BDNF/TrkB signaling endosomes in axons coordinate CREB/mTOR activation and protein synthesis in the cell body to induce dendritic growth in cortical neurons
Figures
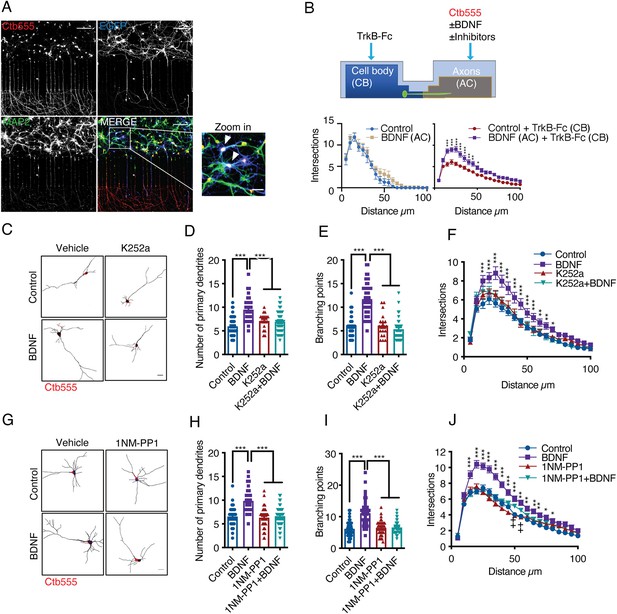
Addition of BDNF to axons promotes dendritic branching in a TrkB-dependent manner.
(A) Representative images of compartmentalized microfluidic chambers (400 µm long and 15 µm wide microgrooves) after addition of BDNF/Ctb-555. Ctb-555 (upper left panel), EGFP (upper right panel), and MAP2 (lower left panel) were visualized by confocal microscopy (Scale bar = 50 µm). The lower right panel is a merged image of the three signals. The white arrows in the zoomed-in panel indicate neurons costained with EGFP/MAP2/Ctb-555 (Scale bar = 20 µm), that is, neurons that project axons through the microgrooves. (B) Upper panel, experimental design used to study retrograde BDNF signaling. DIV six neurons were transfected with a plasmid expressing EGFP. TrkB-Fc (100 ng/mL) was applied to the CB. BDNF (50 ng/mL) in addition to fluorescently labeled Ctb-555 was added to the AC in the presence or absence of different inhibitors. BDNF and TrkB-Fc were administered for 48 hr to evaluate dendritic arborization. Finally, the neurons were fixed, and immunofluorescence for MAP2 was performed. Lower panels, graphs showing Sholl analysis of dendritic arborization of compartmentalized rat cortical neurons stained with Ctb-555. Left graph, BDNF (50 ng/mL) was applied or not to the AC for 48 hr (n=10–15 neurons, from two independent experiment). Right graph, TrkB-Fc (100 ng/mL) was added to the CB, and BDNF (50 ng/mL) was applied or not to the AC for 48 hr (n=37 neurons, from two independent experiment). The results are expressed as the mean ± SEM. ***p<0.001. The Sholl analysis data were analyzed by two-way ANOVA followed by Bonferroni’s multiple comparisons post-hoc test. (C) Representative images of the CB (red indicates Ctb-555) of compartmentalized rat cortical neurons whose axons were treated with DMSO (control), K252a (200 nM), BDNF (50 ng/mL) or BDNF following preincubation with K252a (D–F). Quantification of primary dendrites (D) and branching points (E) and Sholl analysis (F) for neurons labeled with EGFP/MAP2/Ctb-555 under each experimental condition. n=40–45 neurons from three independent experiments. (G) Representative images of the CB (red indicates Ctb-555) of compartmentalized mouse TrkBF616A cortical neurons whose axons were treated with DMSO (control), 1NM-PP1 (1 µM), BDNF (50 ng/mL), or BDNF following preincubation with 1NM-PP1. (H–J) Quantification of primary dendrites (H) and branching points (I) and Sholl analysis (J) for neurons labeled with EGFP/MAP2/Ctb-555 under the four different experimental conditions described in G. Scale bar = 20 µm. n=40–45 neurons from three independent experiments. *p<0.05, **p<0.01, ***p<0.001,++p < 0.01, the 1NM-PP1 group vs. the 1NM-PP1+BDNF group. The data were analyzed by one-way ANOVA followed by Bonferroni’s multiple comparisons post-hoc test (D, E, H and I). Two-way ANOVA followed by Bonferroni’s multiple comparisons post-hoc test (F and J). The results are expressed as the mean ± SEM.
-
Figure 1—source data 1
Addition of BDNF to axons promotes dendritic branching in a TrkB-dependent manner.
- https://cdn.elifesciences.org/articles/77455/elife-77455-fig1-data1-v1.xlsx
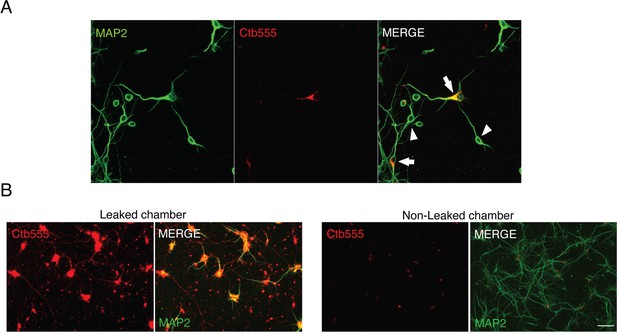
Experimental design used to study retrograde BDNF signaling in compartmentalized cortical neurons.
(A) Representative images of compartmentalized neurons whose axons were incubated with Ctb-555 for 48 hr. Neurons were immunostained for MAP2 (green). The arrows indicate neurons that projected axons into the AC. The arrowheads indicate neurons with axons that do not project to the AC and therefore are not labeled with Ctb-555. (B) Representative images of a microfluidic chamber that were not well compartmentalized (leaking, left panel) and one in which compartmentalization was achieved (no leaking, right panel). Ctb-555 is visualized in red, and MAP2 is visualized in green. Scale bar = 50 μm.
