Adaptor linked K63 di-ubiquitin activates Nedd4/Rsp5 E3 ligase
Figures
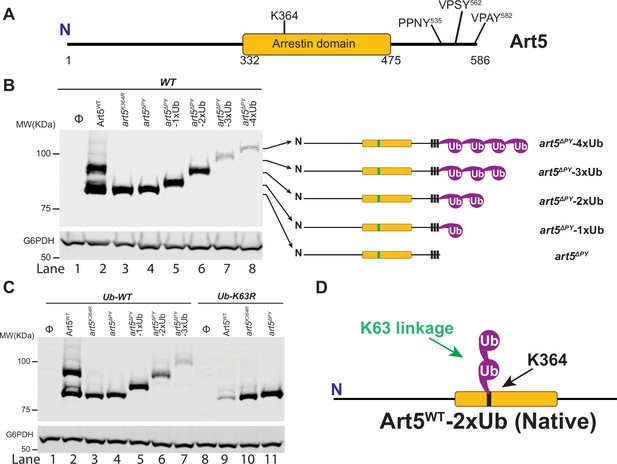
Art5 undergoes K63-linked di-ubiquitination.
(A) Schematic representation of the domain architecture of Art5. (B) A di-ubiquitin (di-Ub) is conjugated at K364 residue of Art5. Western blot analysis of Art5, art5K364R, art5∆PY, art5∆PY-1xUb, art5∆PY-2xUb, art5∆PY-3xUb, and art5∆PY-4xUb in the wild-type (WT) strain. (C) Art5 is di-ubiquitinated in a K63 linkage at the residue K364. Western blot analysis of Art5, art5K364R, art5∆PY in both the Ub-WT and Ub-K63R mutant strains. (D) Model depicting the K63-linked di-ubiquitination of Art5 at the K364 residue. The lanes with Φ symbol indicate negative controls of empty vector. The whole cell lysate protein samples were resolved on 7% SDS-PAGE gels and the blot was probed with FLAG and GAPDH antibodies.
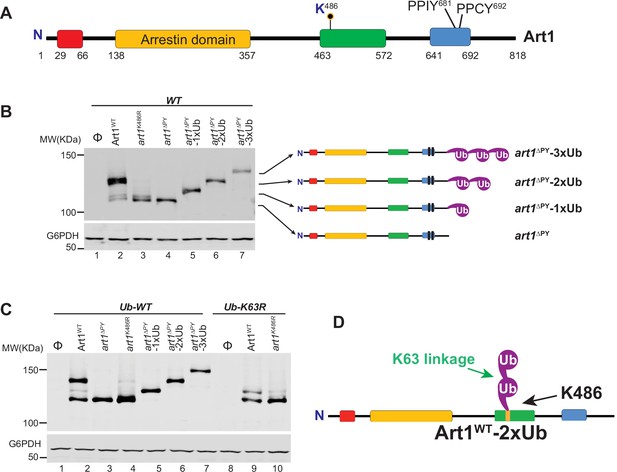
Art1 undergoes K63-linked di-ubiquitination.
(A) Scheme of the Art1 domains. (B) Immunoblot analysis of Art1, art1K486R, art1∆PY, art1∆PY-1xUb, art1∆PY-2xUb, and art1∆PY-3xUb in the wild-type strain. (C) Immunoblot analysis of Art5, art5K364R, art5∆PY in both the Ub-WT and Ub-K63R mutant strains. (D) Art1 is di-ubiquitinated in a K63 linkage at the residue K486. The lanes with Φ symbol indicate negative controls lack of Art1 expression vectors. The protein samples from the whole cell lysates were separated on a 7% SDS-PAGE gel and the blot was probed with FLAG and GAPDH antibodies.
-
Figure 1—figure supplement 1—source data 1
- https://cdn.elifesciences.org/articles/77424/elife-77424-fig1-figsupp1-data1-v2.pdf
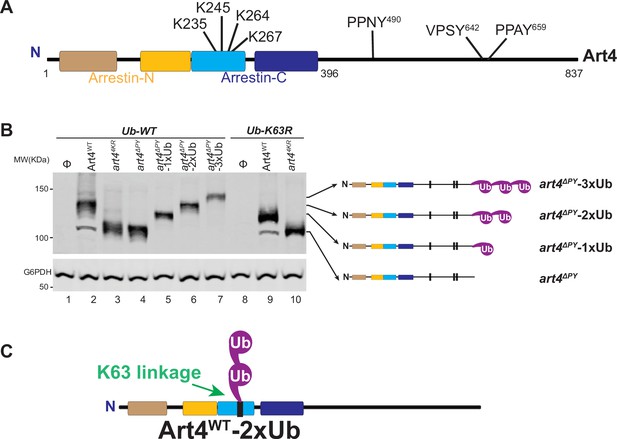
Art4 undergoes K63-linked di-ubiquitination.
(A) Art4 domain architecture. (B) Immunoblot analysis of immunoprecipitated Art4, art4K364R, art4∆PY, as well as in art4∆PY-1xUb, art4∆PY-2xUb, and art4∆PY-3xUb in both the Ub-WT and Ub-K63R mutant strains. (C) Art4 is di-ubiquitinated in a K63 linkage. The lanes with Φ symbol indicate negative controls lack of Art4 expression vectors. The protein samples were resolved on 7% SDS-PAGE gels and the blot was probed with FLAG and GAPDH antibodies.
-
Figure 1—figure supplement 2—source data 1
- https://cdn.elifesciences.org/articles/77424/elife-77424-fig1-figsupp2-data1-v2.pdf
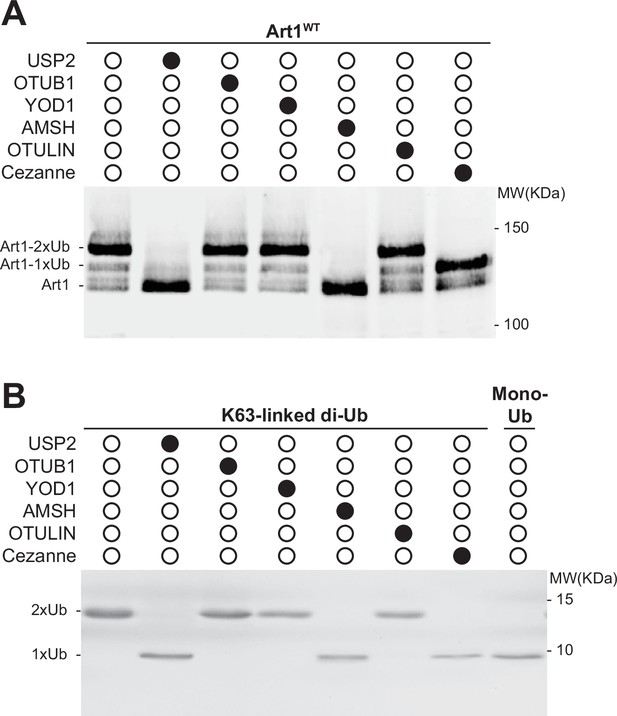
UbiCRest analysis of Art1 and K63-linked di-ubiquitin (di-Ub).
