A regeneration-triggered metabolic adaptation is necessary for cell identity transitions and cell cycle re-entry to support blastema formation and bone regeneration
Figures
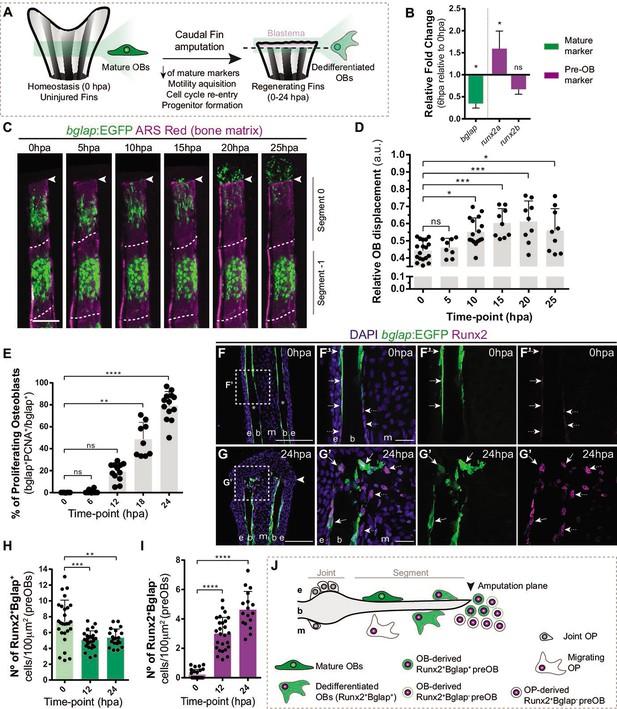
Osteoblast dedifferentiation time-window during caudal fin regeneration.
(A) Biological traits of OB dedifferentiation process. (B) Relative gene expression of mature (green) and pre-OB (magenta) markers, at 6 hpa relative to 0 hpa. Statistical analysis on graph corresponds to paired t-test with Welch’s correction. Mean ± SD are displayed (n=4 biological replicates). (C) Live imaging analysis of OB motility in bglap:EGFP fish (green) during the first 25 hpa, highlighted in the segment bellow amputation (segment 0) and segment –1. Bony-rays are labeled with Alizarin red (magenta). White dashed lines delineate the intersegment region. (D) Quantification of the relative OB displacement in segment 0. Statistical analysis displayed on graph corresponds to Kruskal-Wallis test with Mean ± SD (n=9–18 bony-rays). (E) Percentage of proliferating OBs through immunofluorescence against PCNA in bglap:EGFP fish. Statistical analysis displayed on graph corresponds to Kruskal-Wallis test with Mean ± SD (n=9–13 cryosections). (F-G’) Representative cryosection images of bglap:EGFP (green) fins immunostained for Runx2 (magenta) and counterstained with DAPI (blue), in (F, F’) uninjured fish and (G, G’) at 24 hpa; arrows indicate Runx2 +Bglap + cells and dashed arrows indicate Runx2 +Bglap cells. (H, I) Quantification of (H) Runx2 +Bglap + and (I) Runx2 +Bglap cells during the first 24hpa. Statistical analysis displayed on graph corresponds to Mann-Whitney test with Mean ± SD (n=18–27 cryosections). (J) Cellular sources that contribute for new pre-OBs formation after injury include mature osteoblasts and potentially joint OP. White arrowhead indicates amputation plane and dashed squares represent magnified panels in F’ and G’. E: epidermis; b: bone; m: mesenchyme; ns: not significative; *p<0,05; **p<0,01; ***p<0,001; ****p<0,0001. Scale bars represent 100 µm and 30 µm in magnified panels. See Figure 1—source data 1.
-
Figure 1—source code 1
MatLab scripts quantify the relative osteoblast displacement after caudal fin amputation.
- https://cdn.elifesciences.org/articles/76987/elife-76987-fig1-code1-v1.zip
-
Figure 1—source data 1
Spreadsheets detailing the results regarding the characterization of osteoblast dedifferentiation through caudal fin regeneration, specifically (B) the relative gene expression analysis, (E) the percentage of proliferating osteoblast, and (H,I) the quantification of Runx2+Bglap + and Runx2+Bglapcells during the first 24hpa.
- https://cdn.elifesciences.org/articles/76987/elife-76987-fig1-data1-v1.xlsx
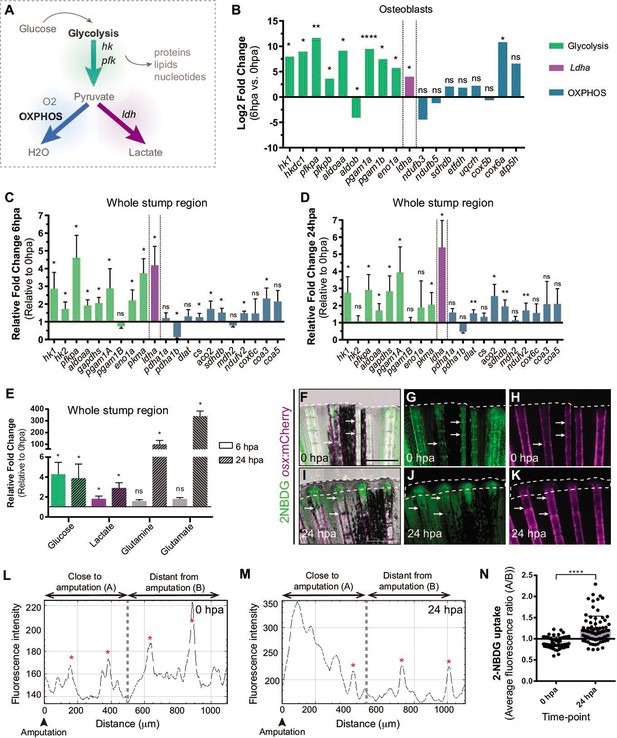
Metabolic adaptation is triggered during zebrafish caudal fin regeneration.
(A) Schematic representation of glucose metabolism. (B) OB gene expression profile of glycolytic enzymes (green), ldha (magenta) and OXPHOS components (blue) at 6 hpa relative to uninjured conditions (0 hpa) obtained from the OB ArrayXS. The horizontal axis represents the log2 fold-change and p-values on a negative log10 scale. Statistical analysis with t test and Welch’s correction (n=3 biological replicates), Mean ± SD are displayed. (C, D) Relative gene expression of glycolytic enzymes (green), ldha (magenta) and OxPhos components (blue), in the whole fin stump, at (C) 6 hpa and at (D) 24 hpa in comparison to uninjured conditions (0 hpa). Statistical analysis with paired t test (n=5 (C) and 4 (D) biological replicates). (E) Metabolite measurements at 6 hpa (clean columns) and 24 hpa (stroked columns) in relation to uninjured conditions (0 hpa), in the whole fin stump. Statistical analysis with Mann-Whitney test (n=4 biological replicates). (F–K) Live imaging of 2NDBG uptake (green) in osx:mCherry fish (magenta) at (F–H) 0 hpa and (I–K) 24 hpa. Arrows indicate uptake of 2NBDG in the intersegment regions. White dashed line delineates the regenerated tissue. Scale bar represents 500 µm. (L–M) Intensity of 2NBDG uptake in regions close and distant to the amputation site, at (L) 0 hpa and (M) 24 hpa. Red * indicate peaks of 2NDBG uptake in the intersegments. (N) Ratio of 2NBDG uptake at 0 hpa and 24 hpa. Statistical analysis on graph corresponds to Mann-Whitney test. Mean ± SD are displayed (n=54 and 83 bony-rays). ns: not significative; *p<0,0001. See Figure 2—source data 1.