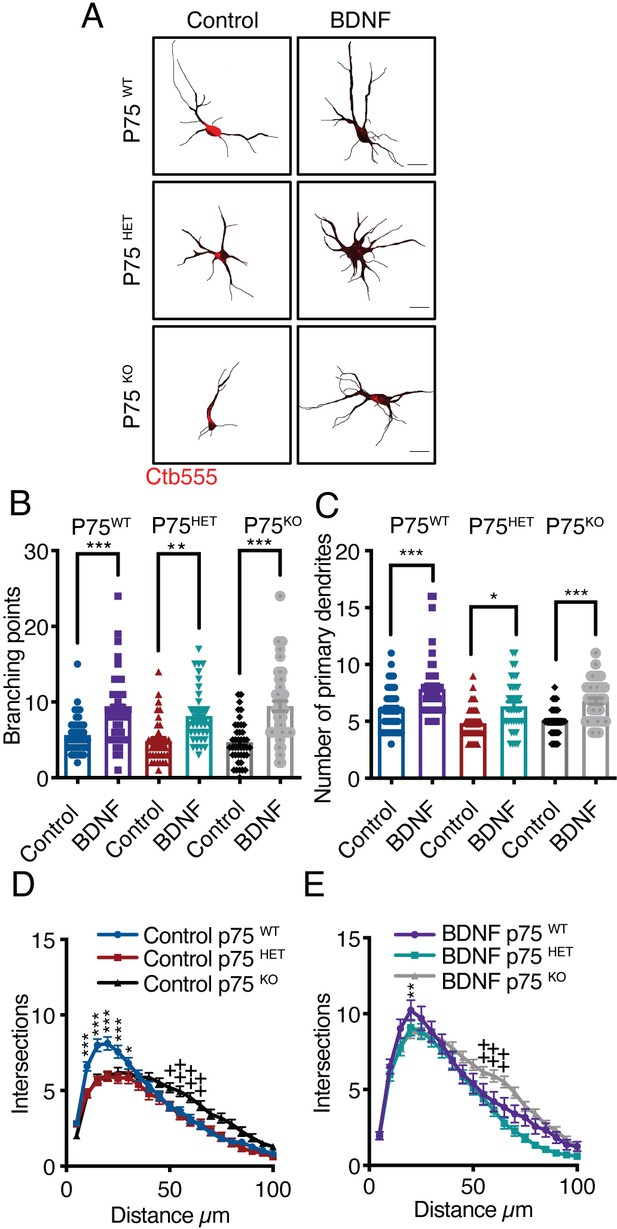
Axonal BDNF increases dendritic arborization in a p75-independent manner.
(A) Representative images of the CB (red indicates Ctb-555) of compartmentalized p75WT, p75HET, or p75KO mouse cortical neurons whose axons were treated with BDNF (50 ng/mL). Quantification of branching points (B) and primary dendrites (C) in MAP2/Ctb-555-labeled neurons from mice of the different genotypes described in A. (D) Sholl analysis of cultured neurons from mice of each genotype following application of TrkB-Fc to the CB (nonstimulated). (E) Sholl analysis of cultured neurons from mice of each genotype following addition of TrkB-Fc to the CB and BDNF to the AC. n=40–50 neurons from two independent experiments (3 mice per experiment). Scale bar = 10 µm *p<0.05, **p<0.01, ***p<0.001,++p < 0.01, neurons from p75KO mice vs. neurons from p75WT mice. The data were analyzed by one-way ANOVA followed by Bonferroni’s multiple comparisons post-hoc test (B and C). Two-way ANOVA followed by Bonferroni’s multiple comparisons post-hoc test (D and E). The results are expressed as the mean ± SEM.
-
Figure 2—source data 1
Axonal BDNF increases dendritic arborization in a p75-independent manner.
- https://cdn.elifesciences.org/articles/77455/elife-77455-fig2-data1-v1.xlsx
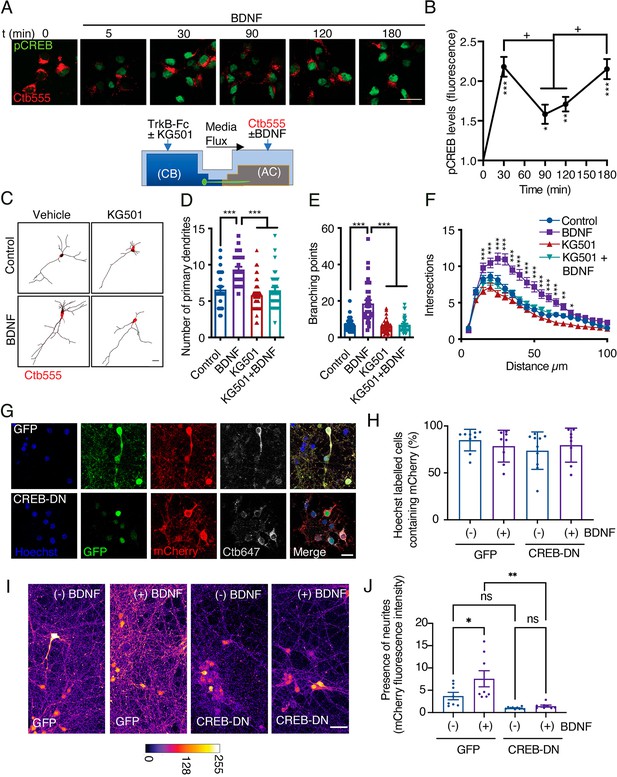
CREB activity is required for long-distance axonal BDNF-induced dendritic arborization.
(A) Lower panel, schematic representation of the protocol used to evaluate CREB phosphorylation induced by axonal BDNF and the requirement of CREB for dendritic arborization induced by BDNF using the drug KG501(panels C, D, E and F). Upper panel, representative figures of nuclear activated CREB (pCREB, S133) immunostaining in control neurons and neurons whose axons were stimulated with BDNF for different times. Ctb-555 was added to the AC of compartmentalized neurons overnight, and after 90 min of serum deprivation, BDNF (50 ng/mL) was added to the AC for 5, 30, 90, 120, or 180 min. pCREB is shown in green and Ctb-555 is shown in red. Scale bar = 20 µm. (B) Quantification (arbitrary units, A.U.) of pCREB fluorescence in the nucleus in the presence and absence of BDNF. Only neurons positive for Ctb-555 were included in the analysis. n=30–40 neurons from two independent experiments. *p<0.05, **p<0.01, ***p<0.01 vs. the control group;+p < 0.05 vs. the 90 min and 120 min BDNF treatment groups. The results are expressed as the mean ± SEM. The data were analyzed by one-way ANOVA followed by Bonferroni’s multiple comparisons post-hoc test. (C) Representative images of the CB (red indicates Ctb-555) of compartmentalized neurons whose cell bodies were treated with DMSO (control), KG501, BDNF, or BDNF in the presence of KG501. TrkB-Fc (100 ng/mL) and KG501 (10 µM) were applied to the CB for 1 hr Then, BDNF (50 ng/mL) was added to the AC for 48 hr in the presence of Ctb-555, and the CB was treated with or without KG501. Finally, the neurons were fixed, and immunofluorescence for MAP2 was performed. Scale bar = 20 µm. (D–F) Quantification of primary dendrites (D) and branching points (E) and Sholl analysis (F) for neurons labeled with EGFP/MAP2/Ctb-555 under each experimental condition. n=30–35 neurons from three independent experiments. *p<0.05, **p<0.01, ***p<0.001. The data were analyzed by one-way ANOVA followed by Bonferroni’s multiple comparisons post-hoc test (D and E). Two-way ANOVA followed by Bonferroni’s multiple comparisons post-hoc test was used to analyze the Sholl analysis data (F). The results are expressed as the mean ± SEM. (G) Representative images of compartmentalized cortical neurons transduced at DIV 4 with AAV1 expressing EGFP/mCherry (upper figures) or CREB-DN-EGFP/mCherry (lower figures). At DIV 7, neurons were treated with Ctb-647 in the AC. After 16–24 hr, neurons were fixed and mounted on Mowiol containing Hoechst. The level of cotransduced cells was about 75%. Scale bar = 20 µm. (H) Quantification of the percentage of cells containing Hoechst that also contained mCherry in the different treatments. EGFP(GFP)/mCherry or CREB-DN-EGFP(CREB-DN)/mCherry transduced neurons were treated (+) or not (-) with BDNF for 4 days since DIV 4 in the AC. Then, the percentage of cells containing Hoechst that also contained mCherry was calculated. Non-significant different were found between groups. The data were analyzed by two-way ANOVA followed by Turkey’s multiple comparisons test. (I) Representative images of compartmentalized cortical neurons treated as explained in G and H. The fluorescence associated with mCherry is observed using the fire LUT from ImageJ. Scale bar = 20 µm. (J) Quantification of mCherry-associated fluorescence in neurites standardized by the mean value of the group of chambers transduced with CREB-DN/mCherry and non-treated with BDNF (-). n=8–9 chambers from three independent experiments. *p<0.05, **p<0.01. The data were analyzed by two-way ANOVA followed by Turkey’s multiple comparisons test. The results are expressed as the mean ± SEM.