(A) Deubiquitination (DUB) treatment of Art1 by linkage specific DUBs. Art1-HTF was immunoprecipitated from cell line YMB1005 using FLAG-M2 affinity gel and divided into 7 aliquots. Art1 was digested with USP2, OTUB1, YOD1, AMSH, OTULIN, and Cezanne at 37°C for 2 hr, respectively. The resulting products were resolved on an 8% SDS-PAGE gel and the blot was probed with FLAG antibody. (B) DUB treatment of K63-linked di-Ub by linkage specific DUBs. Purified K63-linked di-Ub were incubated at 37°C for 2 hr with USP2, OTUB1, YOD1, AMSH, OTULIN, and Cezanne, respectively. The reactions were quenched by 2 × sample buffer and the products were resolved on a 15% SDS-PAGE gel then stained with Coomassie blue R-250.
-
Figure 1—figure supplement 3—source data 1
- https://cdn.elifesciences.org/articles/77424/elife-77424-fig1-figsupp3-data1-v2.pdf
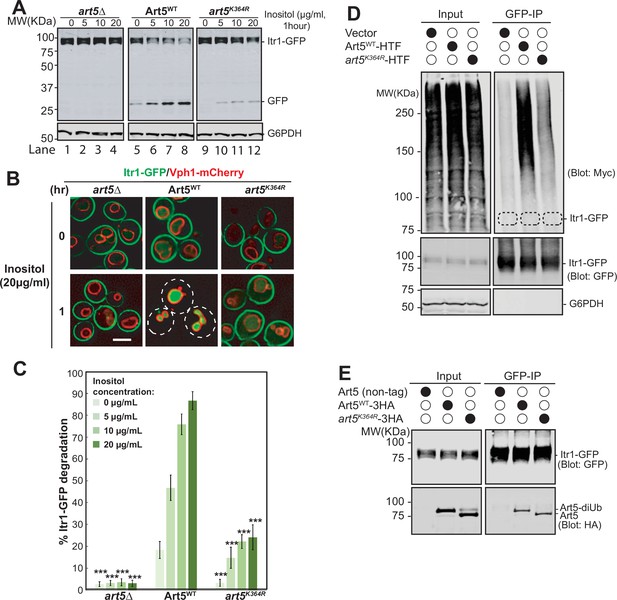
Ubiquitinated Art5 promotes cargo protein Itr1 ubiquitination.
(A) Immunoblot analysis of Itr1-GFP degradation induced with indicated concentration of inositol for 60 min. (B) Fluorescence microscopy of art5∆, Art5WT, or art5K364R cells expressing Itr1-GFP and vacuole membrane marker Vph1-mCherry with or without inducing endocytosis by treating with serial dilution of inositol. Scale bar = 2 µm. (C) Band densities of blots in (A) were quantified and expressed as the mean% Itr1-GFP degradation. Error bars indicate 95% CI, n=3. ***, p<0.005 vs Itr1-GFP degradation in Art5WT at different inositol concentrations. (D) doa4∆pep4∆art5∆ cells expressing Itr1-GFP and Art5WT or art5K364R were grown to mid-log phase in synthetic medium at 30°C. Cells were pretreated with 0.1 µM CuCl2 for 4 hr to induce the Myc-Ub expression before treated with 20 µg/mL of inositol. Cells were collected before and after 15 min of inositol treatment. Itr1-GFP was immunoprecipitated by GFP-Trap nanobody resin. The empty strain (doa4∆pep4∆art5∆) is used as a negative control here. The dashed circles highlight the positions of non-ubiquitinated Itr1-GFP. The whole cell lysate proteins in the left gels represent the loading control and the co-immunoprecipitated protein samples were resolved in right gels. (E) Immunoprecipitation (IP) of Itr1-GFP and blotting for Art5WT or art5K364R. Whole cell lysate and the IP reaction was resolved on 10% SDS-PAGE gels and the blots were probed with both GFP and Myc antibodies.
-
Figure 2—source data 1
- https://cdn.elifesciences.org/articles/77424/elife-77424-fig2-data1-v2.pdf
-
Figure 2—source data 2
- https://cdn.elifesciences.org/articles/77424/elife-77424-fig2-data2-v2.xlsx
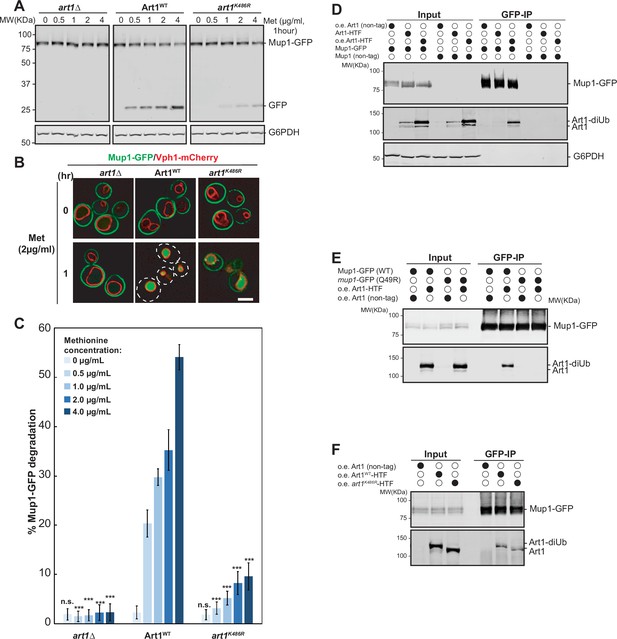
Ubiquitinated Art1 is required for efficient Mup1 ubiquitination.
