Phenotyping single-cell motility in microfluidic confinement
Figures
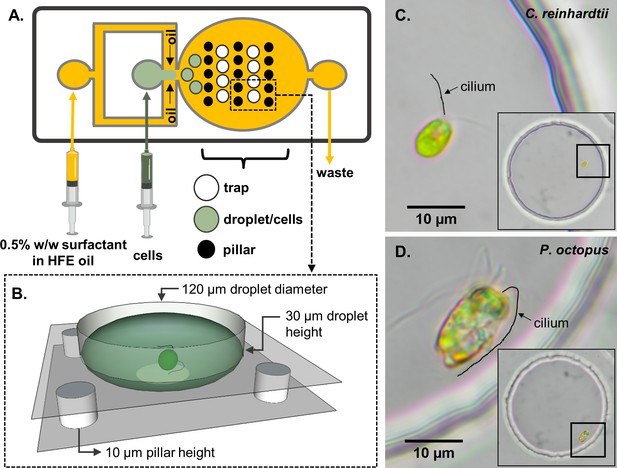
Schematic of the experimental set-up.
(A) A two-layer microfluidic device with embedded single-cell traps, and syringes used for perfusion of the carrier oil phase and an aqueous suspension containing live motile cells. (B) 3D rendering of a single trap in which a cell can be stably trapped and imaged for hours. To demonstrate variability in swimming behaviour, we studied two species of motile algae, images show respectively: (C) a single Chlamydomonas reinhardtii (CR) cell, and (D) a single Pyramimonas octopus (PO) cell, in each case trapped within a 120 μm-diameter circular well. (Cilia positions are highlighted by manual tracings.).
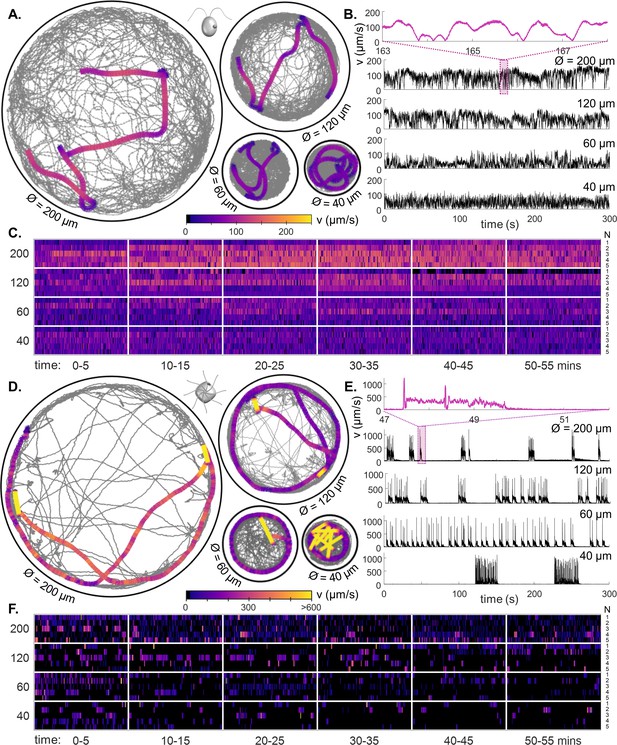
Behavioural ethograms of motile algae.
(A) High-speed trajectories of single C. reinhardtii cells in circular traps of varying sizes (illustrating coverage over 5 min), overlaid with a 5-second representative trajectory (colour-coded by speed). (B) Instantaneous swimming speeds for the sample trajectories shown in (A). Inset: speed fluctuations over short timescales. (C) Heatmaps of cell swimming speed over time indexed by cell number N (rows), and ordered by trap size, using data from all experimental runs. (D–F) Similar, but for PO. All trajectories viewed from above the sample in the lab frame.