-
Figure 3—source data 1
CREB activity is required for long-distance axonal BDNF-induced dendritic arborization.
- https://cdn.elifesciences.org/articles/77455/elife-77455-fig3-data1-v1.xlsx
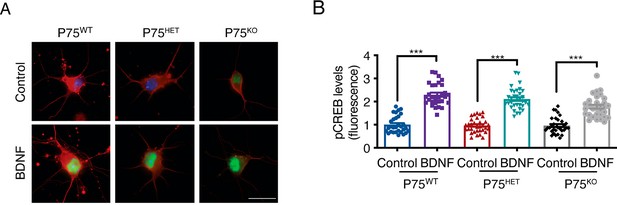
Evaluation of CREB activation in different p75 strains.
DIV seven cortical neurons from p75WT, p75HET, or p75KO mice were treated with BDNF (50 ng/mL) for 30 min, washed and fixed for immunostaining for pCREB (S133), as indicated in the Materials and methods section. (A) Representative image of pCREB (green) and βIII-tubulin (red) in BDNF-stimulated neurons from mice of each p75 genotype. Scale bar = 20 µm. (B) Quantification of the pCREB fluorescence intensity in the nuclei of neurons derived from mice of the three different p75 genotypes treated with or without BDNF. n=30 neurons from two independent experiments. ***p<0.01. The data were analyzed by one-way ANOVA followed by Bonferroni’s multiple comparisons post-hoc test. The results are expressed as the mean ± SEM.
-
Figure 3—figure supplement 1—source data 1
Evaluation of CREB activation in different p75 strains.
Graph B’s raw data.
- https://cdn.elifesciences.org/articles/77455/elife-77455-fig3-figsupp1-data1-v1.xlsx
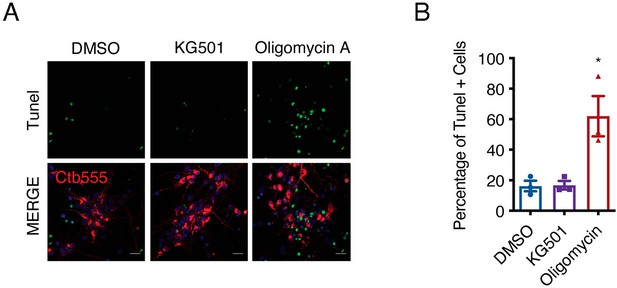
Evaluation of neuronal survival after KG501 treatment.
(A) To control cell death induced by KG501, we performed the experiments as described in Figure 3C–F and visualization of DNA fragmentation in fixed cells by TUNEL staining (green) was performed. Only neurons labeled with Ctb-555 were analyzed. The neurons were treated with oligomycin (10 μM) as a positive control as indicated in the methods section. (B) Quantification of TUNEL-positive cells. Scale bar = 20 μm. n=2 chambers per condition from three independent experiments. *p=0.05. The data were analyzed by one-way ANOVA followed by Bonferroni’s multiple comparisons post-hoc test. The results are expressed as the mean ± SEM.
-
Figure 3—figure supplement 2—source data 1
Evaluation of neuronal survival after KG501 treatment.
Graph B´s raw data.
- https://cdn.elifesciences.org/articles/77455/elife-77455-fig3-figsupp2-data1-v1.xlsx
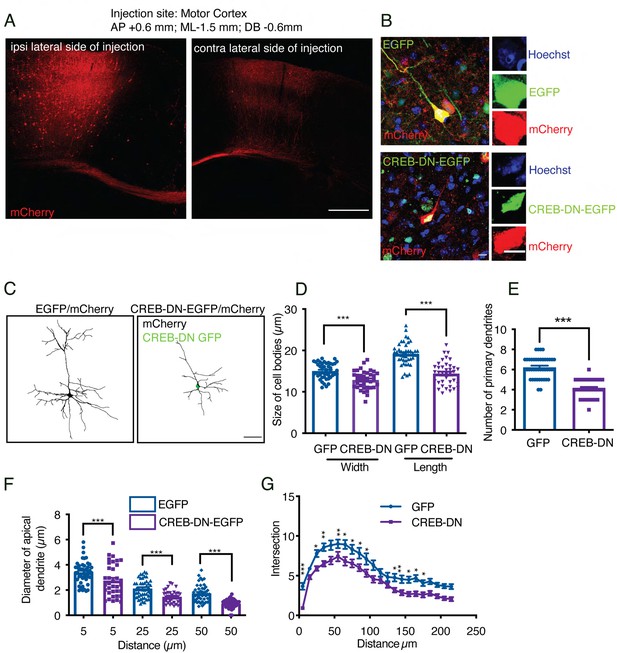
Expression of a dominant-negative mutant of CREB (CREB-DN-EGFP) in the pyramidal neurons of the sensory-motor cortex of mouse brain induced dendritic retraction and shrinkage of neuronal body size.
(A) Representative image of a mouse brain injected unilaterally with AAV1 into layer II/III of the motor cortex. Scale bar = 500 µm. (B) Representative image of neurons expressing EGFP/mCherry or CREB-DN-EGFP/mCherry. Right panel, Hoechst staining was used to visualize nuclear morphology. (C) Representative pictures of pyramidal neurons from cortical layers II-III of the sensory-motor cortex cotransduced with AAV1 expressing CREB-DN-EGFP or EGFP and mCherry. mCherry was amplified by immunostaining for the fluorescent protein. Scale bar = 50 µm. Neurons expressing CREB-DN-EGFP showed reductions in the size of cell bodies (D), the number of primary dendrites (E), the apical dendrite diameter (F), and branching, as measured by Sholl analysis (G). The data were analyzed by one-way ANOVA followed by Bonferroni’s multiple comparisons post-hoc test (D–G). Two-way ANOVA followed by Bonferroni’s multiple comparisons post-hoc test was used to analyze the Sholl analysis data (K). n=35–41 neurons per experimental group derived from five different mice. The results are expressed as the mean ± SEM.
-
Figure 3—figure supplement 3—source data 1
Expression of a dominant-negative mutant of CREB (CREB-DN-EGFP) in the pyramidal neurons of the sensory-motor cortex of mouse brain induced dendritic retraction and shrinkage of neuronal body size.
Graph D-G´s raw data.