(A) Mup1 degradation in the yeast mutant art1∆ expressing empty vector, tetO7-Art1WT or tetO7-art1K486R. The protein samples were resolved on a 11% SDS-PAGE gel and the blot was probed with anti-GFP and anti-G6PDH antibodies. (B) Fluorescence microscopy of yeast mutant art1∆ expressing empty vector, tetO7-Art1WT or tetO7-art1K486R with or without methionine treatment. Scale bar = 2 µm. (C) Quantification of full length Mup1-GFP of the blots in (A). Error bars indicate 95% CI, n=3. ***, p<0.005 vs Mup1-GFP degradation in Art1WT at different methionine concentrations. (D) Co-IP(co-immunoprecipitation) of Mup1-GFP and Art1-HTF. (E) Co-IP of Art1-HTF with Mup1WT and mup1Q49R. (F) Co-IP of Mup1-GFP with Art1WT and art1K486R. The IPed protein samples from (D–F) were resolved on 7% SDS-PAGE gels and the blots were probed with GFP and FLAG antibodies.
-
Figure 2—figure supplement 1—source data 1
- https://cdn.elifesciences.org/articles/77424/elife-77424-fig2-figsupp1-data1-v2.pdf
-
Figure 2—figure supplement 1—source data 2
- https://cdn.elifesciences.org/articles/77424/elife-77424-fig2-figsupp1-data2-v2.xlsx
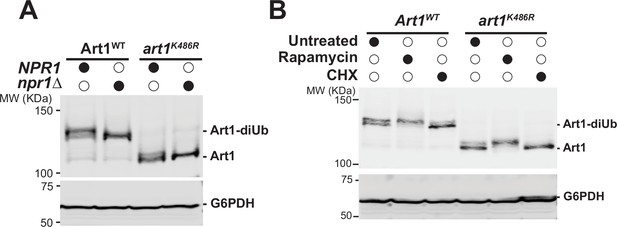
The Npr1-mediated phosphorylation of Art1 is independent of the ubiquitination status of Art1.
(A) Western blot analysis of Art1WT and art1K486R in both NPR1 and npr1∆ conditions. (B) Western blot analysis of Art1WT and art1K486R in NPR1 cells with rapamycin (1 µg/mL) or cycloheximide (50 µg/mL) treatment for 1 hr. The whole cell lysate samples were separated on 7% SDS-PAGE gels and the blots were probed with FLAG and GAPDH antibodies.
-
Figure 2—figure supplement 2—source data 1
- https://cdn.elifesciences.org/articles/77424/elife-77424-fig2-figsupp2-data1-v2.pdf
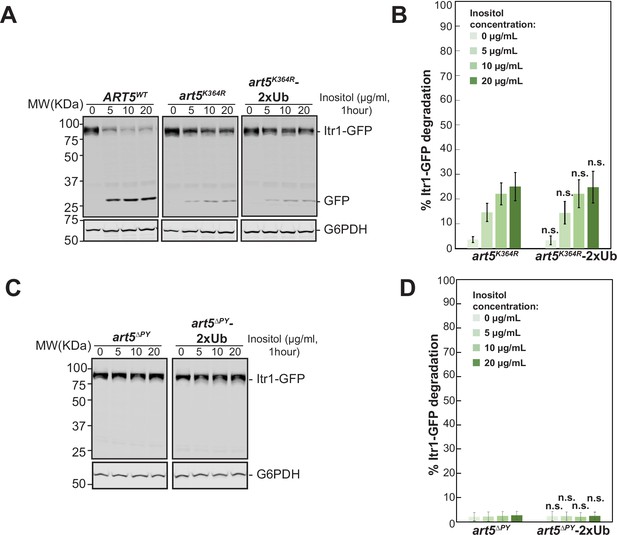
2xubiquitin (2xUb) fusion with art5K364R or art5∆PY mutants do not rescues their cargo protein Itr1 sorting defects.
(A–B) Immunoblot analysis and quantification of Itr1-GFP degradation induced with indicated concentration of inositol for 60 min in the Art5WT, art5K364R, and art5K364R-2xUb conditions. (C–D) Western blot analysis and quantification of Itr1-GFP degradation with exogenous inositol treatment at indicated concentrations for 60 min in the cells expressing art5∆PY and art5∆PY-2xUb. The protein samples were resolved on 11% SDS-PAGE gels and the blots were probed with GFP and GAPDH antibodies. Band densities of blots in (A and C) were quantified and expressed as the mean% Itr1-GFP degradation. Error bars indicate 95% CI, n=3. n.s., not statistically significant vs Itr1-GFP degradation in art5K364R or art5∆PY at different inositol concentrations in (B) and (D), respectively.
-
Figure 2—figure supplement 3—source data 1
- https://cdn.elifesciences.org/articles/77424/elife-77424-fig2-figsupp3-data1-v2.pdf
-
Figure 2—figure supplement 3—source data 2
- https://cdn.elifesciences.org/articles/77424/elife-77424-fig2-figsupp3-data2-v2.xlsx
-
Figure 2—figure supplement 3—source data 3
- https://cdn.elifesciences.org/articles/77424/elife-77424-fig2-figsupp3-data3-v2.xlsx
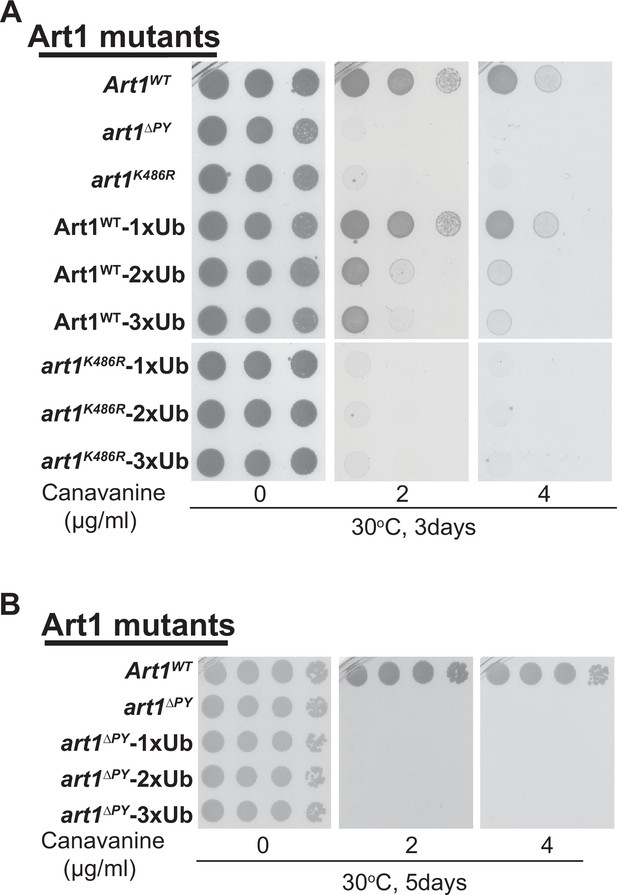
art1K486R or art1∆PY mutant fused with ubitquitin (Ub) variants do not rescues the cargo protein Can1 sorting defects.