-
Figure 2—source data 1
Spreadsheets detailing the results of the metabolic adaptation, specifically the quantification of (C,D) the relative gene expression analysis, (E) the relative metabolite levels, and (N) the average fluorescent levels of 2-NBDG.
- https://cdn.elifesciences.org/articles/76987/elife-76987-fig2-data1-v1.xlsx
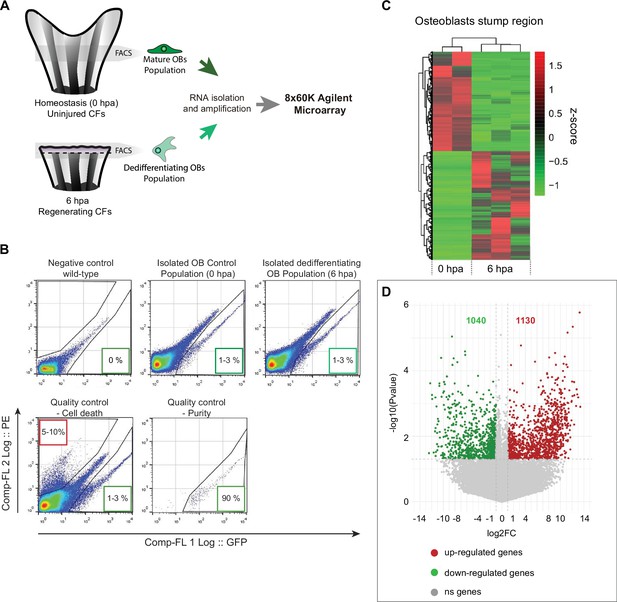
Isolation and gene expression analysis osteoblasts undergoing dedifferentiation.
(A) Schematic representation of the experimental design used to obtain the transcriptional profile of OBs undergoing dedifferentiation, using bglap:EGFP reporter line. OBs from caudal fin tissue corresponding approximately to one bony-ray segment (gray areas), from homeostasis (0 hpa, corresponding to mature OBs) and from one segment bellow amputation at 6 hpa (dedifferentiating OBs) were collected, dissociated, and isolated by FACS for RNA extraction and subjected to a microarray chip assay. (B) Representative flow cytometry plots from wild-type (negative control) and bglap:EGFP transgenic fish from homeostasis and 6 hpa caudal fins. Representative examples of sample quality control through the evaluation of cell death, by propidium iodide (PI) staining, and purity. In flow cytometry plots, GFP fluorescence intensity is given by the x axis (Comp-FL 1 Log::GFP) and PE fluorescence intensity (used to identify PI-positive cells) is given by the y axis (Comp-FL2 Log::PE). Numbers in the lower right boxes indicate relative percentages of GFP + cells and numbers in the upper left boxes indicate the relative percentage of PI + cells. (C) Heatmap depicting hierarchical clustering of Z score transformed expression profiles of all genes from isolated osteoblasts at 0 and 6 hpa. (D) Volcano plot showing the differentially expressed transcripts between homeostatic (6 hpa) and dedifferentiating OBs (0 hpa). The horizontal axis represents the log2 fold-change between 6 and 0 hpa and the vertical axis represents the p-value (on a negative log10 scale). Upregulated transcripts are shown in red and downregulated genes are shown in green. Significant changes were considered for a log2 FC >1 or <-1 for a p-value >0.05.
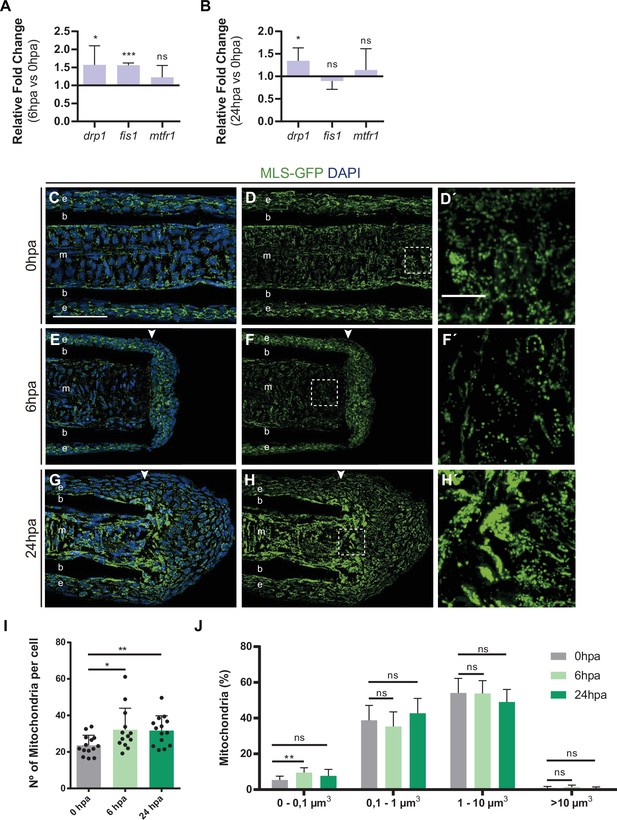
Changes in metabolism are accompanied by alterations in mitochondria dynamics.
(A, B) Relative gene expression of genes related to mitochondrial fission, at (A) 6 hpa and (B) 24 hpa, in comparison to uninjured condition (0 hpa). Statistical analysis corresponds to Paired t-test (B, n=6 biological replicates; C, n=4 biological replicates). (C-H’) Representative cryosection images of MSL:GFP (green) caudal fins, stained for DAPI (blue) at different time-points: (C-D’) 0 hpa (uninjured condition); (E-F’) 6 hpa; and (G-H’) 24 hpa. Dashed boxes delineate amplified panels in D’, F’ and H’. Arrowheads indicate the amputation plane. Scale bar represents 50 µm and 10 µm in amplified panels. (I) Graph showing the quantification of the number of mitochondria per cell, at 0 hpa, 6 hpa, and 24 hpa. Statistical analysis corresponds to Mann-Whitney test, Mean and SD are displayed (n=13 blastemas from 5 fish). (J) Graph displaying the percentage of mitochondria at 0 hpa (gray), 6 hpa (light green), and 24 hpa (dark green), with volumes smaller than 0.1 µm3, between 0.1 and 1 µm3, between 1 and 10 µm3, and larger than 10 µm3. Statistical analysis corresponds to Mann-Whitney test. Mean and SD are displayed (n=12 blastemas from 4 fish). e, epidermis; b, bone; m, mesenchyme; ns, not significative; *, p-value <0.01. See Figure 2—figure supplement 2—source data 1.
