Self-organization of songbird neural sequences during social isolation
Figures
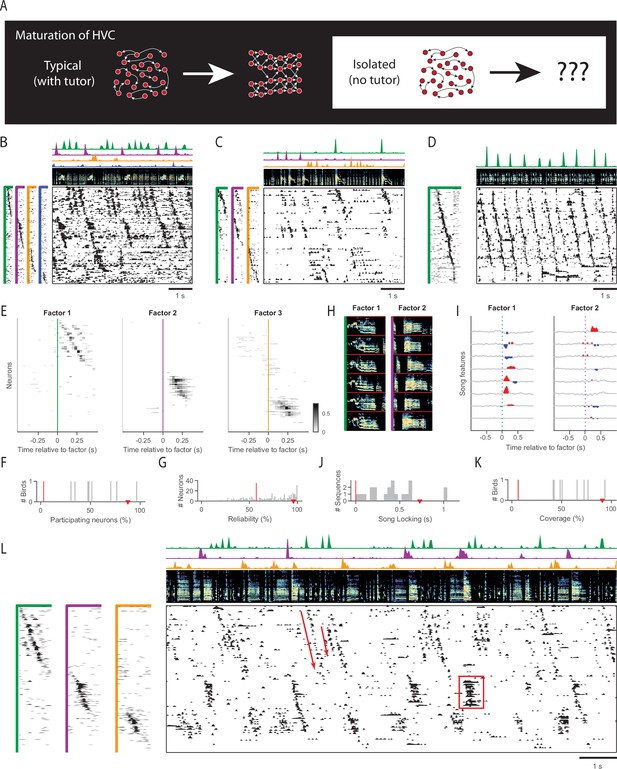
Sequences in isolated birds.
(A) Diagram of HVC maturation. In typically tutored birds, HVC sequences appear to grow and differentiate over time. (B) Example neural sequences recorded in a singing isolated bird (older juvenile, 61 dph). Main panel (lower right), functional calcium imaging recordings from 98 neurons. Rows (neurons) are sorted according to sequences (factors) extracted by unsupervised algorithm seqNMF (see Methods). (Above) Song spectrogram (0–10kHz). The four sequence factor exemplars and timecourses are shown to the left and above, in corresponding colors. Duration of factor exemplars: 0.5s. (C) Same as B, for another example isolated bird (adult, 117 dph). (D) Same as in B, for a typically tutored bird (adult, 217 dph). (E) Time-lagged cross-correlation between each neuron and each of the three extracted factors recorded in a singing isolated bird (older juvenile, 68 dph). Only significant bins in the cross correlation are shown (p<0.05, Bonferroni corrected, compared to a circularly-shifted control). (F, G, J, K) Sequence properties in isolated birds. For reference, the median for a typically tutored bird in D is shown by a red triangle, and the median for a control dataset where each row was circularly shifted by a random amount is shown by a red line. (F) Percent of neurons participating in at least one extracted sequence. (G) Reliability of participating neurons across sequence renditions. Note that in the control dataset, relatively few neurons participate. (H) Example song spectrograms (0.5s) extracted at moments when neural sequences were detected in an isolated bird (older juvenile, 64 dph) (I) Correlation of these sequences with eight song features (top to bottom: amplitude, entropy, pitch goodness, aperiodicity, mean frequency, pitch, frequency modulation, amplitude modulation). (J) Strength of song locking (see Methods). (K) Percent of the song covered by some sequence. (L) Example of sequence abnormalities in an isolated bird (same as in E). Sequences of inconsistent length (8/8 isolated birds) and ensemble persistent activity (7/8 isolated birds) are annotated in red.
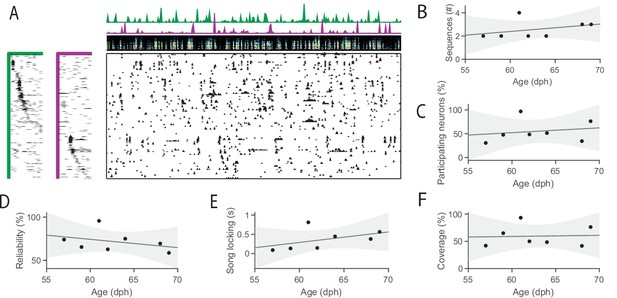
HVC sequences exist even in young isolated birds.