- https://cdn.elifesciences.org/articles/77455/elife-77455-fig3-figsupp3-data1-v1.xlsx
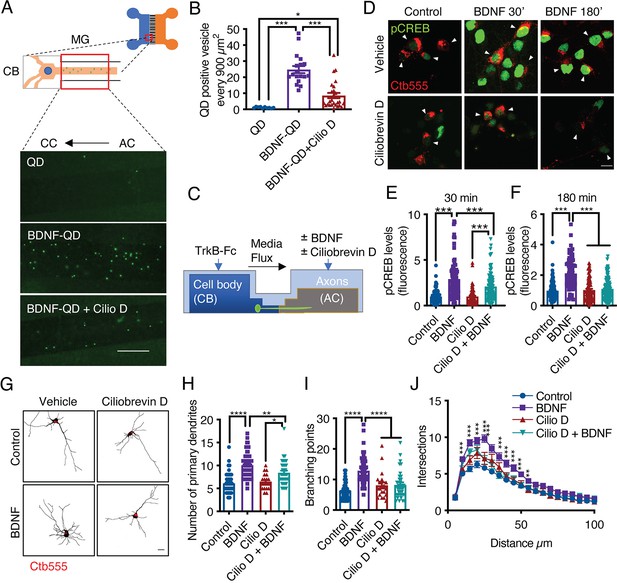
Dynein activity is required for phosphorylation of CREB and dendritic arborization induced by BDNF.
(A) Schematic representation of the protocol used to evaluate the retrograde transport of BDNF. Biotinylated BDNF conjugated to streptavidin QD-655 (2 nM) was added to AC for 4 hr in the presence or absence of CilioD (20 µM). Then, after washing, the neurons were fixed, and the accumulation of BDNF-QDs in the proximal compartment of the microgrooves (MG) was evaluated. Unconjugated QDs were used as controls. Representative images of BDNF-QDs (green) in the proximal region of the MG under each condition. Scale bar = 10 µm (B) Quantification of QD-positive vesicles in every 900 µm2 area of the proximal region of MG under each condition. N=36 MG from three independent experiments were analyzed. (C) Experimental design used to study dynein-dependent axonal BDNF signaling in compartmentalized cortical neurons. DIV five cortical neurons were retrogradely labeled with Ctb-555 (red) overnight. The next day, the culture medium was replaced with serum-free medium for 90 min, and CilioD was applied to the AC. Then, BDNF (50 ng/mL) was added to the AC for 30 or 180 min in the presence or absence of CilioD. Finally, the cultures were fixed, and pCREB (S133) was visualized by immunofluorescence. (D) Representative images of nuclear pCREB in neurons. Scale bar = 10 µm. (E–F) Quantification of (arbitrary units, A.U.) pCREB immunostaining in the nuclei of neurons labeled with Ctb-555 and stimulated with BDNF for 30 min (E) or 180 min (F). n=83–111 neurons from three independent experiments. (G) DIV six cortical neurons were transfected with EGFP, TrkB-Fc (100 ng/mL) was applied to the CB, and Ctb-555 and BDNF (50 ng/mL) with or without CilioD (20 µM) were added to the AC for 48 hr. Finally, the neurons were fixed, and immunofluorescence for MAP2 was performed. Representative images of the morphology of compartmentalized neurons that were labeled with Ctb-555 (red) and whose axons were treated with DMSO (control), CilioD, BDNF, or BDNF following pretreatment with CilioD for 48 hr. Scale bar = 20 µm. (H–J) Quantification of primary dendrites (H) and branching points (I) and Sholl analysis (J) for neurons labeled with EGFP/MAP2/Ctb-555 under each experimental condition described in G. n=34–65 neurons from three independent experiments. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. The data were analyzed by one-way ANOVA followed by Bonferroni’s multiple comparisons post-hoc test (E, F, H, and I). The Sholl analysis data were analyzed by two-way ANOVA followed by Bonferroni’s multiple comparisons post-hoc test. The results are expressed as the mean ± SEM.
-
Figure 4—source data 1
Dynein activity is required for phosphorylation of CREB and dendritic arborization induced by BDNF.
- https://cdn.elifesciences.org/articles/77455/elife-77455-fig4-data1-v1.xlsx
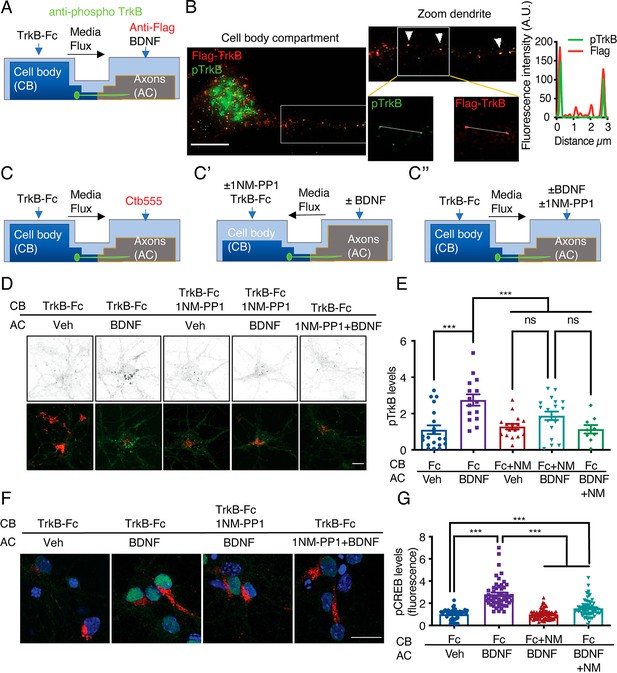
Somatodendritic activity of axonal TrkB is required for long-distance BDNF signaling.
(A) Schematic representation of the protocol used to label axonal TrkB. DIV five compartmentalized cortical neurons were transfected with Flag-TrkB for 48 hr. At DIV seven, anti-Flag antibody was added to AC for 40 min at 4 °C. Then, neurons were incubated with or without BDNF (50 ng/mL) at 37 °C. Finally, immunofluorescence for pTrkB (Y816, pTrkB) (green) and anti-Flag (red) was performed. (B) Immunofluorescence of pTrkB and internalized Flag-TrkB in compartmentalized neurons whose axons were treated with BDNF, as shown in A. Right panel, representative images of neurons whose axons were treated with BDNF. Left panel, a magnified image of a proximal neuronal dendrite. The arrows indicate colocalization of Flag (red) and pTrkB (green). Scale bar, 5 μm. Right panel, graphs showing the fluorescence intensity pixel by pixel along the white lines shown in the right panels in B. The green line indicates pTrkB fluorescence, and the red lines indicate Flag-TrkB fluorescence. (C-C’’) Schematic representation of the protocol used to stimulate neurons in D-G. (C) At DIV 6, Ctb-555 was added to the AC of cortical neurons from TrkBF616A mice overnight. The next day (DIV 7), the neurons were subjected to two treatments: (C’) depletion of B27 supplement for 1 hr in the presence of 1NM-PP1 (1 μM) in the CB, with flux toward the CB. Then, TrkB-Fc (100 ng/mL) and 1NM-PP1 were added to the CB, and BDNF (50 ng/mL) was applied or not to the AC for 3 hr. (C’’) Neurons were depleted of B27 supplement for 1 hr in the presence or absence of 1NM-PP1 (1 μM) in the AC, with flux toward the AC. Then, TrkB-Fc was applied to the CB, and BDNF was added or not to the AC in the presence or absence of 1NM-PP1 for 3 hr. Finally, the neurons were fixed, and immunofluorescence for pTrkB (Y816) or pCREB (S133) was performed. (D) Representative images of pTrkB in the CB compartment of Ctb-positive neurons treated as described in A. Scale bar = 5 µm. Quantification of pTrkB levels in the cell body in each treatment group described in B. n=3 independent experiments. (F) Representative image of pCREB immunostaining in cortical neurons whose axons were stimulated with BDNF for 3 hr in the presence or absence of 1NM-PP1 in the CB (1NM-PP1/BDNF) or AC (1NM-PP1+1NM-PP1). Scale bar, 10 µm. (G) Quantification of pCREB levels in Ctb-positive neurons under each condition. n=78–86 neurons from three independent experiments. **p<0.01, ***p<0.001. vs. the control group. The data were analyzed by one-way ANOVA followed by Bonferroni’s multiple comparisons post-hoc test. The results are expressed as the mean ± SEM.