-
Figure 2—figure supplement 2—source data 1
Spreadsheets detailing the results of the mitochondrial dynamics, specifically (A,B) the relative gene expression analysis, and the quantification of (I) the number of mitochondria per cell and (J) the percentage of each mitochondria volume.
- https://cdn.elifesciences.org/articles/76987/elife-76987-fig2-figsupp2-data1-v1.xlsx
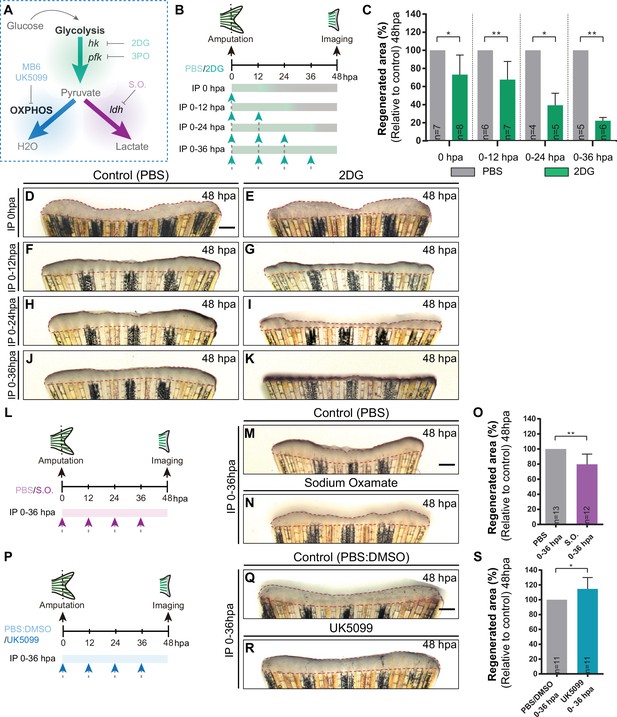
Inhibition of glycolysis, but not OXPHOS, impairs blastema formation.
(A) Schematic representation of the compounds used to manipulate glucose metabolism. (B) Experimental design used to inhibit the glycolytic influx during fin regeneration. Control and treated fish are administered, via IP injection, with vehicle (PBS) or glycolytic inhibitor, 2DG, respectively, every 12 hr, from fin amputation (0 hpa) until 48 hpa. Different time-intervals were used for injections: (0 hpa) IP injection at (0 hpa; 0–12 hpa) IP injection at 0 and 12 hpa; (0–24 hpa) IP injection at 0, 12, and 24 hpa; (0–36 hpa) IP injection at 0, 12, 24, and 36 hpa. (C) Quantification of the total fin regenerated area at 48 hpa, after vehicle (PBS) or 2DG injection, at specific time-intervals during regeneration. (D–K) Representative images of 48 hpa fins treated with (D,F,H,J) vehicle (PBS) or (E,G,I,K) 2DG during different time-intervals. (L) Experimental design used to inhibit the lactate formation during fin regeneration. Fish are administered, via IP injection, with vehicle (PBS) or S.O. every 12 hr, from fin amputation (0 hpa) until 48 hpa. (M, N) Representative images of 48 hpa caudal fin treated with (M) vehicle (PBS) or (N) S.O. (O) Quantification of the total fin regenerated area at 48 hpa, after vehicle (PBS) or with S.O. injection. (P) Experimental design used to inhibit pyruvate translocation to mitochondria during fin regeneration. Fish are administered, via IP injection, with vehicle (PBS) or MPC inhibitor, UK5099, every 12 hr from fin amputation (0 hpa) until 48 hpa. (Q, R) Representative images of 48 hpa fins treated with (Q) PBS:DMSO (control) or (R) UK5099. (S) Quantification of the total fin regenerated area at 48 hpa, after vehicle (PBS) or with UK5099 injection. For all graphs, statistical analysis corresponds to Mann-Whitney test with Mean ± SD, sample number is displayed on each column and corresponds to single fish. Scale bar represents 500 µm. Dashed lines define the regenerated tissue. * p<0.05, ** p<0.01. See Figure 3—source data 1.
-
Figure 3—source data 1
Spreadsheets detailing the results of glycolysis and OXPHOS inhibitory assays, specifically the quantification of (C) the percentage of regenerated fin area after 2DG treatment, (O) the percentage of regenerated fin area after S.O. treatment, and (S) the percentage of regenerated fin area after UK5099 treatment.
- https://cdn.elifesciences.org/articles/76987/elife-76987-fig3-data1-v1.xlsx
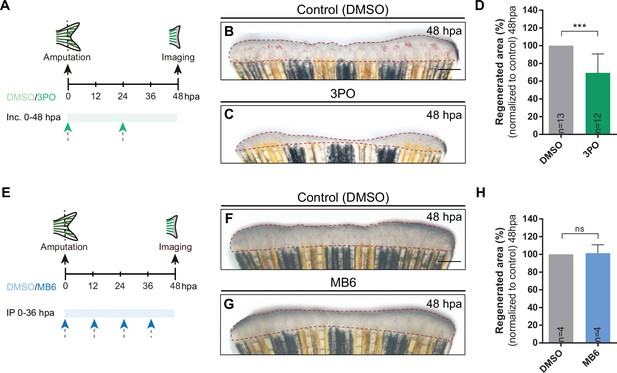
Inhibition of glycolysis impairs blastema formation.
(A) Experimental design used to inhibit glycolysis during fin regeneration. Fish are incubated with control vehicle (PBS) or with the glycolytic inhibitor 3PO, every 24 hr from caudal fin amputation (0 hpa) until 48 hpa. (B–C) Representative images of 48 hpa fins treated with (B) vehicle (DMSO) or (C) 3PO. (D) Quantification of the total caudal fin regenerated area at 48 hpa of fish treated with vehicle (DMSO) or with 3PO. (E) Experimental design used to inhibit pyruvate mitochondrial import during fin regeneration. Fish are incubated with vehicle (DMSO) or MB6, every 24 hr from fin amputation (0 hpa) until 48 hpa. (F–G) Representative images of 48 hpa caudal fins treated with (F) vehicle (DMSO) or (G) MB6. (H) Quantification of the total caudal fin regenerated area at 48 hpa of fish treated with vehicle (DMSO) or with MB6. Statistical analysis displayed on the graphs corresponds to Mann-Whitney test with Mean ± SD, sample number is displayed on each column and corresponds to single fish. Scale bar represents 500 µm. Dashed lines define the regenerated area. ns: not significative; ***p<0.001. See Figure 3—figure supplement 1—source data 1.
-
Figure 3—figure supplement 1—source data 1
Spreadsheets detailing the results of alternative glycolysis and OXPHOS inhibitory assays, specifically the quantification of the percentage of the regenerated fin area after (D) 3PO and (H) MB6 treatment.