(A) Example HVC sequences recorded in a young isolated bird (59 dph) (B–F) Sequence properties as a function of age in seven juvenile isolated birds (five birds recorded prior to the closing of the traditional critical period (<65dph), and two older juvenile birds (65 dph-90 dph)). Line denotes least squares fit, gray area 95% confidence interval. (B) Number of HVC sequences extracted. (C) Percent of neurons participating in at least one sequence. (D) Reliability of neural participation across sequence renditions. (E) Song locking. (F) Percent of the song covered by at least one sequence.
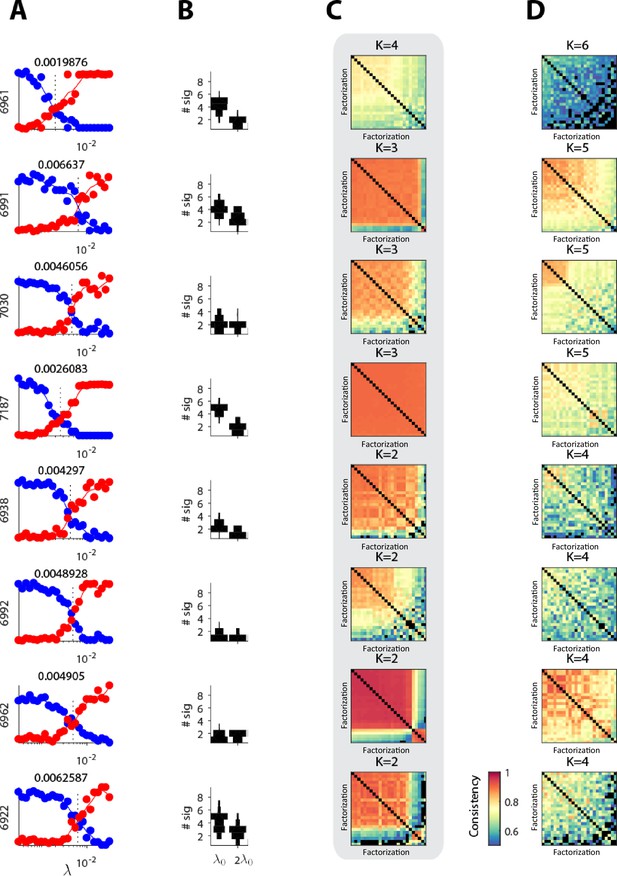
Estimating the number of significant sequences in each dataset.
(A) Reconstruction cost (red) and correlation cost (blue) as a function of (with K=10, L=0.5s) for eight datasets (pre-tutoring data from eight different birds). The crossover point, , is stated and marked by a dashed line. (B) Histogram of the number of significant sequences at and for these datasets. (C) For the chosen K, and , consistency across 25 runs of seqNMF from different random initializations. Factorizations are sorted from most to least consistent. (D) Consistency matrix for 25 runs at K above the estimated K.
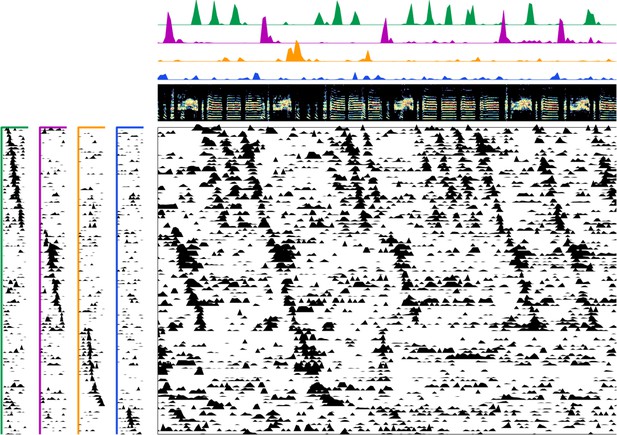
Enlarged data, bird 6961.
Enlarged panel showing spectrogram, neural data, and seqNMF factorization for 6 s of pre-tutoring data.
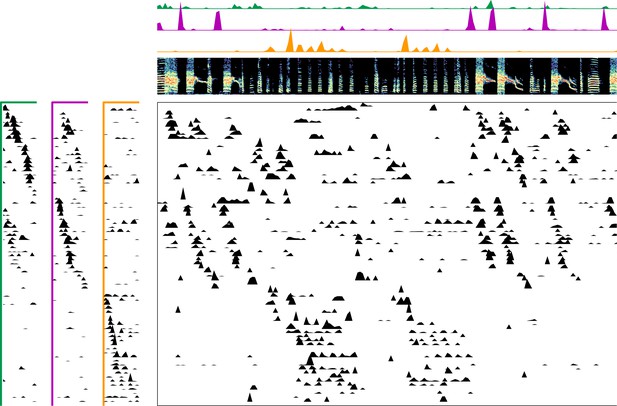
Enlarged data, bird 6991.