(A) Cell growth assay of art1∆ mutant expressing art1∆PY, art1K486R, Art1-1xUb, Art1-2xUb, Art1-3xUb, art1K486R-1xUb, art1K486R-2xUb, or art1K486R-3xUb grown at 30°C for 3 days on synthetic media containing canavanine. (B) Cell growth assay of art1∆ mutant expressing art1∆PY-1xUb, art1∆PY-2xUb, or art1∆PY-3xUb grown in synthetic media with canavanine at 30°C for 5 days.
-
Figure 2—figure supplement 4—source data 1
- https://cdn.elifesciences.org/articles/77424/elife-77424-fig2-figsupp4-data1-v2.pdf
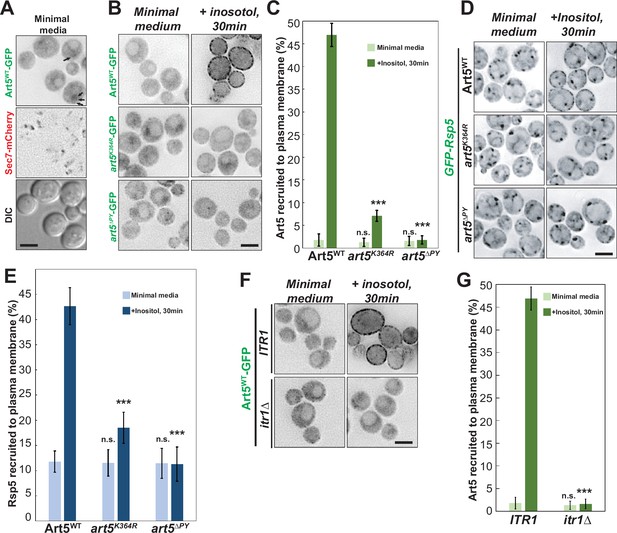
Rsp5 plasma membrane (PM) recruitment is enhanced by Art5 ubiquitination.
(A) Fluorescent microscopy of Art5-GFP with Sec7-mCherry in the WT cells. Black arrows represent occasional cytosolic Art5-GFP dots. (B) Fluorescence microscopy of cells expressing Art5WT, art5K364R, and art5∆PY with C-terminal GFP fusion proteins in minimal media and after inositol treatment (20 µg/mL) for 30 min. (C) Quantification of PM localization of the indicated Art5WT, art5K364R, and art5∆PY mutants in (B). (D) Localization of GFP-Rsp5 in the presence of Art5WT, art5K364R, and art5∆PY mutants before and after inositol treatment (20 µg/mL) for 30 min. (E) Quantification of the PM localized Rsp5 in the Art5WT, art5K364R, and art5∆PY conditions in (D). (F–G) Fluorescence microscopy and quantification analysis of the Art5-GFP recruited to PM (%) in the ITR1 and itr1∆ mutant condition. Error bars indicate 95% CI, n=40 cells. ***, p<0.005; n.s., not significant vs Art5 PM recruitment in Art5WT condition in (C), vs Rsp5 PM recruitment in Art5WT condition in (E), or vs Art5 PM recruitment of ITR1 condition in (F). Scale bar = 2 µm.
-
Figure 3—source data 1
- https://cdn.elifesciences.org/articles/77424/elife-77424-fig3-data1-v2.xlsx
-
Figure 3—source data 2
- https://cdn.elifesciences.org/articles/77424/elife-77424-fig3-data2-v2.xlsx
-
Figure 3—source data 3
- https://cdn.elifesciences.org/articles/77424/elife-77424-fig3-data3-v2.xlsx
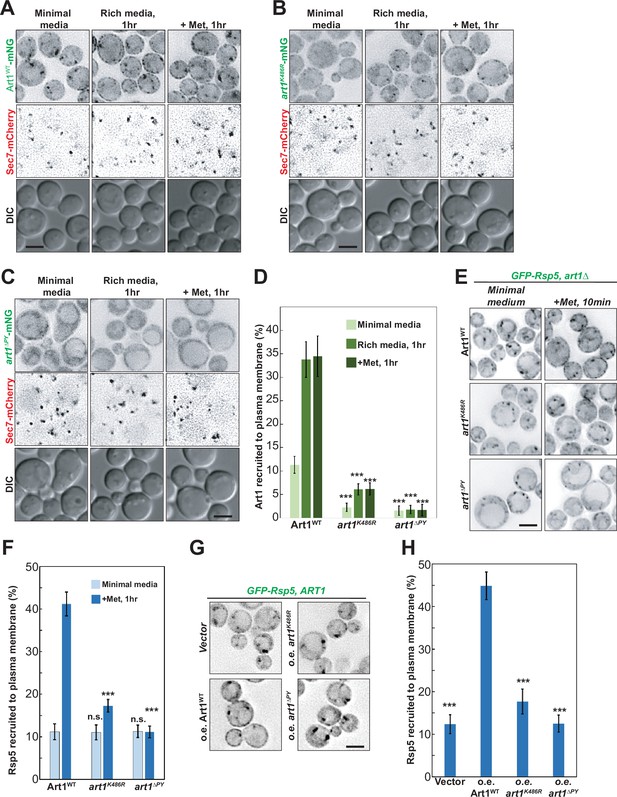
The Art1 di-ubiquitination facilitates Rsp5 plasma membrane (PM) recruitment upon methionine treatment.
(A–C) Fluorescence microscopy of Art1-mNeonGreen (mNG) in wild-type (WT), K486R and PY motif mutants conditions when treated with methionine (20 μg/mL) or shifted from minimal media to rich media for 1 hr. Scale bar = 2 µm. (D) Quantification of Art1 recruited to PM (%) in the experiment of (A–C). (E) Localization of GFP-Rsp5 in art1∆ mutant bearing Art1-WT, art1∆PY, or art1K486R before and after methionine (20 μg/mL) treatment for 10 min. (F) Quantification of PM recruitment of Rsp5 in the experiment (E). (G) Localization of GFP-Rsp5 in WT cells with overexpression of vector control, Art1, art1K486R, or art1∆PY. Error bars indicate 95% CI, n=40 cells. ***, p<0.005; n.s., not statistically significant vs Art1 PM recruitment in (D), vs Rsp5 PM recruitment of Art1 condition in (F), or vs Rsp5 PM recruitment of Art1 overexpression (o.e.) condition in (H), respectively.