- https://cdn.elifesciences.org/articles/76987/elife-76987-fig3-figsupp1-data1-v1.xlsx
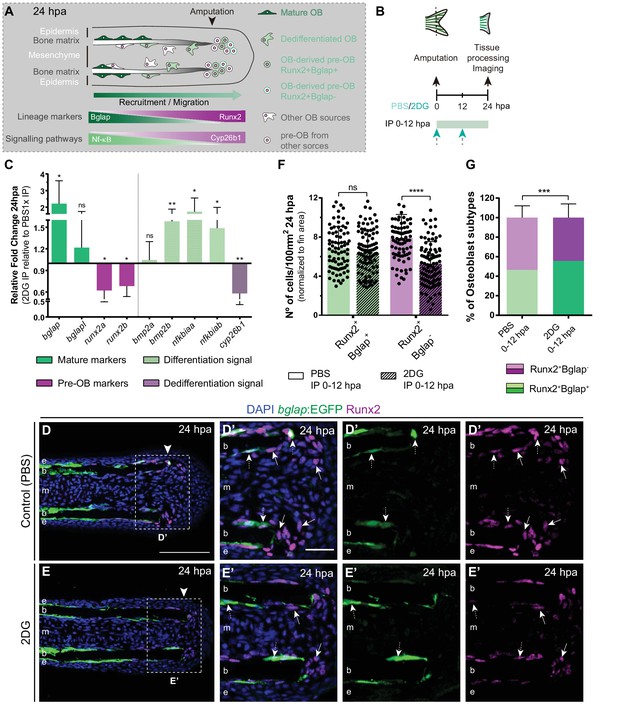
Inhibition of glycolysis impairs osteoblast dedifferentiation.
(A) Schematic representation of pre-OBs formation during regeneration. Pre-OBs arise from OB dedifferentiation and potentially from the joint OP niche. OB dedifferentiation is correlated with inactivation of NF-ΚB and increase in Cyp26b1 activity. (B) Experimental design used to inhibit glycolysis. Fish are administered, via IP injection, with vehicle (PBS) or 2DG, from fin amputation (0 hpa) until 24 hpa. (C) Relative gene expression of mature and pre-OBs markers, and differentiation and dedifferentiation pathways, in the whole fin stump at 24 hpa, in 2DG treated fins compared to control condition (0 hpa). Statistical analysis with paired t-test (n=10 biological replicates). (D-E’) Representative cryosection images of 24 hpa bglap:EGFP (green) caudal fins immunostained for Runx2 (magenta) and counterstained with DAPI (blue), in fish treated with (D,D’) vehicle (PBS) or (E,E’) 2DG. White dashed boxes delineate magnified panels in D’ and E’. Arrows indicate Runx2 +Bglap cells. Dashed arrows indicate Runx2 +Bglap + cells. Arrowhead indicates amputation plane. E: epidermis; b: bone; m: mesenchyme. Scale bar represents 100 µm and 30 µm in magnified panels. (F) Total number of Runx2 +Bglap + and Runx2 +Bglap cells per area at 24 hpa. (G) Percentage of Runx2 +Bglap + and Runx2 +Bglap OBs subtypes. Statistical analysis displayed on each graph corresponds to Mann-Whitney test with Mean ± SD (n=79 (PBS) and 89 (2DG) cryosections). ns: not significant; ** p<0.01; *** p<0.0001. See Figure 4—source data 1.
-
Figure 4—source data 1
Spreadsheets detailing the results of the impaired osteoblast dedifferentiation after glycolysis inhibition with 2DG, specifically (C) the relative gene expression analysis, and the quantification of (F) the number of Runx2 +Bglap + and Runx2 +Bglap cells, and (G) the percentage of osteoblasts subtypes (Runx2 +Bglap + and Runx2 +Bglap-).
- https://cdn.elifesciences.org/articles/76987/elife-76987-fig4-data1-v1.xlsx
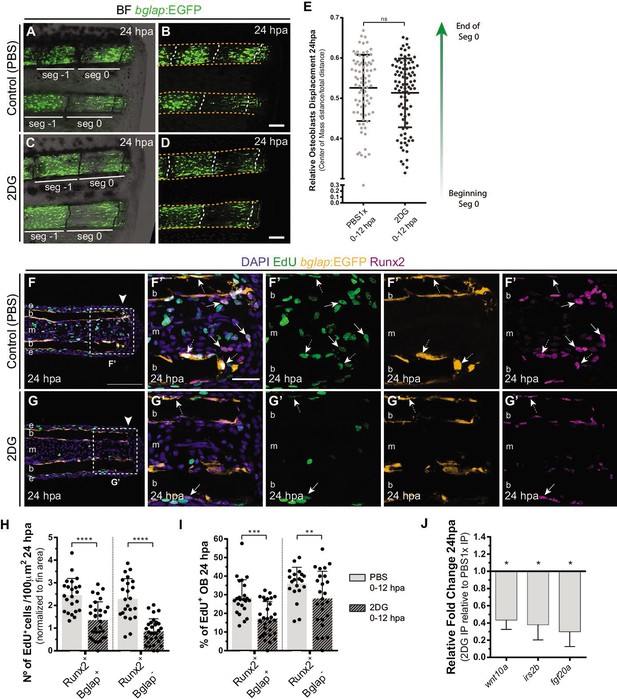
Inhibition of glycolysis impairs osteoblast cell cycle-entry.
(A–D) Representative images of bglap:EGFP caudal fins at 24 hpa, treated with (A–B) vehicle (PBS) or (C–D) 2DG. Double white arrows indicate the anterior (A) and posterior (P) axis. White dashed lines indicate intersegment regions. Orange dashes lines delineate the bony-ray surface. (E) Measurement of relative OB displacement along segment 0, below the amputation plane, at 24 hpa in fins treated with vehicle (PBS) or 2DG. Statistical analysis on graph corresponds to Mann-Whitney test with Mean ± SD (PBS = 90, 2DG = 82 bony-rays). (F-G’) Representative cryosection images of 24 hpa bglap:EGFP (orange) caudal fins immunostained for Runx2 (magenta), labeled with EdU (green) and counterstained with DAPI (blue), in fish treated with (F) control (PBS) or (G) 2DG. Dashed boxes delineate amplified panels in F’ and G’. Arrows indicate proliferative EdU +Runx2+Bglap cells. Dashed arrows indicate proliferative EdU +Runx2+Bglap + cells. Arrowhead indicates amputation plane. Scale bar represents 100 µm and 30 µm in amplified panels. (H) Total number of Runx2 +Bglap + and Runx2 +Bglap cells at 24hpa, in fins treated with vehicle (PBS) or 2DG. (I) Percentage of proliferative Runx2 +Bglap + and Runx2 +Bglap cells at 24hpa, in fins treated with vehicle (PBS) or 2DG. Statistical analysis displayed on each graph corresponds to Mann-Whitney test with Mean ± SD (n=23–30 cryosections). (J) Relative gene expression at 24 hpa in 2DG treated fins, compared to control. Statistical analysis with unpaired t test and Welch’s correction (n=5 biological replicates). ns: not significant; *p<0.0001. See Figure 5—source data 1.