Enlarged panel showing spectrogram, neural data, and seqNMF factorization for 6 s of pre-tutoring data.
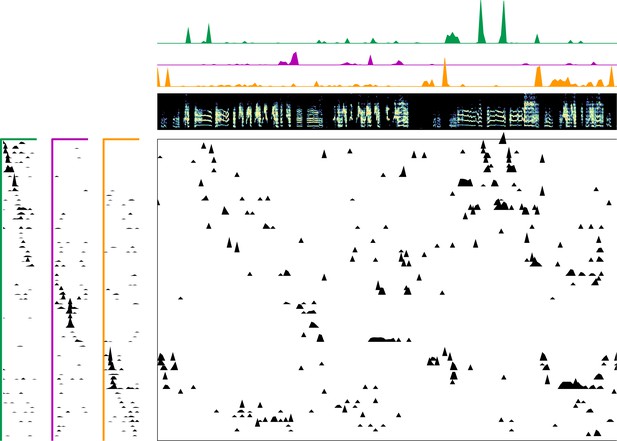
Enlarged data, bird 7030.
Enlarged panel showing spectrogram, neural data, and seqNMF factorization for 6 s of pre-tutoring data.
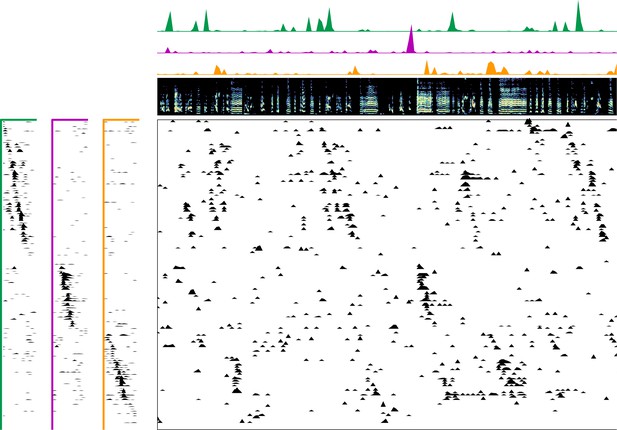
Enlarged data, bird 7187.
Enlarged panel showing spectrogram, neural data, and seqNMF factorization for 6 s of pre-tutoring data.
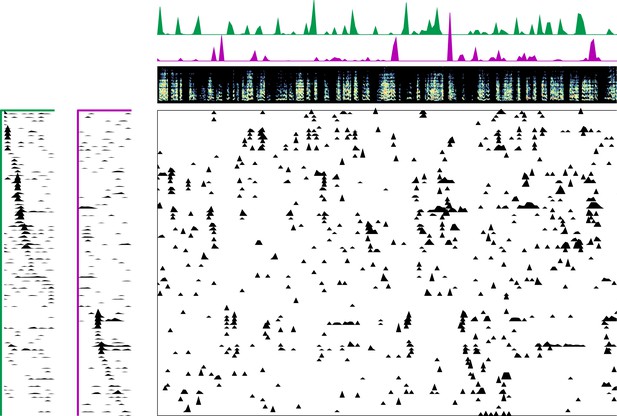
Enlarged data, bird 6938.
Enlarged panel showing spectrogram, neural data, and seqNMF factorization for 6 s of pre-tutoring data.
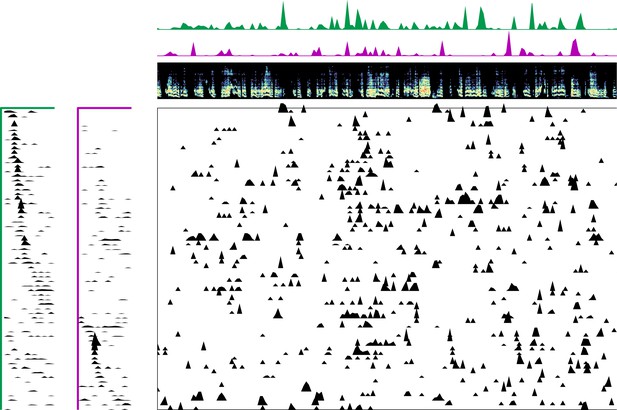
Enlarged data, bird 6992.