-
Figure 3—figure supplement 1—source data 1
- https://cdn.elifesciences.org/articles/77424/elife-77424-fig3-figsupp1-data1-v2.xlsx
-
Figure 3—figure supplement 1—source data 2
- https://cdn.elifesciences.org/articles/77424/elife-77424-fig3-figsupp1-data2-v2.xlsx
-
Figure 3—figure supplement 1—source data 3
- https://cdn.elifesciences.org/articles/77424/elife-77424-fig3-figsupp1-data3-v2.xlsx
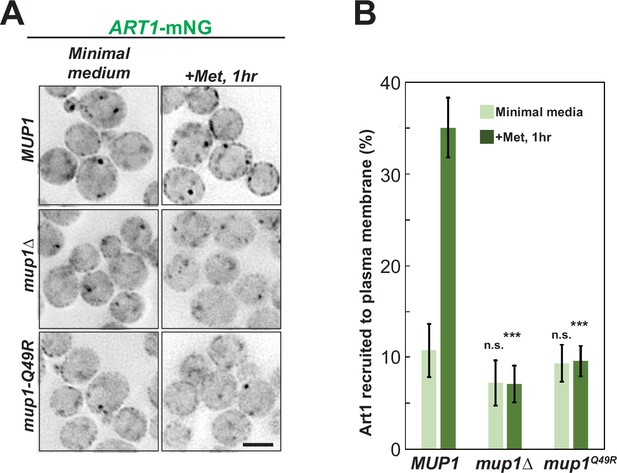
Substrate dependent plasma membrane (PM) recruitment of adaptor protein Art1.
(A) GFP-Rsp5 PM recruitment in the yeast cells expressing MUP1, mup1∆, or mup1-Q49R mutant after treatment of methionine for 1 hr. Scale bar = 2 µm. (B) Quantification of the Rsp5 PM recruitment in the experiment (A). Error bars indicate 95% CI, n=40 cells. ***, p<0.005; n.s., not statistically significant vs Art1 recruitment to plasma membrane in minimal media or methionine treated condition of MUP1 in (A).
-
Figure 3—figure supplement 2—source data 1
- https://cdn.elifesciences.org/articles/77424/elife-77424-fig3-figsupp2-data1-v2.xlsx
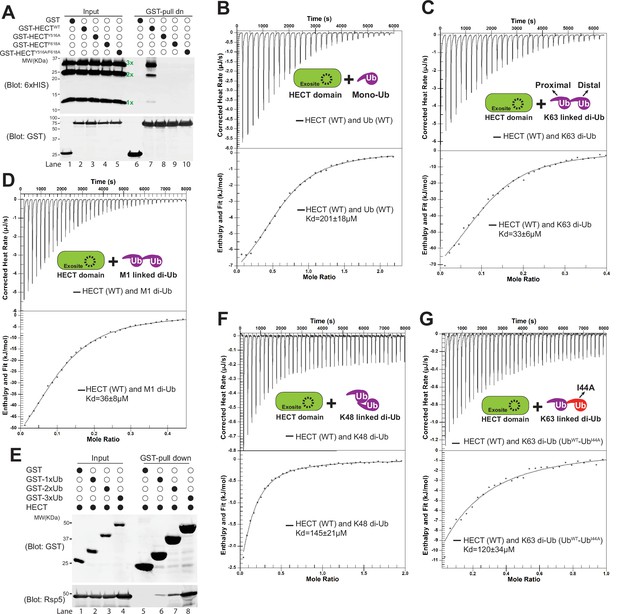
K63-linked di-ubiquitin (di-Ub) binds with Rsp5 HECT domain.
Rsp5 exosite is required for K63-linked di-Ub binding with HECT domain. (A) Glutathione-S-transferase (GST) pull down assay between HECT-wild-type (HECT-WT), Y516A, F618A, or Y516A/F618A mutant and K63-linked Ub ladder. (B) Example isothermal titration calorimetry (ITC) titration curves showing the binding of mono-Ub-WT or I44A mutant to Rsp5 HECT domain. (C) ITC-based measurements of the bindings between K63 di-Ub and Rsp5 HECT domain. (D) The representative ITC curves of showing the binding of M1-linked di-Ub and Rsp5 HECT domain. (E) GST pull down assay between GST only, GST-1xUb, 2xUb, or 3xUb, and Rsp5 HECT domains. (F) Measurement of affinity between K48 di-Ub and Rsp5 HECT domain by ITC. (G) ITC-based measurements showing that the K63 di-Ub with a distal end ubiquitin mutant (I44A) partially disrupts the binding affinity with Rsp5 HECT domain.
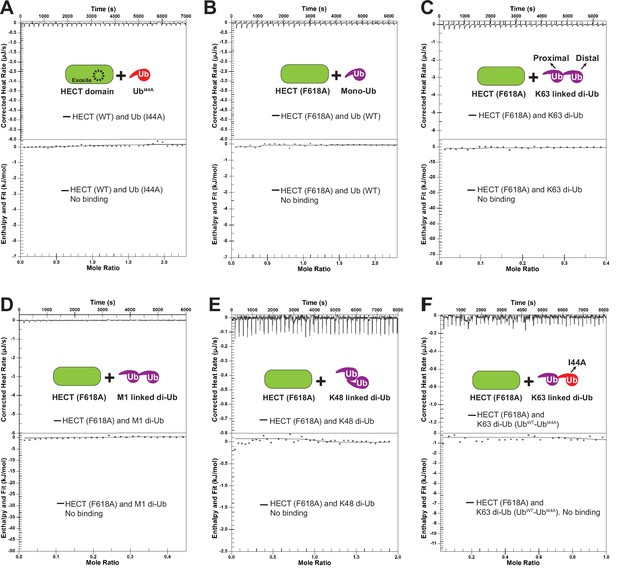
Rsp5 exosite is required for K63-linked di-ubiquitin (di-Ub) to bind with HECT domain.
(A) An isothermal titration calorimetry (ITC)-based measurement of the binding between mono-Ub (I44A) mutant and Rsp5 HECT domain. (B) An ITC-based measurement of the binding between mono-Ub (WT) and Rsp5 HECT (F618A) mutant. (C) An example ITC titration curve showing the binding of K63-linked di-Ub to Rsp5 HECT (F618A) mutant. (D) An example ITC titration curve showing the binding of M1-linked di-Ub to Rsp5 HECT (F618A) mutant. (E) An ITC-based measurement of the binding between K48-linked di-Ub and Rsp5 HECT domain. (F) The representative ITC curve of showing the binding of K63 di-Ub with a distal end mutant Ub (UbWT-UbI44A) and Rsp5 HECT (F618A) mutant.