-
Figure 5—source code 1
MatLab scripts to quantify the relative osteoblast displacement after caudal fin amputation in controls and after 2DG treatment.
- https://cdn.elifesciences.org/articles/76987/elife-76987-fig5-code1-v1.zip
-
Figure 5—source data 1
Spreadsheets detailing the results of impaired osteoblasts cell cycle re-entry after glycolysis inhibition with 2DG, specifically the quantification of (H) the number of Runx2 +Bglap + and Runx2 +Bglap- EdU + cells, (I) the percentage of Runx2 +Bglap + and Runx2 +Bglap- EdU + cells and (J) the relative gene expression analysis.
- https://cdn.elifesciences.org/articles/76987/elife-76987-fig5-data1-v1.xlsx
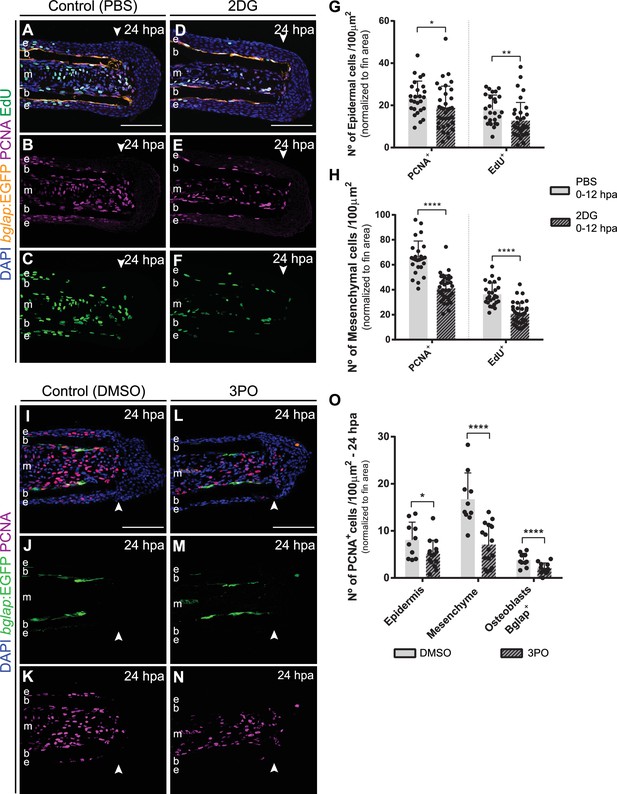
Inhibition of glycolysis prevents cell cycle re-entry.
(A–F) Representative 24 hpa cryosection images of bglap:EGFP (orange) caudal fins immunostained for PCNA (magenta), EdU (green) and counterstained with DAPI (blue), in fish treated with (A–C) vehicle (PBS) or (D–F) 2DG. (G–H) Quantification of the number of PCNA +proliferating cells and EdU +proliferating cells at 24 hpa, in the (G) epidermis and (H) mesenchyme, in caudal fins from control and 2DG-treated fish. Statistical analysis displayed on the graphs corresponds to Mann-Whitney test (n=26 (PBS) and 35 (2DG) cryosections). (I–N) Representative 24 hpa cryosection images of bglap:EGFP (green) caudal fins immunostained for PCNA (magenta) and counterstained with DAPI (blue), in fish treated with (I–K) vehicle (DMSO) or with (L–N) 3PO. (O) Quantification of the number of PCNA +proliferating cells in the epidermal and mesenchymal compartments, and PCNA +Bglap + cells, at 24 hpa in fins from control and 3PO treated fish. Statistical analysis displayed on the graphs corresponds to Mann-Whitney test with Mean ± SD (n=10 (DMSO) and 14 (3PO) cryosections). Arrowheads indicate amputation plane. Scale bars represents 100 µm. e: epidermis; b: bone; m: mesenchyme. *p<0.0001. See Figure 5—figure supplement 1—source data 1.
-
Figure 5—figure supplement 1—source data 1
Spreadsheets detailing the results of impaired cell cycle re-entry after glycolysis inhibition on individual fin tissues, specifically the quantification of the number of PCNA +and EdU + cells in the (G) epidermis and (H) mesenchyme after 2DG treatment, and (I) the number of PCNA + cells in the epidermis, mesenchyme, and osteoblasts after 3PO treatment.
- https://cdn.elifesciences.org/articles/76987/elife-76987-fig5-figsupp1-data1-v1.xlsx
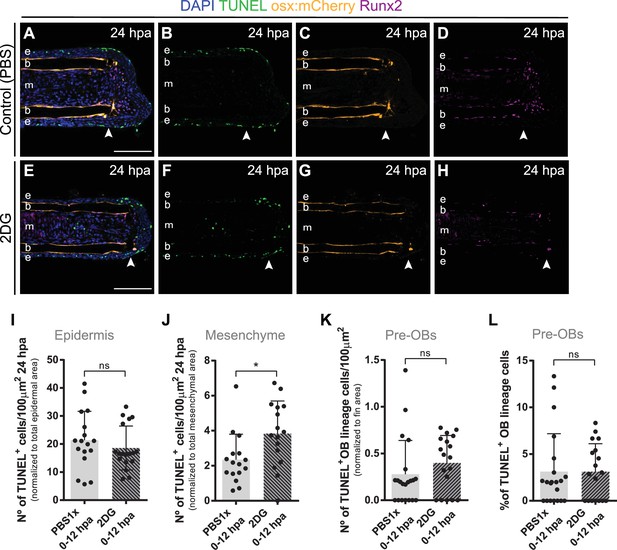
Inhibition of glycolysis has no effect on pre-osteoblasts cell death.
(A–H) Representative 24 hpa cryosection images of osx:mCherry (orange) caudal fins immunostained for Runx2 (magenta), TUNEL (green) and counterstained with DAPI (blue), in fish treated with (A–D) vehicle (PBS) or (E–H) 2DG. Arrowheads indicate amputation plane. Scale bar represents 100 µm. e, epidermis; b, bone; m, mesenchyme. (I–K) Quantification of the total number of TUNEL + cells in the (I) epidermis, (J) mesenchyme and (K) and in the pre-OBs at 24 hpa, in caudal fins from controls and 2DG-treated fish. (L) Percentage of TUNEL +pre OBs at 24 hpa, in fins from controls and 2DG-treated fish. Statistical analysis displayed on all graphs corresponds to Mann-Whitney test with Mean ± SD (n=16–21 cryosections). ns: not significant; *p<0.05. See Figure 5—figure supplement 2—source data 1.
-
Figure 5—figure supplement 2—source data 1
Spreadsheets detailing the results of the impact of glycolysis inhibition, after 2DG treatment, on pre-osteoblasts cell death, specifically the quantification of the number of TUNEL + cells in the (I) epidermis, (J) mesenchyme and (K) pre-osteoblasts, and (L) the percentage of TUNEL +osteoblasts.