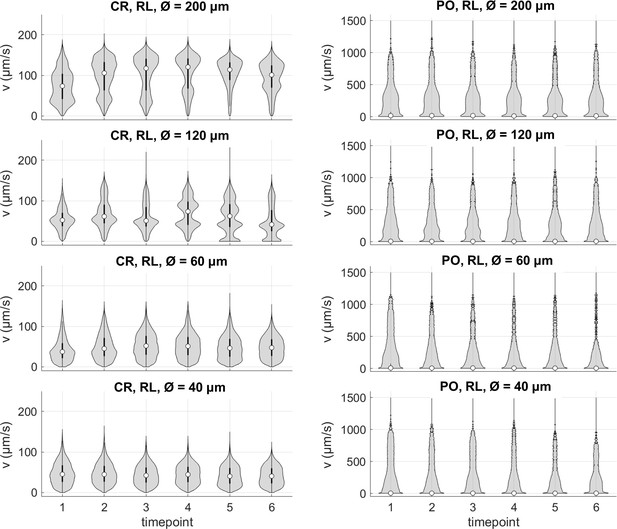
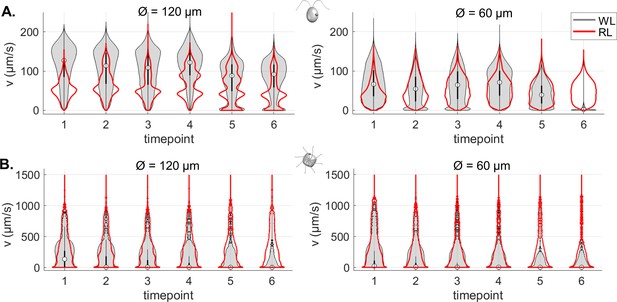
Summary violin plots of the speed data for different trap sizes, over the 6 timepoints (respectively, 0-5, 10-15, 20-25, 30-35, 40-45, and 50-55 min).
Note that the scaling of the probability density function is linear for CR and logarithmic for PO.
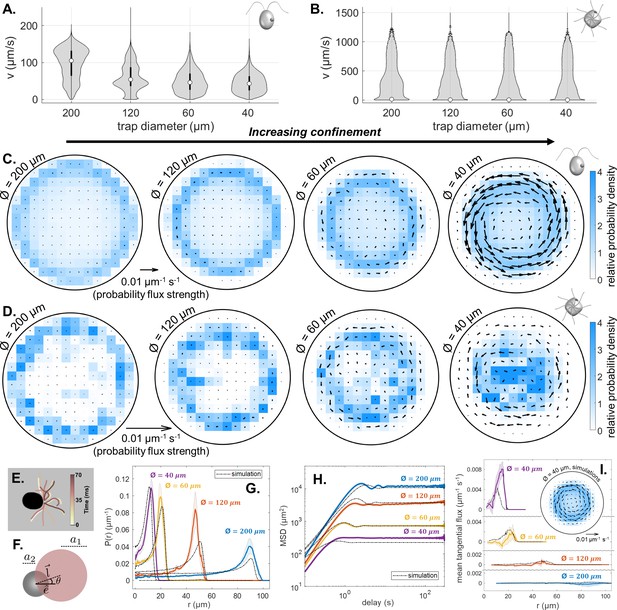
Effect of trap size on single cell motility.
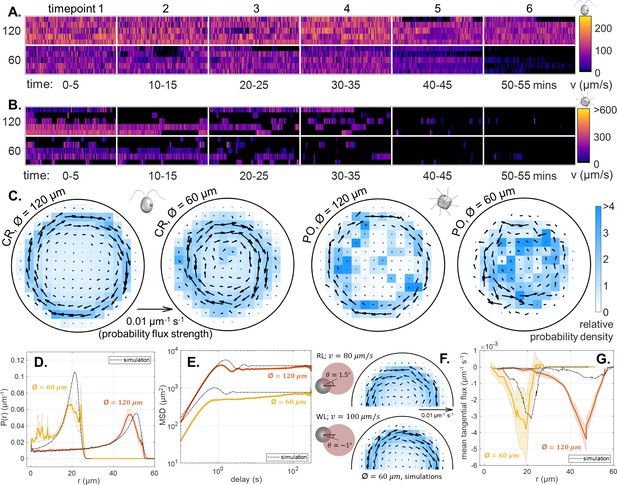
(A,B) Violin plots of pdfs of speed () across all cells, and all time points, for CR and PO. (Note the scaling of the pdfs is linear for CR, but logarithmic for PO.) (C,D) Time-averaged heatmaps of the cells’ centroid positions, for each of the four different trap sizes, overlaid with arrows showing the direction and strength of steady-state non-equilibrium fluxes computed from trajectory statistics, for CR and PO. (E) Traced cilia waveforms for CR showing the asymmetric beat pattern of the two cilia under red light. (F) The asymmetric dumbbell model used for CR with a smaller circle (radius a2) for the cell body and a larger circle (radius a1) representing the area covered by the cilia beat, which is offset from the swimming direction by angle . Comparison of the experimental mean and standard error () of the radial probability densities (G), mean-squared displacement curves (H) and mean tangential flux (I) with the simulation results for each trap size. The inset in (I) shows the time-averaged heatmap of centroid position overlaid with arrows showing the probability flux results for the simulation of the 40 μm diameter trap.