Enlarged panel showing spectrogram, neural data, and seqNMF factorization for 6 s of pre-tutoring data.
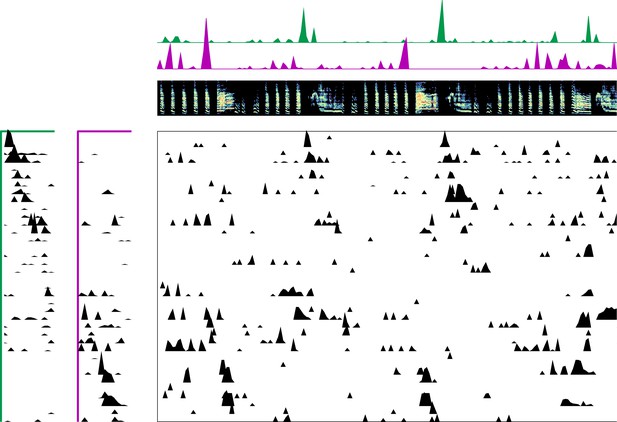
Enlarged data, bird 6962.
Enlarged panel showing spectrogram, neural data, and seqNMF factorization for 6 s of pre-tutoring data.
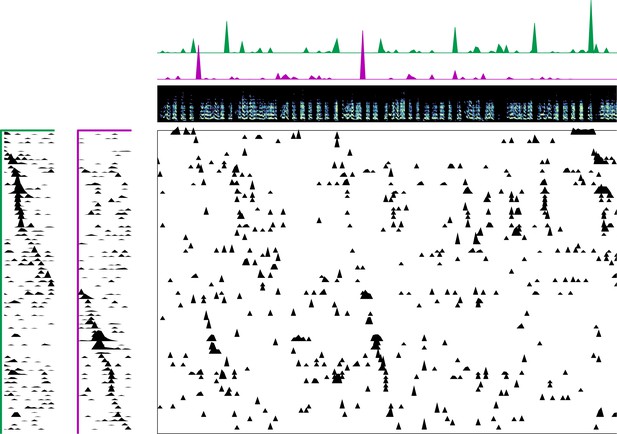
Enlarged data, bird 6922.
Enlarged panel showing spectrogram, neural data, and seqNMF factorization for 6 s of pre-tutoring data.
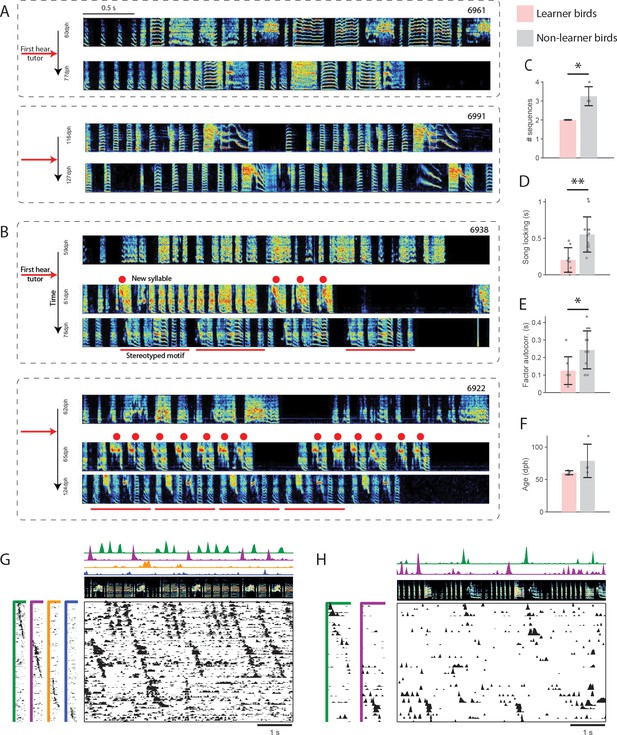
Relation between HVC sequence maturity and subsequent song learning.