- https://cdn.elifesciences.org/articles/76987/elife-76987-fig5-figsupp2-data1-v1.xlsx
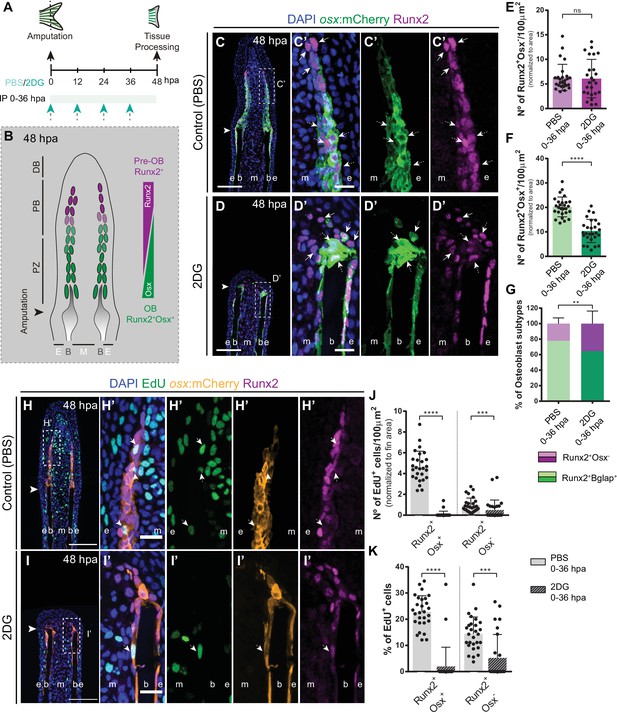
Inhibition of glycolysis affects formation of osteoblast subtypes and proliferation within the blastema.
(A) Experimental design used to inhibit glycolysis. Fish are administered, via IP injection, with control (PBS) or 2DG every 12 hr, from fin amputation (0 hpa) until 48 hpa. (B) Schematic representation of the distribution of OBs subtypes along the blastema. (C–D) Representative cryosection images of 48 hpa osx:mCherry (green) caudal fins immunostained for Runx2 (magenta) and counterstained with DAPI (blue), in fish treated with (C,C’) PBS and (D,D’) 2DG. Dashed boxes represent magnified panels in C’ and D’. Arrows indicate Runx2 +Osx- pre-OBs. Dashed arrows indicate Runx2 +Osx + immature OBs. (E–F) Total number of (E) Run2 +Osx and (F) Runx2 +Osx + subtypes in 48 hpa fins treated with PBS or 2DG (PBS = 27, 2DG = 25 cryosections). (G) Percentage of Runx2+/Osx- and Runx2 +Osx + subtypes in 48 hpa fins treated with PBS or 2DG. (H-I’) Representative cryosection images of 48 hpa osx:mCherry (orange) caudal fins immunostained for Runx2 (magenta), EdU (green) and counterstained with DAPI (blue), in fish treated with (H,H’) PBS and (I,I’) 2DG. Dashed boxes represent magnified panels in H’ and I’. Arrows indicate proliferative Edu +Runx2+Osx- pre-OBs. Dashed arrows indicate proliferative Edu +Runx2+Osx + immature OBs. Arrowheads indicate amputation plane. Scale bar represents 100 µm and 20 µm in magnified panels. (J) Total number of Runx2 +Osx + and Runx2 +Osx- proliferative OBs subtypes at 48 hpa fins, treated with PBS or 2DG (PBS = 28, 2DG = 27 cryosections). (K) Percentage of proliferative Runx2 +Osx + and Runx2 +Osx OBs subtypes in 48 hpa caudal fins , treated with PBS or 2DG. E: epidermis; B: bone; M: mesenchyme. For all graphs, statistical analysis corresponds to Mann-Whitney and Mean ± SD are displayed. ns: not significant; **p<0.0001. See Figure 6—source data 1.
-
Figure 6—source data 1
Spreadsheets detailing the results of the formation of osteoblast subtypes and proliferation within the blastema after glycolysis inhibition with 2DG, specifically the quantification of the number of (E) Runx2 +Osx- pre-osteoblasts and (F) Runx2 +Osx + osteoblasts, (G) the percentage of osteoblasts subtypes, (K) the total number of Runx2 +Osx + and Runx2 +Osx- EdU +osteoblast subtypes, and (K) the percentage of proliferative Runx2 +Osx + and Runx2 +Osx- osteoblast subtypes.
- https://cdn.elifesciences.org/articles/76987/elife-76987-fig6-data1-v1.xlsx
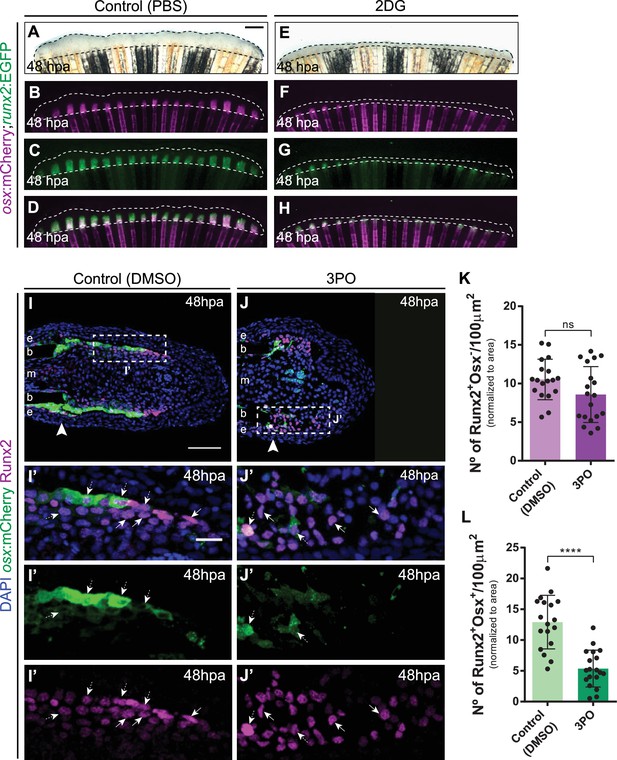
Inhibition of glycolysis affects distribution of osteoblast subtypes in the blastema.
(A–H) Representative 48 hpa caudal fin images of osx:mCherry (magenta) and runx2:EGFP (green) double transgenics treated with (A–D) vehicle (PBS) or (E–H) 2DG. Dashed lines delineate regenerated area. Scale bar represents 500 µm. (I-J’) Representative 48 hpa cryosection images of osx:mCherry (green) caudal fins immunostained for Runx2 (magenta) and counterstained with DAPI (blue), in fish treated with (I,I’) vehicle (DMSO) or (J,J’) 3PO. Dashed boxes represent magnified panels in I’ and J’. Arrowheads indicate amputation plane. Scale bar represents 100 µm and 20 µm for magnified panels. (K–L) Quantification of the number of (K) Runx2 +Osx and (L) Run2 +Osx + OB subtypes at 48hpa, in caudal fins from control and 3PO treated fish. Statistical analysis displayed on all graphs corresponds to Mann-Whitney test with Mean ± SD (n=18 (DMSO) and 19 (3PO) cryosections) ns: not significant; ****p<0.0001. See Figure 6—figure supplement 1—source data 1.