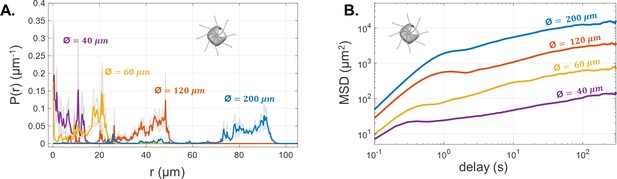
The experimental mean and standard error (N = 5) of the radial probability densities (A) and mean-squared displacement curves (B) for each of the trap sizes, for PO.
Compared with CR, the curves are more irregular and the MSD curves show a more gradual increase toward the maximum.
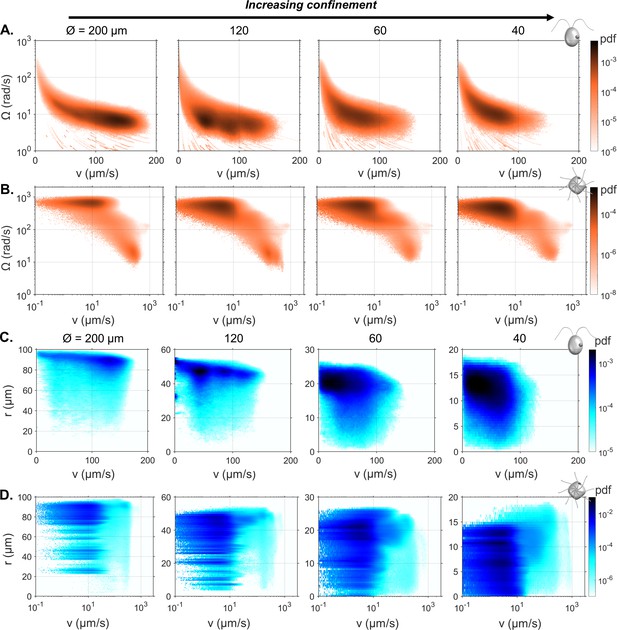
Bivariate histograms of linear and angular speed () across all cells, and all time points for CR (A), and PO (B).
Bivariate histograms of speed and radial position () across all cells, and all time points, for CR (C), and PO (D).
Effect of white light on single cell motility.
(A,B) Heatmaps of cell swimming speed over time, for CR and PO. (C) Time-averaged heatmaps of the cells’ centroid position overlaid with arrows showing the direction and strength of the probability flux computed from trajectory statistics for both trap sizes under white light conditions. Comparison of the experimental mean and standard error () of the radial probability densities (D) and mean-squared displacement curves (E) with the simulation results for each trap size. (F) An illustration of the different dumbbell model parameters used for RL and WL conditions together with semi-circles showing the time-averaged heatmaps of the centroid position overlaid with probability flux arrows for the 60 μm diameter trap size. A change in the direction of the offset angle changes the direction of the non-equilibrium flux. (G) Comparison of the simulations results with the experimental mean and standard error () of the mean tangential flux as a function of radial displacement for each trap size.
Violin plots of the pdfs of speed over time for all cells, for CR (A) and PO (B).
Note that the pdfs are scaled linearly for CR and logarithmically for PO.
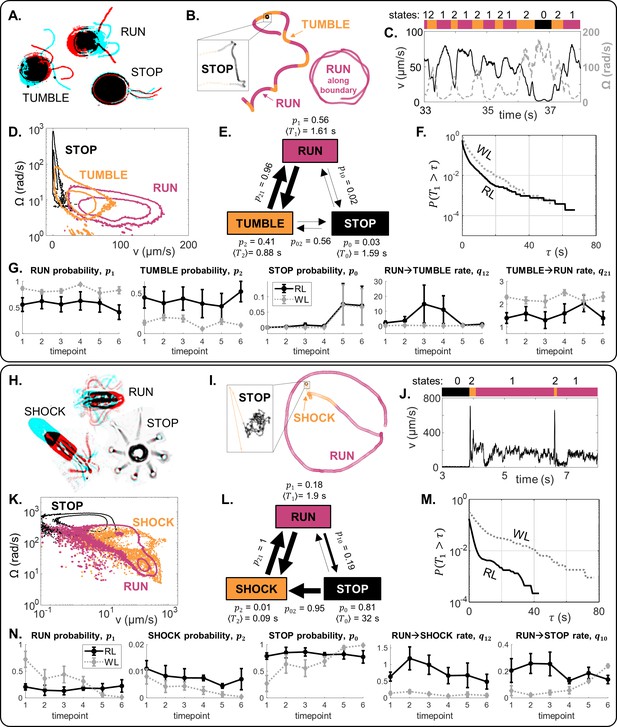
The locomotor behaviour of single-cells is controlled by a trio of motility macrostates and their transition probabilities.