(A) Example spectrograms for two non-learner birds, prior to tutoring and several weeks later (at least 77dph). (B) Example spectrograms for two learners, prior to tutoring, shortly after tutoring, and several weeks later. Red dots mark the new syllable. Red bars mark a stereotyped motif. (C–E) Three measures of HVC sequence maturity for learners (pink) and non-learners (gray). Error bars denote standard deviation (*p<0.05, *p<0.01). (C) Number of sequences in HVC. (D) Fraction of neurons that participate in a sequence. (E) Autocorrelation of sequence factor timecourses. (F) Age of first tutoring for learners and non-learners. (G–H) Example pre-tutoring data from two birds that were brothers. (G) A non-learner first tutored at 61 dph. (H) A learner first tutored at 64 dph.
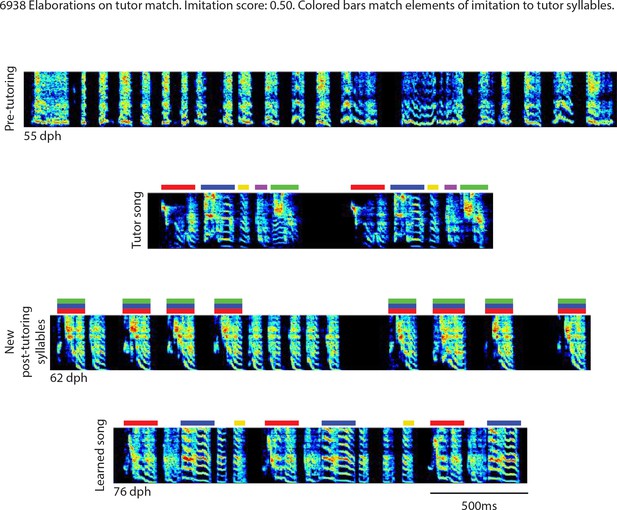
Elaborations on tutor match, bird 6938.
Song spectrograms at different stages of development, and comparison to tutor song spectrogram. Colored bars match elements of the imitation to tutor song syllables.
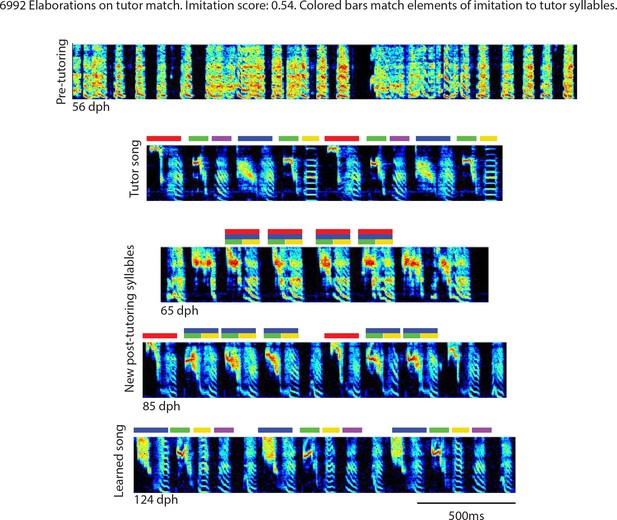
Elaborations on tutor match, bird 6992.
Song spectrograms at different stages of development, and comparison to tutor song spectrogram. Colored bars match elements of the imitation to tutor song syllables.
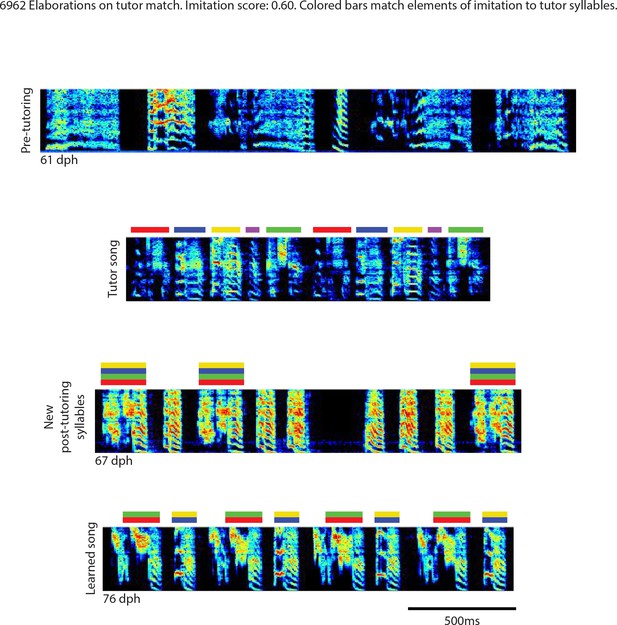
Elaborations on tutor match, bird 6962.
Song spectrograms at different stages of development, and comparison to tutor song spectrogram. Colored bars match elements of the imitation to tutor song syllables.