-
Figure 6—figure supplement 1—source data 1
Spreadsheets detailing the results of the distribution of osteoblast subtypes in the blastema after glycolysis inhibition with 3PO, specifically the quantification of the number of (K) Runx2 +Osx and (L) Runx2 +Osx + cells.
- https://cdn.elifesciences.org/articles/76987/elife-76987-fig6-figsupp1-data1-v1.xlsx
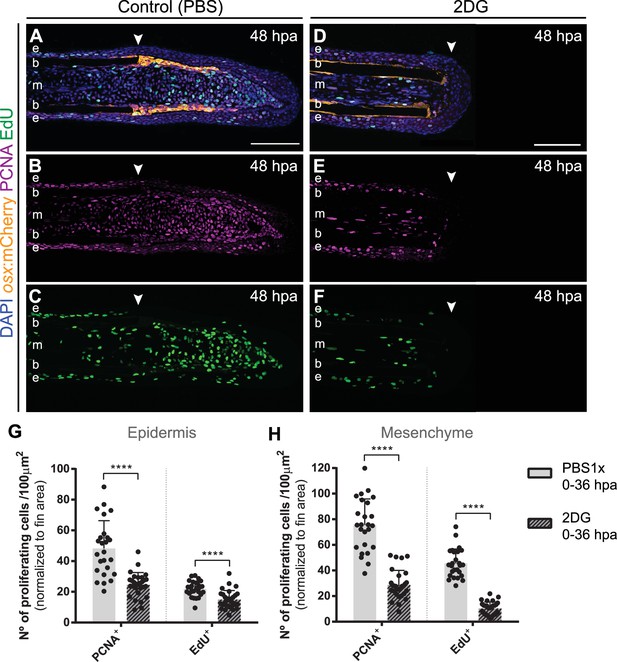
Inhibition of glycolysis impairs proliferation during blastema formation.
(A–F) Representative 48 hpa cryosection images of osx:mCherry (orange) caudal fins immunostained for PCNA (magenta), EdU (green) and counterstained with DAPI (blue), in fish treated with (A–C) vehicle (PBS) or (D–F) 2DG. Arrowheads indicate amputation plane. Scale bar represents 100 µm. e: epidermis; b: bone; m: mesenchyme. (G–H) Quantification of the number of proliferative PCNA +and EdU + cells in the (G) epidermal and (H) mesenchymal compartments at 48 hpa, in caudal fins from controls and 2DG-treated fish. Statistical analysis displayed on all graphs corresponds to Mann-Whitney test with Mean ± SD (n=25 (DMSO) and 29 (3PO) cryosections). ****p<0.0001. See Figure 6—figure supplement 2—source data 1.
-
Figure 6—figure supplement 2—source data 1
Spreadsheets detailing the results of cell proliferation after glycolysis inhibition with 2DG, specifically the quantification of the number of EdU +and PCNA + cells in the (G) epidermis and (H) mesenchyme.
- https://cdn.elifesciences.org/articles/76987/elife-76987-fig6-figsupp2-data1-v1.xlsx
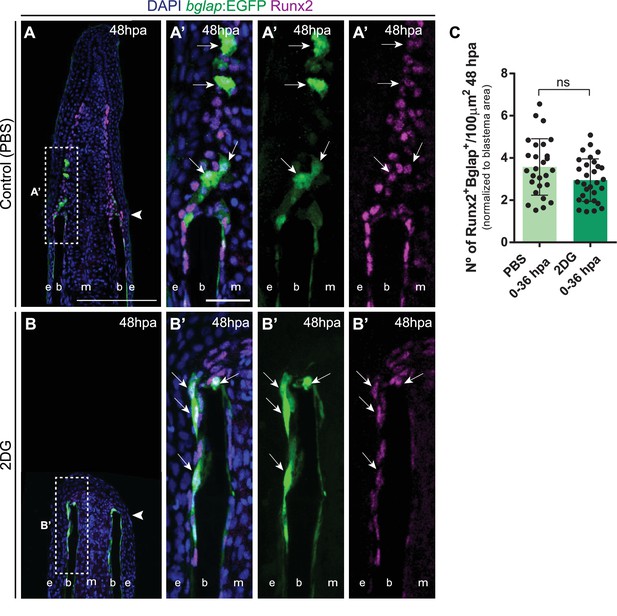
Mature osteoblasts accumulate at stump region after glycolysis inhibition.
(A,B) Representative cryosection images of 48 hpa bglap:EGFP (green) caudal fins immunostained for Runx2 (magenta) and counterstained with DAPI (blue), in fish treated with (A, A) PBS and (B, B’) 2DG. Dashed boxes represent magnified panels in A’ and B’. Arrows indicate Runx2 +Bglap + OBs. Arrowheads indicate amputation plane. E: epidermis; B: bone; M: mesenchyme. Scale bar represents 200 µm and 30 µm in magnified panels. (C) Total number of Runx2 +Bglap + OBs at 48 hpa fins, treated with PBS or 2DG. Statistical analysis on graph corresponds to Mann-Whitney test. Mean ± SD are displayed (n=26 (PBS) and 32 (2DG) cryosections). ns: not significant. See Figure 6—figure supplement 3—source data 1.
-
Figure 6—figure supplement 3—source data 1
Spreadsheets detailing the results of mature osteoblasts accumulation in the stump region after inhibition of glycolysis with 2DG, specifically (C) the quantification of the number of Runx2 +Bglap + cells.
- https://cdn.elifesciences.org/articles/76987/elife-76987-fig6-figsupp3-data1-v1.xlsx
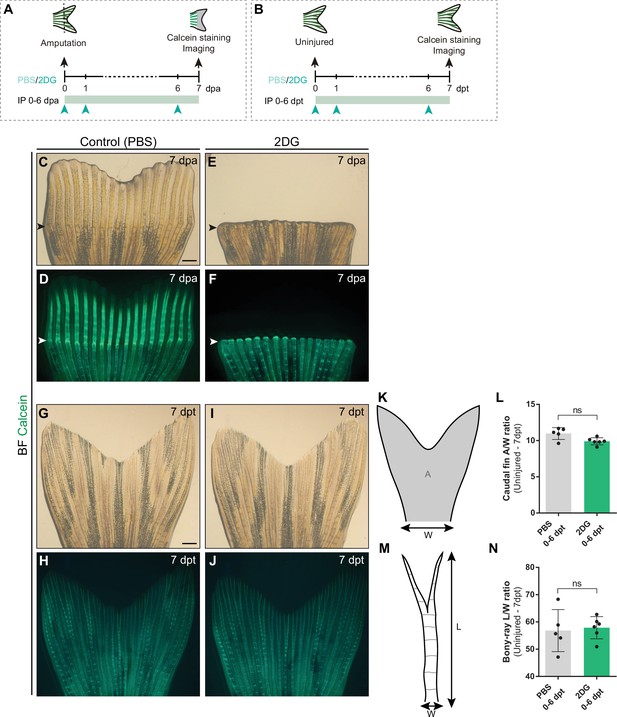
Inhibition of glycolysis impairs bone regeneration, but not fin and bony-ray integrity in uninjured conditions.