(A–G) For CR, distinct ciliary beat patterns and coordination states (A) produce different swimming trajectories (B), and a discrete sequence (C) of states (run, stop, tumble). For cells in 120 μm traps, RL, the states occupy distinct regions in -space (D), and a time-averaged reaction network with characteristic transition parameters (E). Network parameters are different in WL for the same trap size, as evidenced in (F): survival probability of run states, and (G): mean and standard error (N = 5) for selected state probabilities and transition rates. (H–N) Similarly for PO.
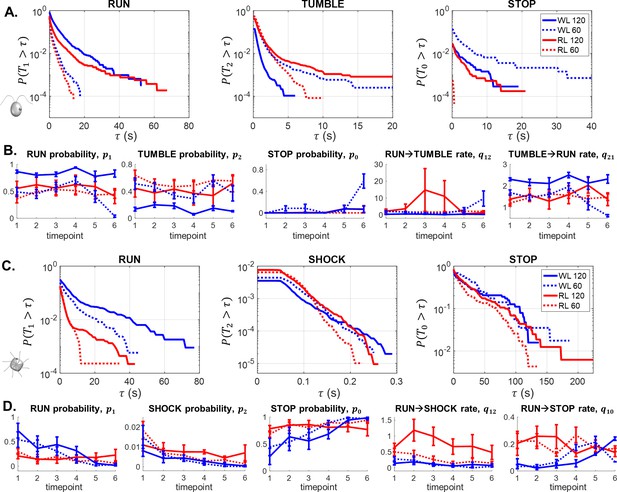
Survival probabilities and selected reaction network parameters comparing the 120 μm and 60 μm traps in RL and WL for CR (A–B) and PO (C–D).
Error bars correspond to the standard errors (N = 5).
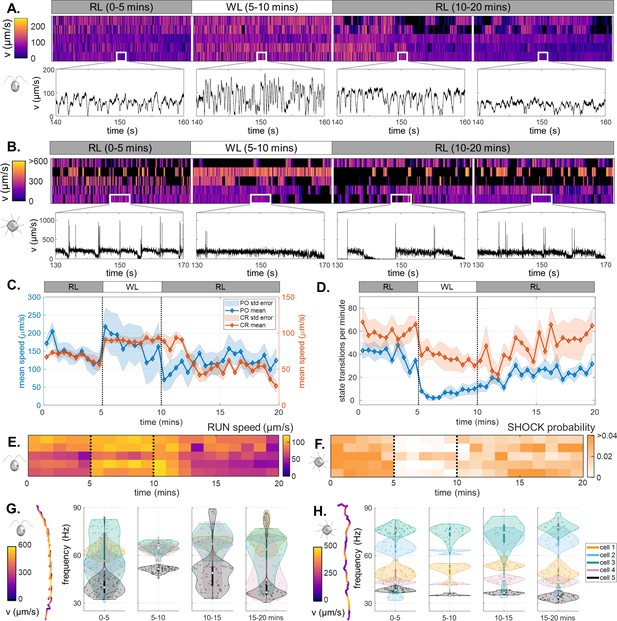
Light-switchable motility of green algae.
(A) Heatmaps of CR swimming speed over the course of a 20-min photoresponse assay, insets show short, representative timeseries in each phase. (B) The same for PO. (C) Population-averaged swimming speed, over time. (D) Population-averaged rates of transitions between motility states, over time. (Shaded regions are standard errors.) (E) Mean speed of ‘runs’ in CR, over time. (F) Probability of ‘shocks’ in PO, over time. Violin plots of the most prominent frequencies extracted from helical trajectories (inset), for CR (G) and for PO (H). For CR, this corresponds to the ciliary beat frequency.