-
Figure 5—source data 1
Somatodendritic activity of axonal TrkB is required for long-distance BDNF signaling.
- https://cdn.elifesciences.org/articles/77455/elife-77455-fig5-data1-v1.xlsx
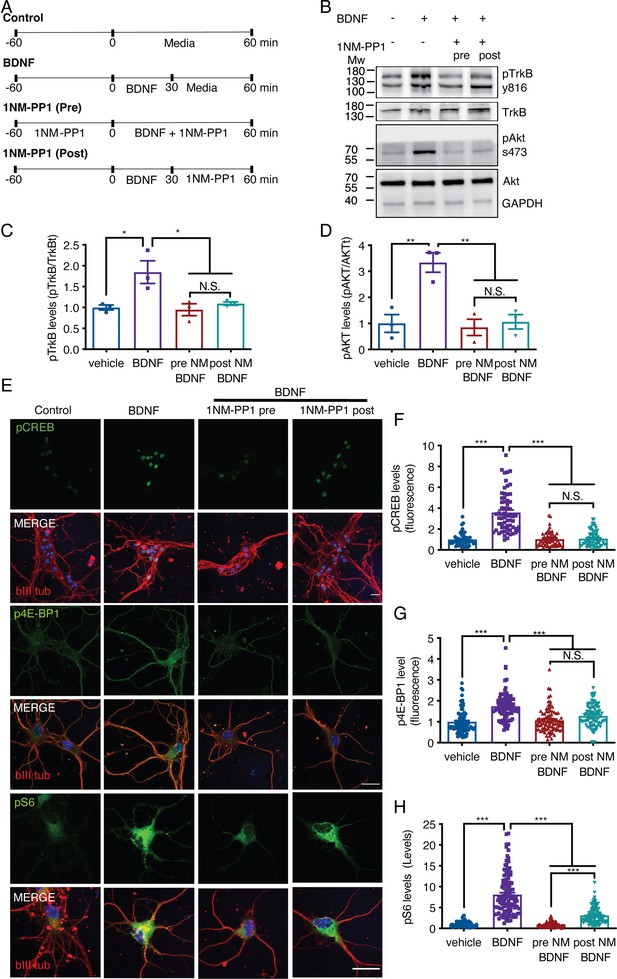
1NM-PP1 can reduce TrkB activation in TrkBF616A mouse cortical neurons after BDNF treatment.
(A) Diagrams of the experimental designs used to stimulate neurons in noncompartmentalized cultures. The medium of DIV 7 cortical neurons was removed for 1 hr, and the neurons were treated with or without 1NM-PP1 (1 μM). Then, the neurons were treated as follows: neurons in the control group were treated with vehicle for 1 hr; neurons in the BDNF group were incubated with BDNF (50 ng/mL) for 30 min, rinsed twice and incubated with vehicle; neurons in the 1NM-PP1 (Pre) group were incubated with BDNF in the presence of 1NM-PP1 for 1 hr; and neurons in the 1NM-PP1 (Post) group were incubated with BDNF for 30 min, rinsed twice and incubated with 1NM-PP1 for 30 min. (B) Immunoblotting of pTrkB (Y816), TrkB, pAkt (S437), Akt and GAPDH in cortical neurons stimulated as described in A. (C) Quantification of pTrkB levels normalized to total TrkB levels. (D) Quantification of pAkt levels normalized to total Akt levels. n=3 independent experiments. (E) Representative image of cortical neurons treated as described in A. pCREB, p4E-BP1, and pS6 are visualized in green, and βIII tubulin is shown in red. Scale bar = 20 µm. (F) Quantification of pCREB fluorescence (arbitrary units, A.U.) in the nuclei of neurons. (G) Quantification of p4E-BP1 fluorescence (arbitrary units, A.U.) in primary dendrites. (H) Quantification of pS6 fluorescence (arbitrary units, A.U.) in primary dendrites. n=49–72 neurons from three independent experiments. *p<0.05, **p<0.01, ***p<0.001. The data were analyzed by one-way ANOVA followed by Bonferroni’s multiple comparisons post-hoc test. The results are expressed as the mean ± SEM.
-
Figure 5—figure supplement 1—source data 1
1NM-PP1 can reduce TrkB activation in TrkBF616A mouse cortical neurons after BDNF treatment.
Original Western blots supporting Figure 5—figure supplement 1. This source data contains the original Western blots that supports the Figure 5—figure supplement 1. The images are separated in folders each one corresponding to independent experiments (N1, N2, N3). In the original pictures, the order of lines is the same than in Figure 5—figure supplement 1B and the name of the file indicates the antibody used to develop the nitrocellulose membrane.
- https://cdn.elifesciences.org/articles/77455/elife-77455-fig5-figsupp1-data1-v1.zip
-
Figure 5—figure supplement 1—source data 2
1NM-PP1 can reduce TrkB activation in TrkBF616A mouse cortical neurons after BDNF treatment.
Graph C, D, F-H´s raw data.
- https://cdn.elifesciences.org/articles/77455/elife-77455-fig5-figsupp1-data2-v1.xlsx
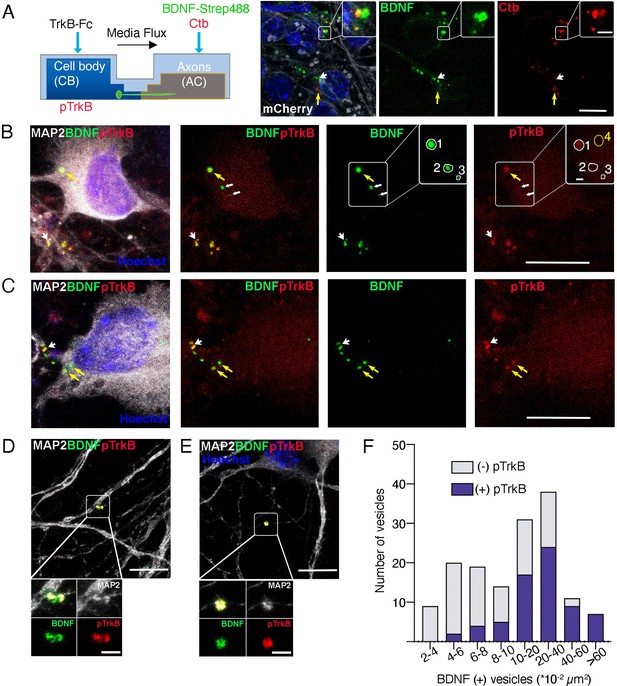
Axonal BDNF colocalizes with active TrkB in signaling endosomes within cell bodies and dendrites of compartmentalized cortical neurons.