(A, B) Experimental design used for extended inhibition of the glycolytic influx during (A) fin regeneration and in (B) uninjured caudal fins. Control and treated fish are administered, via IP injection, with vehicle (PBS) or glycolytic inhibitor, 2DG, daily from 0 to 6 dpa or dpt and fins collected next day and subjected to calcein staining and imaging. (C–F) Representative images of 7 dpa regenerating caudal fins treated with (C, D) vehicle or (E, F) 2DG. (G–J) Representative images of 7 dpt uninjured caudal fins treated with (G, H) vehicle or (I, J) 2DG. (K) Schematic representation of an uninjured caudal fin: gray area represents the caudal fin area and the basal peduncle used as a measure of caudal fin width. (L) Quantification of the uninjured caudal fin area to width ratio after vehicle (PBS) or with 2DG injection. (M) Schematic representation of caudal fin bony-ray length and width. (N) Quantification of the average bony-ray length to width ratio uninjured caudal fin L/W ratio after vehicle (PBS) or with 2DG injection. For all graphs, statistical analysis corresponds to Mann-Whitney test with Mean ± SD displayed, each dot on every column corresponds to a single fish (n=5 (PBS), 6 (2DG)). White arrowhead indicates the amputation plane. Scale bar represents 500 µm. dpa: days post-amputation; dpt: days post-treatment; IP: intraperitoneal injection; A: area; W: width; L: length; ns: not significant. See Figure 6—figure supplement 4—source data 1.
-
Figure 6—figure supplement 4—source data 1
Spreadsheets detailing the results of inhibiting glycolysis with 2DG in uninjured caudal fins, specifically the quantification of (L) fin area to width ratio, and (N) the average of bony-ray length to width ratio.
- https://cdn.elifesciences.org/articles/76987/elife-76987-fig6-figsupp4-data1-v1.xlsx
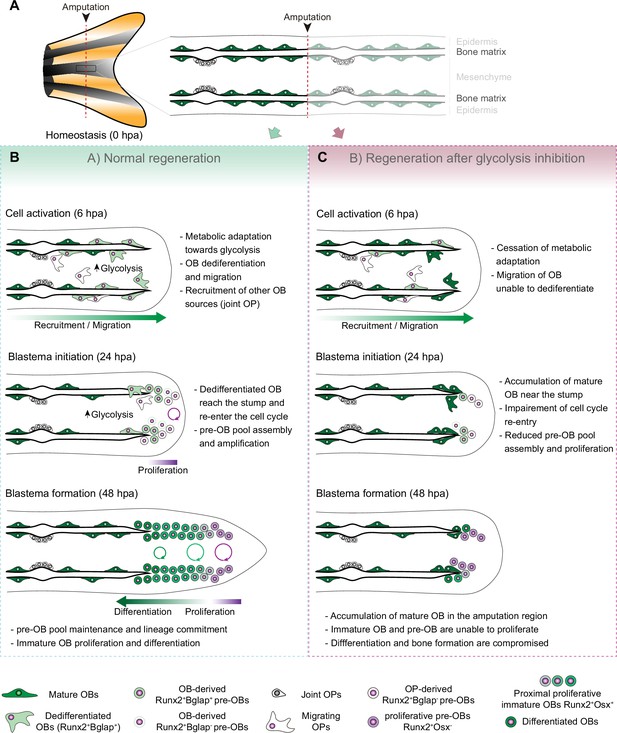
Model for the role of glucose metabolism during caudal fin regeneration.
(A) In homeostasis, mature OBs reside in close contact with the bony-ray surface, secreting the collagenous bone matrix. (B) Upon caudal fin amputation, OBs and other cell types in the regenerating fin respond by undergoing a metabolic adaptation that stimulates glycolysis and is essential for regeneration to proceed. Enhancing glycolytic influx promotes OB dedifferentiation, by releasing Cyp26b1 from NF-ΚB repression, and cell cycle re-entry, by interfering with the master regulation of caudal fin proliferation Fgf20a, thereby enabling OBs to act as progenitor cells. Moreover, glycolysis is necessary to maintain the correct proliferative ability and distribution of OBs populations within the blastema, during its formation. (C) Glycolysis inhibition has a severe impact on OB dedifferentiation and pre-OBs pool assembly, which supports new OB formation and proliferation, ultimately leading to impaired bony-ray regeneration and suppression of blastema formation.
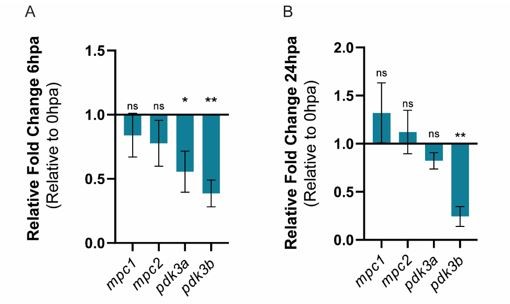
Relative gene expression of mpc1, mpc2, pdk3b, in the whole caudal fin stump, a (A) 6hpa and at (B) 24 hpa in comparison to uninjured conditions (0 hpa).
Statisical analysis with t test (n=5 (A) and 4 (B) biological replicates).
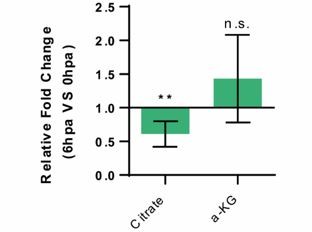
Relative measurement of Citrate and a-KG metabolite levels, in the whole caudel fin stump, at 6 hpa in comparison to uninjured conditions (0 hpa) Statistical analysis with Turkey HSD test (n=4 biological replicates).
Mean and SD displayed on the graphs. n.s., not significative, *, p-value <0,01.
Additional files
-
Supplementary file 1
Spreadsheet detailing the list of significantly differentially expressed genes with p-value lower than the 0.05 threshold and log2 fold change < –1 or >1.
- https://cdn.elifesciences.org/articles/76987/elife-76987-supp1-v1.xlsx
-
Supplementary file 2
Tables detailing primer sequences for q-PCR assay (a), primary antibodies used for immunofluorescence assays(b), secondary antibodies used for immunofluorescence assays (c) and sample size number and statistical test preformed for each quantitative experimental design (d).
- https://cdn.elifesciences.org/articles/76987/elife-76987-supp2-v1.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/76987/elife-76987-transrepform1-v1.docx