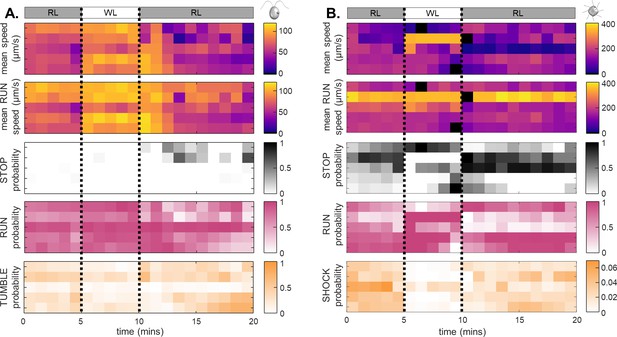
Light switchable states data over time for individual cells of CR (A) and PO (B).
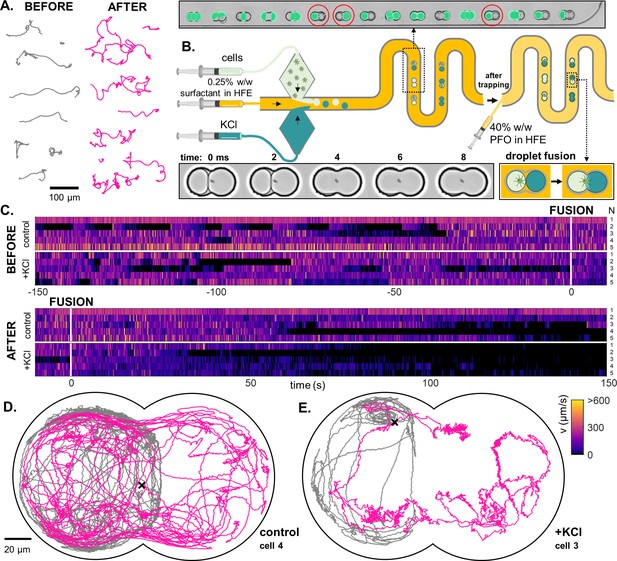
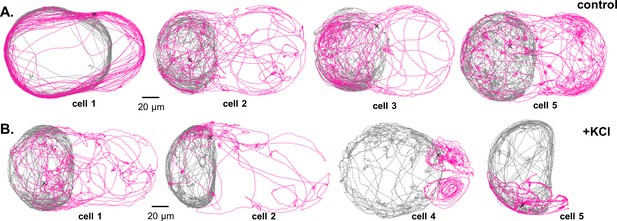
A novel droplet-fusion assay for querying single-cell motility responses to rapid chemical perturbation.
(A) Example tracks from a bulk motility assay (see Materials and methods), immediately before (’normal motility phenotype’) and after addition of 50 mM KCl to the medium (‘chemically perturbed phenotype’). (B) Schematic of the serpentine microfluidic device for generation and pairing of droplets - one containing the trapped cell, and the other loaded with 10 mM KCl (inset, red circles). Fluorescein was added to the KCl phase to identify paired droplets. Fusion was then induced by replacing the 0.5% surfactant in HFE with 40% 1 H,1H,2H,2H-Perfluoro-1-octanol (PFO) in HFE. Two such droplets merge within 1 or 2 ms (inset). (C) Heatmaps of single-cell swimming speed, before and after the moment of fusion to a droplet containing KCl, compared to a control pair where KCl was replaced with inert culture medium. An example single-cell trajectory before (grey) and after (pink) droplet fusion, for a control (D) and a KCl pair (E).
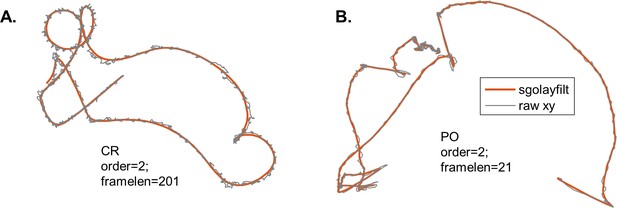
Smoothing tracking data using a Savitzky-Golay filter (sgolayfilt).
Example raw and smoothed trajectories for CR (A) and PO (B).