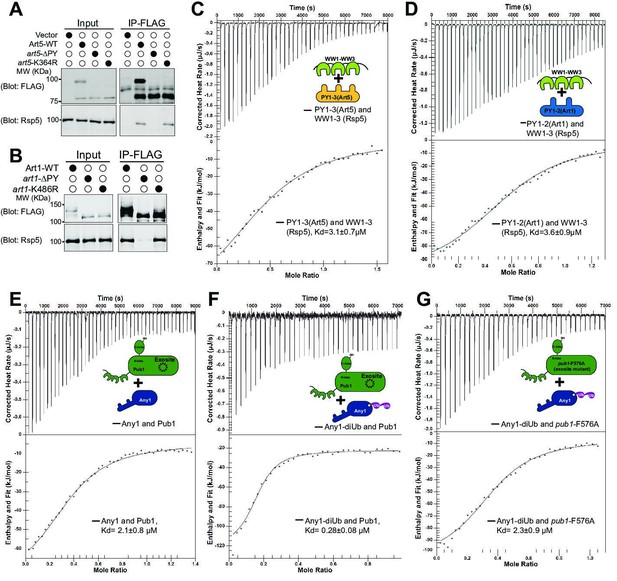
K63-linked di-ubiquitination enhances the interaction between adaptor proteins and Rsp5.
(A–B) Co-IP of Art1 and Art5, WT, KR, and PY motif mutants with Rsp5. (C–D) Isothermal titration calorimetry (ITC) analysis of Art1 or Art5 PY motifs containing domain and Rsp5 WW1-HECT domain. (E) Analysis of binding affinity between Any1 (Art1 ortholog in Schizosaccharomyces pombe) and the Pub1 (Rsp5 orthologue in S. pombe). (F–G) ITC results obtained by titration of Any1 conjugated with K63 di-ubiquitin (di-Ub) into WT or exosite mutant F576A of Pub1.
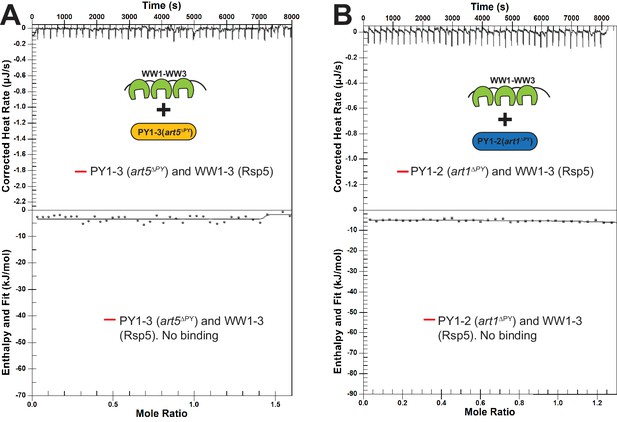
Isothermal titration calorimetry (ITC) analysis between Rsp5 tryptophan-tryptophan (WW) domains and PY motif mutants of Art1 and Art5.
(A–B) A representative ITC measurement of the binding between Art1 or Art5 PY motif mutants and Rsp5 WW1-HECT domain.
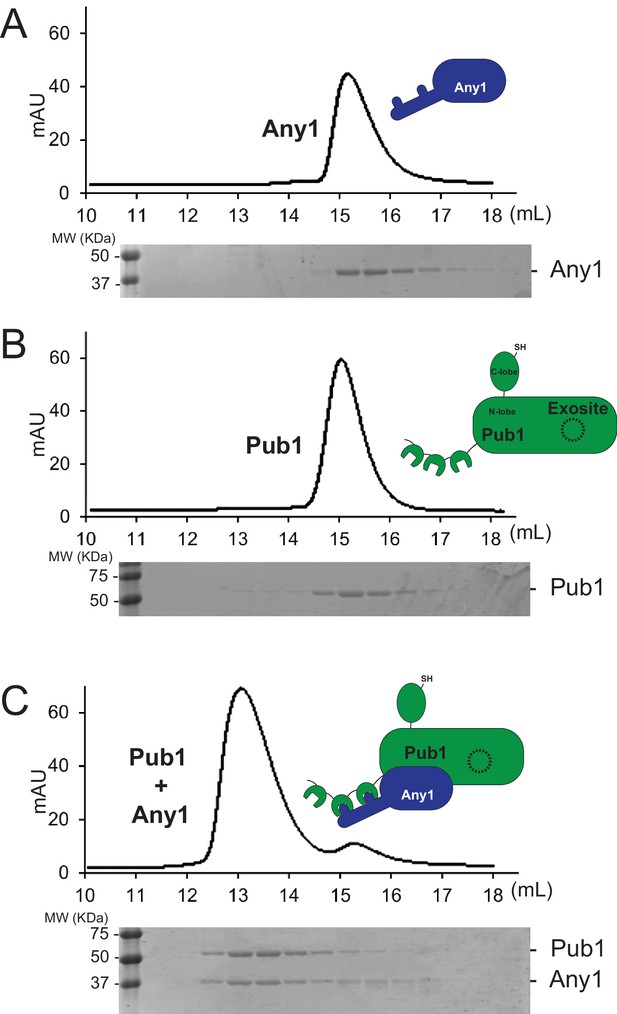
Size exclusion chromatography analysis of the Any1-Pub1 complex.
(A–C) Size exclusion chromatogram profile of purified recombinant protein (top) and the peak fractions visualized by SDS-PAGE followed by Coomassie staining (bottom). (A) Any1; (B) Pub1; (C) Any1 and Pub1.
-
Figure 5—figure supplement 2—source data 1
- https://cdn.elifesciences.org/articles/77424/elife-77424-fig5-figsupp2-data1-v2.pdf
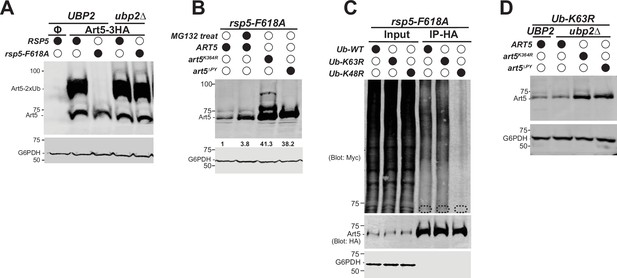
Deubiquitination of Art5 di-ubiquitin (di-Ub) by Ubp2.
