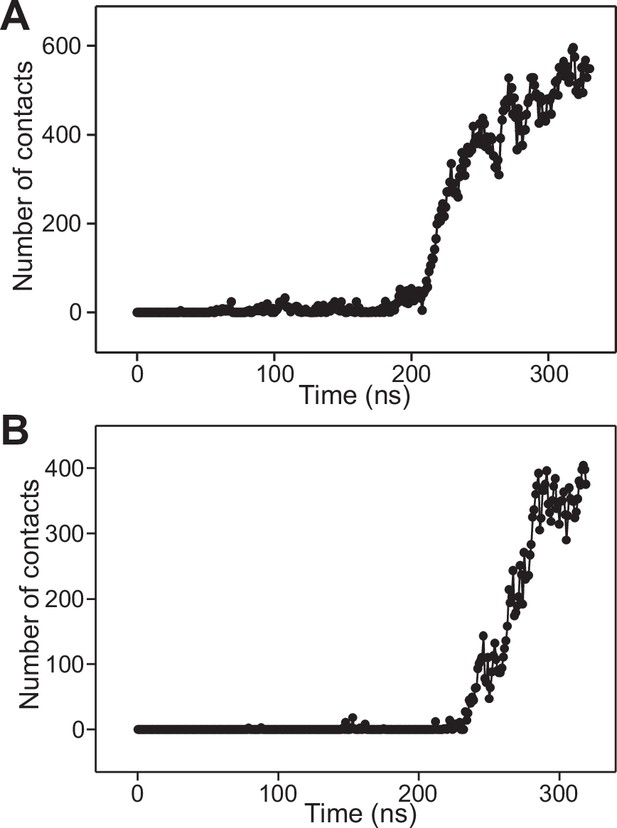
Number of contacts in frames taken at 1 ns steps in the first (A) or second (B) simulation of four primed complexes bridging a vesicle and a flat bilayer.
The number of contacts was defined as the number of distances between oxygen atoms of the vesicle and oxygen atoms of the flat bilayer that were smaller than 1 nm.
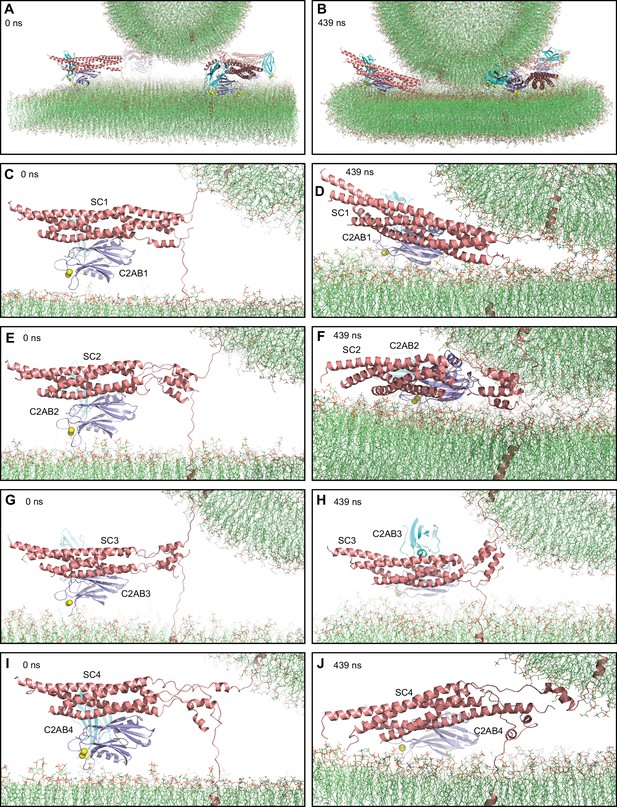
MD simulation of C2AB bound to Ca2+ and to trans-SNARE complexes bridging a vesicle and a flat bilayer.
(A) Close-up view of the four C2AB-SNARE complexes in the initial system. (B) Close-up view of the system after a 439 ns MD simulation. (C–J) Close-up views of the assemblies between C2AB molecules (named C2AB1-4) and SNARE complexes (SC1-SC4) in the initial configuration (C,E,G,I) and after the 439 ns MD simulation (D,F,H,J). In all panels, the SNAREs are represented by ribbon diagrams in salmon and the Syt1 C2AB fragment by ribbon diagrams in cyan (C2A domain) and violet (C2B domain). Ca2+ ions are shown as yellow spheres. The lipids are shown as thin stick models (carbon lime, oxygen red, nitrogen blue, phosphorous orange).