(A) Left panel, schematic representation of the protocol used to stimulate neurons. Right panel, DIV 4 neurons were transduced with AAV1 expressing mCherry. After 4 days, Ctb-647 (Ctb, red) and biotinylated BDNF conjugated to streptavidin Dylight 488 (BDNF-strep488, green) were added to the axonal compartment (150 ng/mL) for 6 hr. Then, cultures were fixed, mounted in Mowiol containing Hoechst, and prepared for visualization. Representative images are shown where yellow arrows indicate BDNF-strep488 only vesicles, and white arrows indicate Ctb only vesicles. Scale bar = 10µm. Inset, magnification of endocytic structures where BDNF-strep488 and Ctb are partially colocalizing (observed in yellow Scale bar = 2 µm). (B–E) DIV 8 neurons were serum-deprived for 90 min and BDNF-strep488 was added to the axonal compartment (150 ng/mL) for 6 hr. Then, neurons were fixed and immunolabelled with an antibody for pTrkB (red) and MAP2 (white). Nuclei were counterstained with Hoechst (blue). (B and C). Yellow arrows indicate BDNF-strep488 colocalizing with pTrkB in cell bodies (signaling endosomes). Big white arrowheads indicate signaling endosomes in dendrites (labeled by MAP2). The small white arrowhead in B indicated BDNF-strep488 vesicles containing no detectable levels of pTrkB Scale bar = 10 µm. Inset in B, four different regions of interest (ROI) are selected. ROI number 1 has BDNF-strep488 and detectable levels of pTrkB. ROI numbers 2 and 3 contain BDNF-strep488-positive endocytic structures with undetectable levels of pTrkB. ROI number 4 indicates a region considered background noise when quantifying pTrkB immunostaining. Scale bar = 1 µm. (D and E) Representative images of MAP2-positive dendrites containing signaling endosomes Scale bar = 10 µm. Inset, magnification of signaling endosomes were BDNF-strep488 (green) and pTrkB (red) are observed in close association with MAP2 immunostaining (white). Scale bar = 2µm. (F), the presence of detectable levels of pTrkB was quantified in 304 BDNF-strep488-positive endocytic structures, as indicated in the methodology section. BDNF-strep488 endosomes were classified by range size and the number of vesicles containing detectable levels of pTrkB was quantified (purple) in each range.
-
Figure 6—source data 1
Axonal BDNF colocalizes with active TrkB in signaling endosomes within cell bodies and dendrites of compartmentalized cortical neurons.
- https://cdn.elifesciences.org/articles/77455/elife-77455-fig6-data1-v1.xlsx
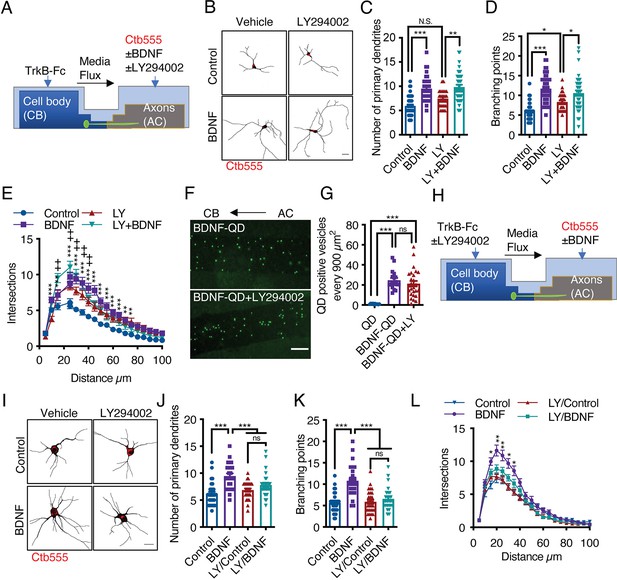
Activation of PI3K activity in the cell body but not the axons is required for dendritic arborization induced by axonal BDNF.
(A) Schematic representation of the protocol used to stimulate compartmentalized cortical neurons. DIV six cortical neurons were transfected with EGFP, and then TrkB-Fc (100 ng/mL) was added to the CB and Ctb-555 and BDNF (50 ng/mL) with or without LY294002 (10 µM) were applied to the AC for 48 hr. Finally, the neurons were fixed, and immunofluorescence for MAP2 was performed. (B) Representative images of the CB (red indicates Ctb-555) of compartmentalized neurons whose axons were treated with DMSO in the presence or absence of BDNF and LY294002. Scale bar 20 µm. (C–E) Quantification of primary dendrites (C) and branching points (D) and Sholl analysis (E) for neurons labeled with EGFP/MAP2/Ctb-555 under the different experimental conditions described in B. n=29–48 neurons from three independent experiments. (F) BDNF-QDs were added to the AC for 4 hr in the presence or absence of LY294002 (10 µM) to promote the retrograde transport of BDNF. Then, the neurons were fixed, and the accumulation of BDNF-QDs in the proximal compartment of the microgrooves (MG) was evaluated. Representative image of accumulated BDNF-QDs (green dots) in the proximal part of MG under each treatment. Scale bar = 10 µm. (G) Quantification of QD-positive vesicles in every 900 µm2 area of the proximal region of MG under each condition. Scale bar = 10 µm. n=36 MG from three independent experiments were analyzed. (H) Schematic representation of the protocol used to stimulate compartmentalized cortical neurons. DIV six cortical neurons were transfected with EGFP. Then, TrkB-Fc (100 ng/mL) was added to the CB in the presence or absence of LY294002 (10 µM), and Ctb-555 and BDNF (50 ng/mL) were applied to the AC for 48 hr. Finally, the neurons were fixed, and immunofluorescence for MAP2 was performed. (I) Representative images of the CB (red indicates Ctb-555) of compartmentalized neurons whose cell bodies were treated with DMSO (control) or LY294002 and whose axons were treated with or without BDNF. Scale bar = 20 µm. (C–E) Quantification of primary dendrites (C) and branching points (D) Sholl analysis (E) for neurons labeled with EGFP/MAP2/Ctb-555. n=25–30 neurons from three independent experiments. *p<0.05, **p<0.01, ***p<0.001,+p < 0.05 the LY294002 group vs. the LY294002/BDNF group. The data were analyzed by one-way ANOVA followed by Bonferroni’s multiple comparisons post-hoc test (C, D, G, J, and K). Statistical analysis of the Sholl analysis data was performed by two-way ANOVA followed by Bonferroni’s multiple comparisons post-hoc test. The results are expressed as the mean ± SEM.
-
Figure 7—source data 1
Activation of PI3K activity in the cell body but not the axons is required for dendritic arborization induced by axonal BDNF.
- https://cdn.elifesciences.org/articles/77455/elife-77455-fig7-data1-v1.xlsx
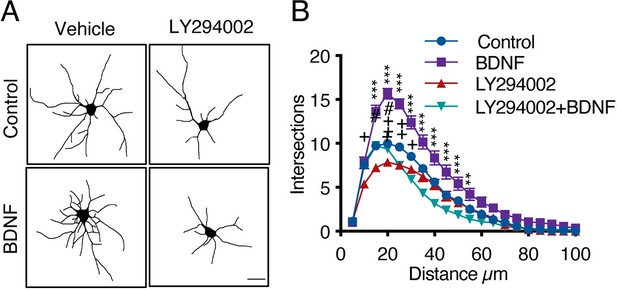
PI3K activation is required for BDNF signaling in cortical neurons.