(A) Immunoblot analysis of Art5-3HA in the indicated yeast strains: RSP5/UBP2, rsp5-F618A/UBP2, RSP5/ubp2∆, and rsp5-F618A/ubp2∆. RSP5/UBP2 cells bearing empty vector is used as a negative control in the first lane. The lanes with Φ symbol indicate negative controls lack of Art5 expression vectors. (B) Immunoblot analysis of Art5-3HA in rsp5-F618A mutant treated with DMSO or MG132 (25 µg/mL) for 60 min. art5K364R-3HA and art5∆PY-3HA are shown as controls. (C) Ub blot of rsp5-F618A yeast cells carrying Art5-3HA, as well as WT, K63R, or K48R Myc-ubiquitin expression vector. Cells were treated with MG132 (25 µg/mL) for 2 hr. Samples were immunoprecipitated (IPed) using anti-HA resin and analyzed by immunoblot. The dashed circles highlight the positions of non-ubiquitinated Art5-3HA. (D) Immunoblot analysis of Art5-3HA, art5K364R-3HA, and art5∆PY-3HA expressed in Ub-K63R and Ub-K63R/ubp2∆.
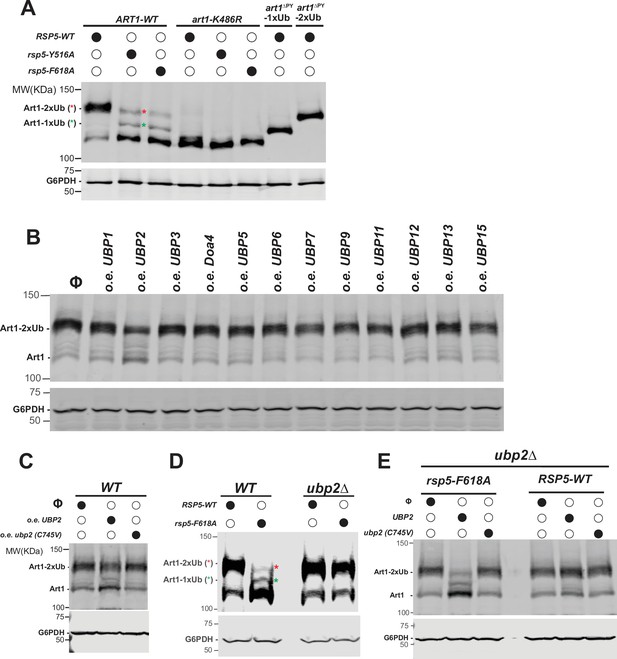
Deubiquitination (DUB) of K63 di-ubiquitin (di-Ub) of adaptor protein Art1 by Ubp2.
(A) Western blot analysis of Art1WT, art1K486R, art1∆PY-1xUb, and art1∆PY-2xUb mutant in the indicated yeast strains: RSP5-WT, rsp5-Y516A, and rsp5-F618A. (B) Western blot analysis of Art1-HTF with overexpression of yeast DUBs proteins individually. The lanes with Φ symbol indicate negative controls lack of Art1 expression vector. (C) Western blot analysis of Art1-HTF in the ubp2∆ mutant bearing an empty vector, or with overexpression of UBP2 or ubp2 (C745V) mutant. (D) Western blot analysis of Art1-HTF in yeast strains: RSP5 (WT), rsp5-F618A, ubp2∆ and rsp5-F618A/ubp2∆. (E) Western blot of Art1-HTF in ubp2∆ and rsp5-F618A/ubp2∆ yeast strains bearing an empty vector, UBP2 or ubp2 (C745V) mutant. The whole cell lysate proteins samples were separated on 7% SDS-PAGE gels and the blots were probed with FLAG and GAPDH antibodies.
-
Figure 6—figure supplement 1—source data 1
- https://cdn.elifesciences.org/articles/77424/elife-77424-fig6-figsupp1-data1-v2.pdf
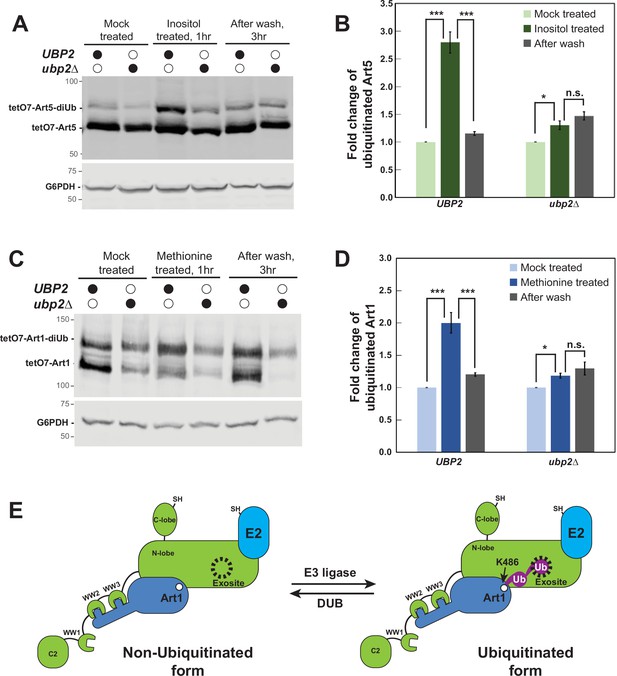
The Ubp2-mediated deubiquitination (DUB) recycle of the ubiquitinated Art1 and Art5.
(A) Western blot analysis of tetO7-Art5-HTF in WT and ubp2∆ mutant with mock treatment or inositol treatment (1 hr). After inositol treatment, cells were washed and grown in fresh media for 3 hr. (B) Quantification for the fold change of ubiquitinated Art5 in UBP2 and ubp2∆ mutant. Fold change = (ratio of ubiquitinated Art5 among total Art5 in ‘inositol treated’ or ‘after wash’ condition)/(ratio of ubiquitinated Art5 among total Art5 in ‘mock treated’ condition), n=3. (C) Western blot analysis of tetO7-Art1-HTF in WT and ubp2∆ mutant with or without methionine treatment (1 hr). The methionine treated cells were then washed and grown in fresh media for 3 hr. (D) Quantification for the fold change of ubiquitinated Art1 in UBP2 and ubp2∆ mutant. Fold change = (ratio of ubiquitinated Art1 among total Art1 in ‘methionine treated’ or ‘after wash’ condition)/(ratio of ubiquitinated Art1 among total Art1 in ‘mock treated’ condition). The protein samples in (A) and (C) were resolved on 7% SDS-PAGE gels and the blots were probed with FLAG and GAPDH antibodies. The significance in (B) and (D) was determined by two-tail t-test, α=0.05 (Bonferroni correction), n=3. n.s., not significant; *, p<0.05; ***, p<0.001. (E) Cartoon model depicting the Art1 is ubiquitinated by E3 ligase upon environmental cue then deubiquitinated by Ubp2. Non-ubiquitinated form of Art1 is ubiquitinated at K486 residue and engaged by Rsp5 for activation. This activated form of Art1 is then deubiquitinated by Ubp2 and the non-ubiquitinated form of Art1 is disengaged from Rsp5.