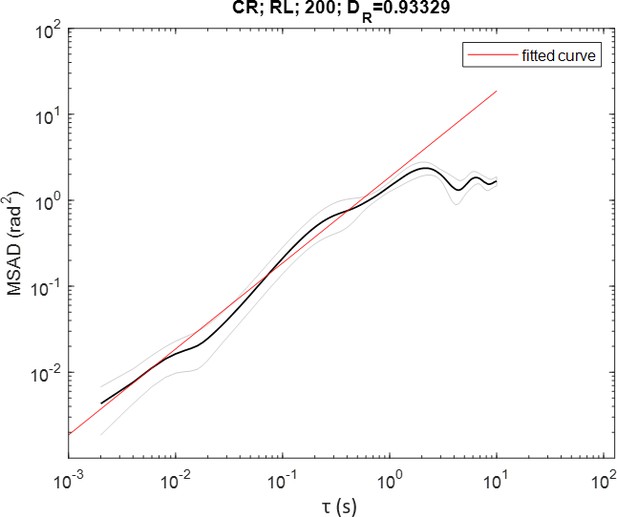
The mean square angular displacement (MSAD) for CR in the 200 µm diameter droplets, fitted to for .
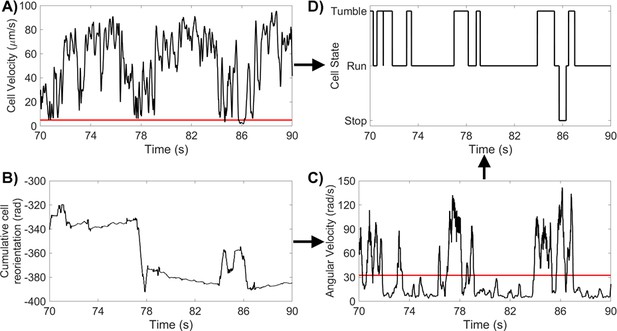
Method For Identifying CR States (A) CR cell velocity for a 20 s segment, with a run/stop threshold of 5 μms-1.
(B) CR cumulative reorientation for the same 20 s segment. (C) CR angular velocity for the same 20 s segment, with a tumble threshold of . (D) CR states for the same 20 s segment.
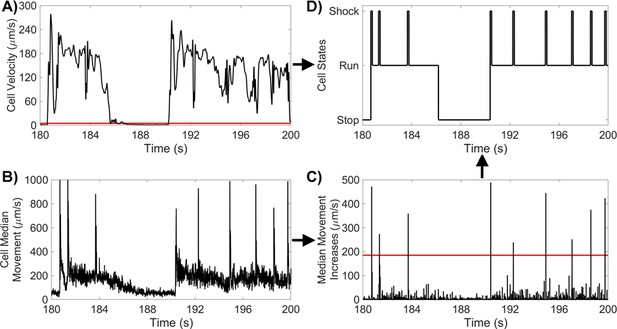
Method For Identifying PO States.
(A) PO cell velocity for a 20 s segment, with a run/stop threshold of 5 μms-1. (B) PO median movement over 9-frame windows for the same 20 s segment. (C) PO increases in cell median movement for the same 20 s segment, with a shock threshold of 185 μms-1. (D) PO states for the same 20 s segment.

In simulations the cell is modelled as an asymmetric dumbbell of two spheres a distance apart.
One sphere represents the cell body and the other models the stroke averaged shape of the flagella. To account for differences in the beating patterns of the two flagella the direction of motion is offset from by an angle .
Videos
Example videos showing single Chlamydomonas reinhardtii cells trapped by droplet microfluidics inside circular arenas of different diameters (∅ = 200, 120, 60, 40 μm), under red light illumination.
Example videos showing single Pyramimonas octopus cells trapped by droplet microfluidics inside circular arenas of different diameters (∅ = 200, 120, 60, 40 μm), under red light illumination.
A single Chlamydomonas reinhardtii cell is trapped inside a circular arena of diameter (∅ = 120 μm), under white light illumination.
Videos show different timepoints for the same individual.
A single Pyramimonas octopus cell is trapped inside a circular arena of diameter (∅ =120 μm), under white light illumination.
Videos show different timepoints for the same individual.
Example video showing a single Pyramimonas octopus cell before and after fusion with KCl solution, under red light illumination.
Tables
A comparison of the concentrations of key ions (in mM), before and after paired droplet fusion.
| Ion | Pre-fusion | Post-fusion (with 10 mM KCl solution) |
|---|---|---|
| Na+ | 395 | 197 |
| K+ | 8.33 | 9.17 |
| Ca2+ | 8.75 | 4.38 |
| Mg2+ | 44.5 | 22.3 |
| Cl- | 459 | 230 |
| SO42- | 23.6 | 11.8 |
| NO3- | 1.1 | 0.6 |
| HPO42- | 0.1 | negligible |