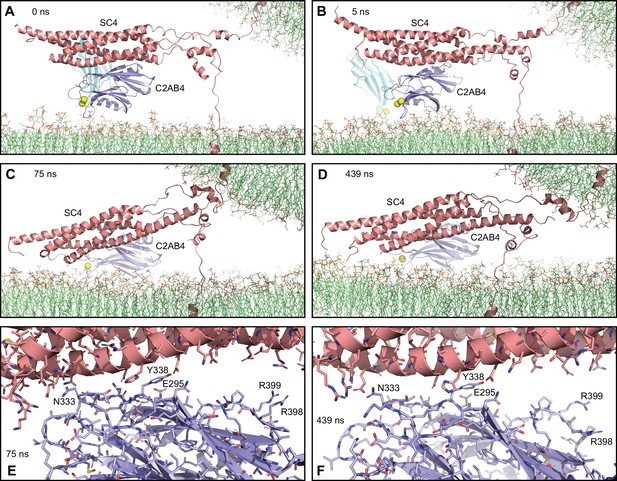
Additional views of the complexes from Figure 5.
(A–D) Close-up views of SC4 and C2AB4 in the MD simulation of C2AB bound to Ca2+ and to trans-SNARE complexes bridging a vesicle and a flat bilayer in the initial configuration (A) and after 5, 75, and 439 ns (B–D), respectively.
The SNAREs are represented by ribbon diagrams in salmon and the Syt1 C2AB fragment by ribbon diagrams in cyan (C2A domain) and violet (C2B domain). Ca2+ ions are shown as yellow spheres. The lipids are shown as thin stick models (carbon lime, oxygen red, nitrogen blue, phosphorous orange). Note that in just the first 5 ns one of the helices that was disrupted was almost fully formed but overall there was no substantial progress toward assembly of the C-terminal part of the SNARE four-helix bundle in 439 ns. (E–F) Close-up views of the primary interface between SC4 and C2AB4 after 75 (E) and 439 (F) ns. The C2B domain and the SNARE complex are illustrated by ribbon diagrams and stick models with oxygen atoms in red, nitrogen atoms in blue, sulfur atoms in light orange and carbon atoms in violet [for the C2B domain] or salmon (for the SNAREs). The positions of selected side chains are indicated.
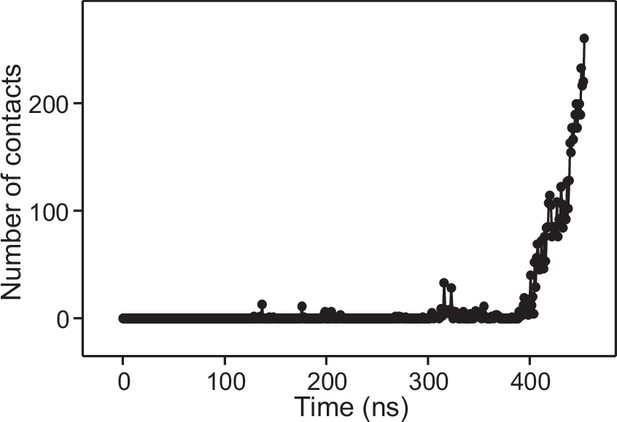
Number of contacts in frames taken at 1 ns steps in the MD simulation of C2AB bound to Ca2+ and to trans-SNARE complexes bridging a vesicle and a flat bilayer.
The number of contacts was defined as the number of distances between oxygen atoms of the vesicle and oxygen atoms of the flat bilayer that were smaller than 1 nm.
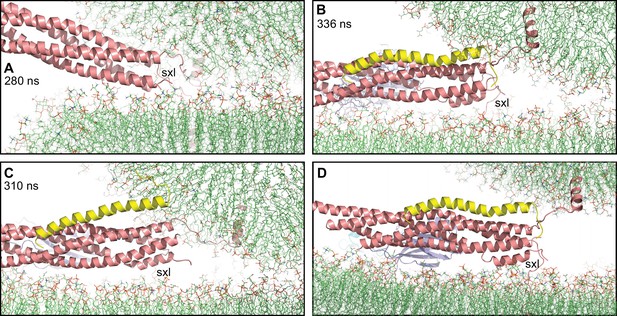
Complexin-1 may hinder the final action of trans-SNARE complexes to bring membranes together.
(A–C) Close-up views of one of the SNARE complexes bridging a vesicle and a flat bilayer after simulation for 280 ns at 310 K (A) (shown in Figure 2D after 520 ns at 310 K and 454 ns at 325 K), of PC1 in the first MD simulation of primed complexes after 336 ns (B) (also shown in Figure 3E) and of PC1 in the second MD simulation of primed complexes after 310 ns (C) (also shown in Figure 4—figure supplement 2E). The complexes are illustrated by ribbon diagrams, with the SNAREs in salmon, Cpx1(27-72) in yellow and the Syt1 C2AB fragment in cyan (C2A domain) and violet (C2B domain). The positions of the syntaxin-1 juxtamembrane linkers (sxl) are indicated. The comparison shows how the SNARE complex with the fully assembled four-helix bundle in (A) drew the two membranes closer than the two primed complexes of (B,C). (D) Close up view of the pose shown in (B) after moving the vesicle upwards so that it’s minimal distance from the flat bilayer is 3 nm.