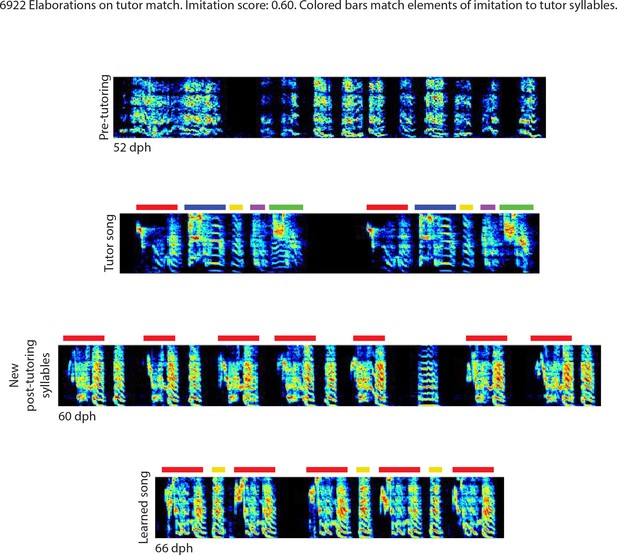
Elaborations on tutor match, bird 6922.
Song spectrograms at different stages of development, and comparison to tutor song spectrogram.Colored bars match elements of the imitation to tutor song syllables.

Tracking HVC sequences as isolated birds rapidly learn a new syllable.
(A) Neurons detected before (left) and after (right) tutoring shown in grayscale (CNMF_E algorithm). Colored contours indicate locations of neurons tracked across five days, from blue to red (B) Sequence in HVC, tracked before and after first tutor exposure, through the development of a new syllable. Each of the five panels shows data from a different day, starting one day before tutoring, through three days after tutoring. (Top) On each recording day, cross-correlation of neurons with the sequence that becomes associated with the new syllable. The sequence was extracted by running seqNMF on neural data recorded two days after tutoring, and selecting the factor associated with the new syllable. Neurons are sorted according to participation in this factor. Significant bins are shown in black, non-significant bins in gray (p=0.05, Bonferroni corrected, compared to circularly-shifted control) (Middle). On each recording day, for example, spectrograms at times when the sequence occurs on each day. Red circles indicate putative newly learned syllables. (Bottom) On each recording day, cross-correlation of sequence with acoustic features (amplitude, entropy, pitch goodness, aperiodicity, mean frequency, pitch, frequency modulation, and amplitude modulation) (C) Same as B for a different example bird. (D) Correlation with song amplitude before (pink) and after (gray) tutoring for all sequences in learner birds extracted data when a new syllable had been learned. (E) Similar to B and C, for a different example bird. Here, sequences are extracted from pre-tutoring data, then tracked forward in time. (F) Song locking (maximum cross-correlation with song amplitude) before and after tutoring for the pre-tutoring sequences that had weaker song locking. (G) Song locking before and after tutoring for the pre-tutoring sequences that started off with stronger song locking.
Tables
Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Software, algorithm | seqNMF | Mackevicius et al., 2019 | github.com/FeeLab/seqNMF | Sequence detection |
| Software, algorithm | CNMF_E | Zhou et al., 2018 | github.com/zhoupc/CNMF_E | Cell extraction |
| Software, algorithm | STAT | Gu et al., 2023 | https://github.com/shijiegu/STAT | Tracking neurons across days |
| Software, algorithm | Chronux | Mitra and Bokil, 2007 | chronux.org/ | Spectrogram computation |
| Software, algorithm | SAP | Tchernichovski et al., 2000 | soundanalysispro.com | Sound analysis |
| Software, algorithm | SI | Mandelblat-Cerf and Fee, 2014 | https://doi.org/10.1371/journal.pone.0096484 | Song imitation |
| Software, algorithm | MATLAB | MathWorks | mathworks.com/products/matlab | Programming language |
| Biological sample (Taeniopygia guttata) | Zebra finches | MIT animal facility | Taeniopygia guttata | |
| Strain, strain background (adeno-associated virus) | AAV9.CAG.GCaMP6f. WPRE.SV40 | Chen et al., 2013 | Addgene viral prep # 100836-AAV9, http://n2t.net/addgene:100836, RRID:Addgene_100836 | |
Commercial assay or kit | Miniature microscope | Inscopix | https://www.inscopix.com/nvista |