(A) Representative images of rat cortical neurons in non-compartmentalized cultures treated with vehicle, LY294002 (10 μM), BDNF (50 ng/mL) or BDNF after preincubation with LY294002. Scale bar = 20 µm. (B) Quantification of total dendritic branching by Sholl analysis. n=25–32 from two independent experiments. **p<0.01 and ***p<0.001, the control group vs. the BDNF group;++p < 0.01, the control group vs. the LY294002 group. The Sholl analysis data were analyzed by two-way ANOVA followed by Bonferroni’s multiple comparisons post-hoc test. The results are expressed as the mean ± SEM.
-
Figure 7—figure supplement 1—source data 1
PI3K activation is required for BDNF signaling in cortical neurons.
Graph B´s raw data.
- https://cdn.elifesciences.org/articles/77455/elife-77455-fig7-figsupp1-data1-v1.xlsx
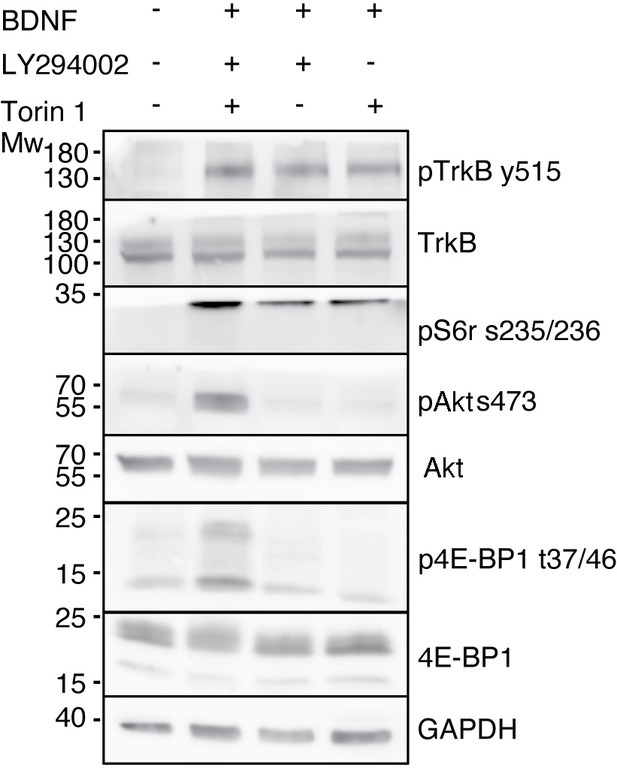
PI3K and mTOR activity is required for activation of downstream TrkB signaling pathways.
Western blot analysis of DIV 7 non-compartentalized neurons treated with BDNF (50 ng/mL) in the presence or absence of LY294002 (10 μM) or Torin 1 (0.25 μM) for 1 hr. pTrkB (Y515), TrkB, pAkt (S473), total Akt, pS6r (S235/236), p4E-BP1 (T37/46), total 4E-BP1 and GAPDH expression was evaluated.
-
Figure 7—figure supplement 2—source data 1
This source data contains the original western blots that supports the Figure 7—figure supplement 2.
In the original pictures, the order of lines is the same than in Figure 7—figure supplement 2 and the name of the file indicates the antibody used to develop the nitrocellulose membrane.
- https://cdn.elifesciences.org/articles/77455/elife-77455-fig7-figsupp2-data1-v1.zip
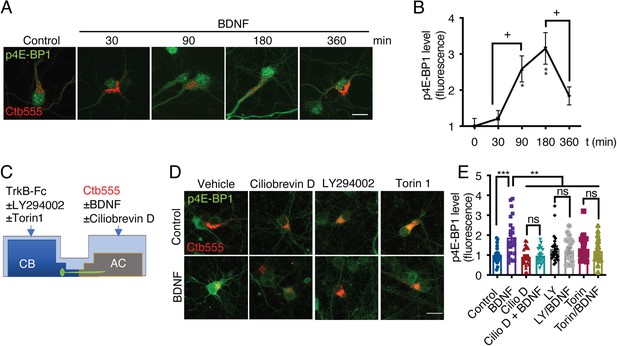
Axonal BDNF promotes mTOR activation in the cell bodies of compartmentalized cortical neurons in a PI3K-dependent manner.
(A) Representative images of p4E-BP1 immunostaining (green) and Ctb-555 (red). DIV six compartmentalized cortical neurons were incubated with Ctb-555 overnight. At DIV 7, BDNF (50 ng/mL) was added to the AC of the neurons for 30, 60, 180, or 360 min. Scale bar = 20 µm. (B) Quantification of somatodendritic p4E-BP1 immunofluorescence in primary dendrites over time. (C) Schematic representation of the protocol used to evaluate the effect of different pharmacological inhibitors on the axonal BDNF-induced phosphorylation of 4E-BP1 in the cell body. DMSO (control), LY294002 (10 µm; LY), and Torin 1 (0.25 µm; Torin) were added to the CB of DIV six cortical neurons, or CilioD (20 µm; CilioD) was applied to the AC for 1 hr. Then, BDNF was added to the AC for 180 min in the presence or absence of these inhibitors. (D) Representative images of p4E-BP1 (green) in the somatodendritic compartment of neurons stimulated with BDNF in the presence or absence of different inhibitors. (E) Quantification of somatodendritic p4E-BP1 expression in neurons labeled with Ctb-555 (red) under each treatment. Scale bar = 20 µm. n=31–36 neurons from three independent experiments. *p<0.05, **p<0.01, ***p<0.001. vs. the control group;+p < 0.05 vs. the 90- and 360 min BDNF treatment groups in B. The data were analyzed by one-way ANOVA followed by Bonferroni’s multiple comparisons post-hoc test. The results are expressed as the mean ± SEM.
-
Figure 8—source data 1
Axonal BDNF promotes mTOR activation in the cell bodies of compartmentalized cortical neurons in a PI3K-dependent manner.
- https://cdn.elifesciences.org/articles/77455/elife-77455-fig8-data1-v1.xlsx
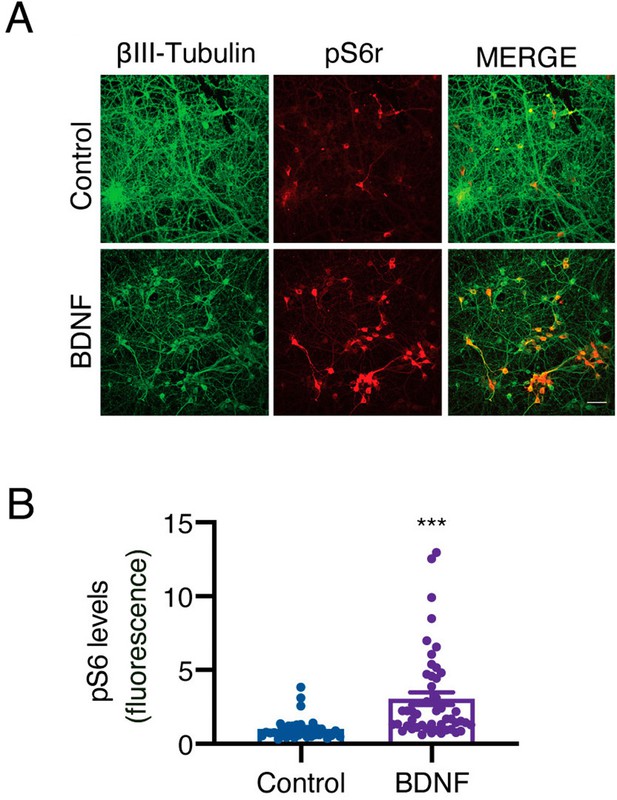
BDNF added to axons increases pS6r in cell bodies.