-
Figure 6—figure supplement 2—source data 1
- https://cdn.elifesciences.org/articles/77424/elife-77424-fig6-figsupp2-data1-v2.pdf
-
Figure 6—figure supplement 2—source data 2
- https://cdn.elifesciences.org/articles/77424/elife-77424-fig6-figsupp2-data2-v2.xlsx
-
Figure 6—figure supplement 2—source data 3
- https://cdn.elifesciences.org/articles/77424/elife-77424-fig6-figsupp2-data3-v2.xlsx
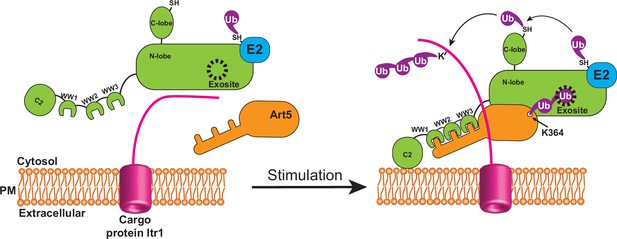
Attachment of K63 di-ubiquitin (di-Ub) to adaptor protein Art5 enables efficient membrane recruitment of Rsp5.
This model depicts that the adaptor protein Art5 forms binary protein complex with E3 ligase Rsp5 via the interaction between the PY motif and tryptophan-tryptophan (WW) domain, following by the stimulation of cargo protein Itr1. The efficient membrane recruitment of Rsp5 is activated by this binding scaffold when adaptor protein Art5 linked with K63 di-Ub is fully engaged into the Rsp5 exosite.
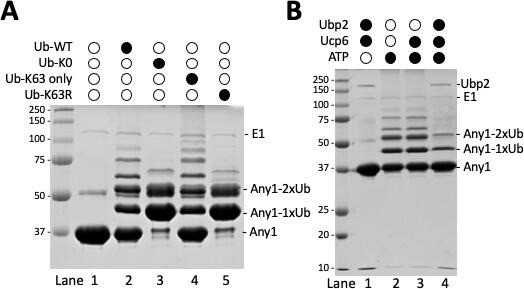
Any1 is ubiquitinated in K63 linkage using in-vitro assay.
(A) E1 (Uba1), E2 (UbcH5), E3 (Pub1), substrate (Any1) and ubiquitin variants were mixed together in the 1x PBDM buffer. (B) E1 (Uba1), E2 (UbcH5), E3 (Pub1), substrate (Any1), WT-Ub, Ubp2 and the adaptor protein Ucp6 in different combination were mixed together in the 1x PBDM buffer. Thereactions were incubated in the room temperature for 30minutes then quenched by 2x sample buffer. The resulting products were solved on 10% SDS-PAGE gel then stained with Coomassie Blue R-250.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Anti-GFP, B2 (mouse monoclonal) | Santa Cruz | sc-9996 | WB (1:2000) |
| Antibody | Anti-Myc (mouse monoclonal) | Santa Cruz | sc-40 | WB (1:2000) |
| Antibody | Anti-GFP (rabbit polyclonal) | Torrey Pines Biolabs | TP401 | WB (1:10,000) |
| Antibody | Anti-HA 12CA5 (mouse monoclonal) | Sigma-Aldrich | 11583816001 | WB (1:5000) |
| Antibody | Anti-FLAG M2 (mouse monoclonal) | Sigma-Aldrich | F1804 | WB (1:5000) |
| Antibody | Anti-G6PDH (rabbit, polyclonal) | Sigma-Aldrich | SAB2100871 | WB (1:30,000) |
| Antibody | IRDye 800CW (Goat anti-Mouse, polyclonal) | LI-COR | 926–32210 | WB (1:10,000) |
| Antibody | IRDye 800CW (Goat anti-Rabbit, polyclonal) | LI-COR | 926–32211 | WB (1:10,000) |
| Antibody | IRDye 680LT (Goat anti-Rabbit, polyclonal) | LI-COR | 926–68021 | WB (1:10,000) |
| Antibody | IRDye 680LT (Goat anti-Mouse, polyclonal) | LI-COR | 925–68070 | WB (1:10,000) |
| Cell line (E. coli) | Competent cells of DH5α | ThermoFisher | 18258012 | Super competent cells. |
| Cell line (E. coli) | Competent cells of BL21, rosetta | Sigma-Aldrich | CMC0016 | Super competent cells. |
| Software, algorithm | ImageJ | NIH | Version: 1.53 n | https://imagej.nih.gov/ij/ |
| Software, algorithm | NanoAnalyze | TA Instruments | Version: 3.12.0 | https://www.tainstruments.com |
| Software, algorithm | SnapGene | GSL Biotech | Version: 6.0.2 | https://www.snapgene.com |
| Other | cOmplete Protease Inhibitor Cocktail | Roche | 11697498001 | Protease Inhibitors for protein purification. |
Additional files
-
Supplementary file 1
All yeast strains and plasmids used in this study.
- https://cdn.elifesciences.org/articles/77424/elife-77424-supp1-v2.docx
-
Supplementary file 2
Coomassie blue stained SDS-PAGE gels showing the purity of proteins used in the ITC experiment.
- https://cdn.elifesciences.org/articles/77424/elife-77424-supp2-v2.pdf
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/77424/elife-77424-transrepform1-v2.pdf
-
Source data 1
Source data combined: the uncropped gels or blots for all the figures.
- https://cdn.elifesciences.org/articles/77424/elife-77424-data1-v2.zip
-
Source data 2
Source data combined: all the uncropped gels in Supplementary file 2.
- https://cdn.elifesciences.org/articles/77424/elife-77424-data2-v2.pdf