Tables
Size in atoms, length of productions MD simulations, temperature, speed of the simulations on Frontera at TACC and lipid composition of the flat bilayers and the vesicle of the different systems.
| qscff | 1699436 atoms | 750 ns | 310 K | 24 ns/day with 16 nodes | |||||
| Upper bilayer | CHL1 | POPC | POPS | SAPE | SDPE | SDPS | total | ||
| Upper leaflet | 540 | 468 | 0 | 72 | 120 | 0 | 1200 | ||
| % | 45 | 39 | 0 | 6 | 10 | 0 | |||
| Lower leaflet | 540 | 146 | 84 | 120 | 240 | 96 | 1226 | ||
| % | 44 | 11.9 | 6.9 | 9.8 | 19.6 | 7.8 | |||
| Lower bilayer | CHL1 | POPC | POPS | SAPI2D | SAPE | SDPE | SDPS | total | |
| Upper leaflet | 540 | 134 | 120 | 60 | 84 | 168 | 120 | 1226 | |
| % | 44 | 11 | 9.8 | 4.9 | 6.9 | 13.7 | 9.8 | ||
| Lower leaflet | 540 | 538 | 0 | 0 | 48 | 84 | 0 | 1200 | |
| % | 45 | 44 | 0 | 0 | 4 | 7 | 0 | ||
| sqscff | 1700475 atoms | 270 ns | 310 K 24 ns/day with 16 nodes | ||||||
| qscv | 3222393 atoms | 520 ns | 310 K | 23 ns/day with 32 nodes | |||||
| 454 ns | 325 K | ||||||||
| Vesicle | CHL1 | POPC | SAGL | SAPE | SAPI2D | SDPE | SDPS | SOPS | total |
| Outer leaflet | 1258 | 296 | 0 | 534 | 0 | 282 | 210 | 199 | 2779 |
| % | 45.3 | 10.6 | 0 | 19.2 | 0 | 10.1 | 7.6 | 7.2 | |
| Inner leaflet | 814 | 668 | 0 | 183 | 0 | 99 | 1 | 1 | 1766 |
| % | 46.1 | 37.8 | 0 | 10.4 | 0 | 5.6 | 0.1 | 0.1 | |
| Flat bilayer | CHL1 | POPC | SAGL | SAPE | SAPI2D | SDPE | SDPS | SOPS | total |
| Upper leaflet | 540 | 96 | 12 | 84 | 60 | 156 | 120 | 120 | 1188 |
| % | 45.4 | 8.1 | 1 | 7.1 | 5.1 | 13.1 | 10.1 | 10.1 | |
| Lower leaflet | 540 | 516 | 12 | 36 | 0 | 84 | 0 | 0 | 1188 |
| % | 45.4 | 43.4 | 1 | 3 | 0 | 7.1 | 0 | 0 | |
| prsg | 5056443 atoms | 336 ns | 310 K | 13 ns/day with 32 nodes | |||||
| Flat bilayer | CHL1 | POPC | SAGL | SAPE | SAPI2D | SDPE | SDPS | SOPS | total |
| Upper leaflet | 830 | 151 | 18 | 132 | 93 | 262 | 182 | 181 | 1849 |
| % | 44.9 | 8.2 | 1 | 7.1 | 5 | 14.2 | 9.8 | 9.8 | |
| Lower leaflet | 810 | 774 | 18 | 72 | 0 | 126 | 0 | 0 | 1800 |
| % | 45 | 43 | 1 | 4 | 0 | 7 | 0 | 0 | |
| prs2 | 5870280 atoms | 310 ns | 310 K | 16 ns/day 48 nodes | |||||
| Flat bilayer | CHL1 | POPC | SAGL | SAPE | SAPI2D | SDPE | SDPS | SOPS | total |
| Upper leaflet | 1035 | 184 | 23 | 161 | 115 | 322 | 230 | 230 | 2300 |
| % | 45 | 8 | 1 | 7 | 5 | 14 | 10 | 10 | |
| Lower leaflet | 1009 | 964 | 22 | 88 | 0 | 154 | 0 | 0 | 2237 |
| % | 45.1 | 43.1 | 1 | 3.9 | 0 | 6.9 | 0 | 0 | |
| prsncpxca | 5870246 atoms | 439 ns | 310 K | 16 ns/day 48 nodes | |||||
-
Vesicle: same as qscv system.
-
Flat bilayer: same as prs2 system.