(A) Representative images of β-III-tubulin (green) and pS6r (red) in cortical neurons whose axons were stimulated with BDNF (50 ng/mL) for 180 min. Scale bar = 50 μm. (B) Quantification of pS6r fluorescence intensity in the soma of neurons treated with or without BDNF. n=36–48 neurons from three independent experiments. ***p<0.01. Student’s t-test was used for statistical analysis. The results are expressed as the mean ± SEM.
-
Figure 8—figure supplement 1—source data 1
BDNF added to axons increases pS6r in cell bodies.
Graph B´s raw data.
- https://cdn.elifesciences.org/articles/77455/elife-77455-fig8-figsupp1-data1-v1.xlsx
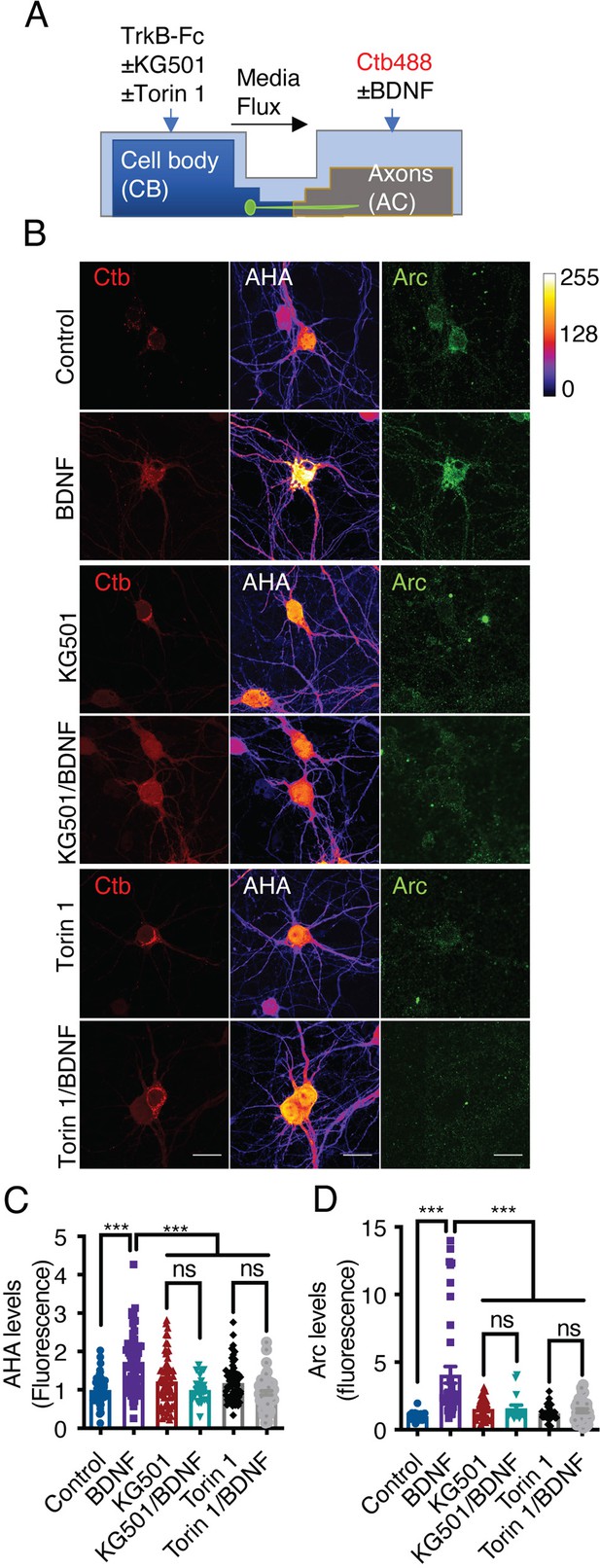
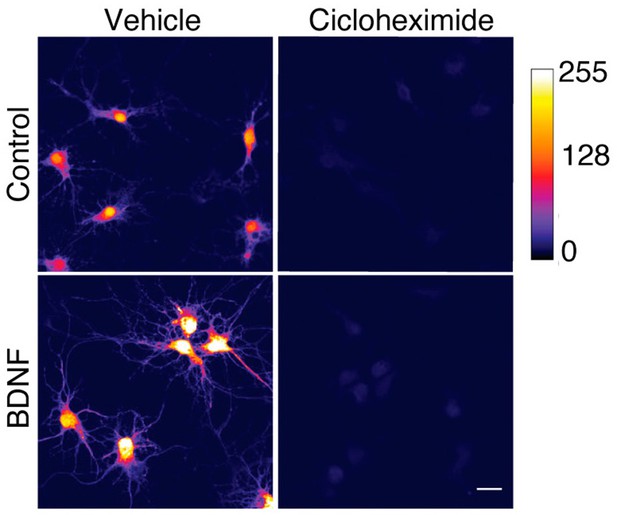
Axonal BDNF promotes somatodendritic protein synthesis in an mTOR- and CREB-dependent manner.
(A) Schematic representation of the protocol used to evaluate protein synthesis. DIV 6–7 cortical neurons were incubated in methionine-free medium for 1 hr, and KG501 (10 µm) or Torin 1 (0.25 µm) was applied or not to the CB. Then, AHA was added to both the CB and AC, and BDNF (50 ng/mL) in the presence of Ctb-488 was added to only the AC for 5 hr. (B) Representative images of control and BDNF-stimulated neurons positive for Ctb-488 (red), AHA (in intensity color code mode, see color bar on the right) and Arc (green) in the presence or absence of KG501 and Torin 1. Scale bar = 20 µm (C) Quantification of AHA fluorescence in the primary dendrites of neurons labeled with Ctb-488 under each treatment. (D) Quantification of Arc fluorescence in the somas of neurons labeled with Ctb-488 under each different treatment. n=30–48 neurons from three independent experiments. ***p<0.001. The data were analyzed by one-way ANOVA followed by Bonferroni’s multiple comparisons post-hoc test. The results are expressed as the mean ± SEM.
-
Figure 9—source data 1
Axonal BDNF promotes somatodendritic protein synthesis in an mTOR- and CREB-dependent manner.
- https://cdn.elifesciences.org/articles/77455/elife-77455-fig9-data1-v1.xlsx
Additional files
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/77455/elife-77455-transrepform1-v1.pdf
-
Supplementary file 1
List of materials and reagents.
- https://cdn.elifesciences.org/articles/77455/elife-77455-supp1-v1.xlsx