The Neurodata Without Borders ecosystem for neurophysiological data science
Figures
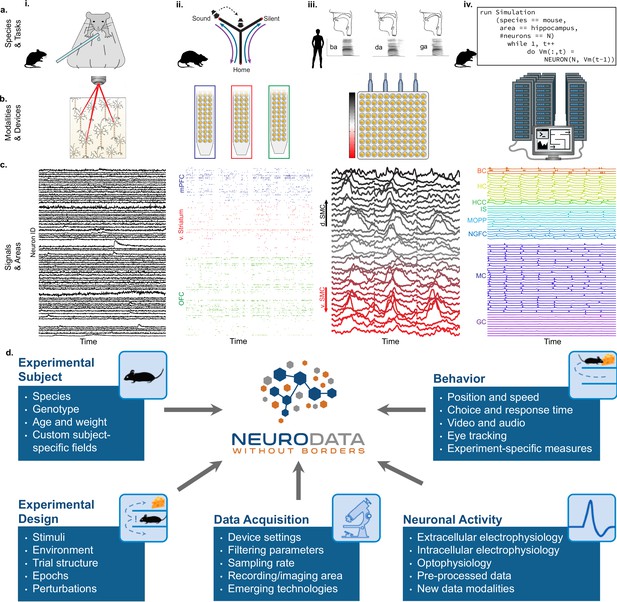
NWB addresses the massive diversity of neurophysiology data and metadata.
(a) Diversity of experimental systems: species and tasks: (i) mice performing a visual discrimination task; (ii) rats performing a memory-guided navigation task; (iii) humans speaking consonant-vowel syllables; (iv) biophysically detailed simulations of mouse hippocampus during memory formation. The corresponding acquisition modalities and signals are shown in the corresponding columns in figure (b and c). (b) Diversity of data modalities and acquisition devices: (i) optophysiological Ca2+ imaging with two-photon microscope; (ii) intra-cortical extracellular electrophysiological recordings with polytrodes in multiple brain areas (indicated by color, see c.ii); (iii) cortical surface electrophysiology recordings with electrocorticography grids; (iv) high-performance computing systems for large-scale, biophysically detailed simulations of large neural networks. (c) Diversity of signals and areas: (i) Ca2+ signals as a function of time from visually identified individual neurons in primary visual cortex (V1) (Mallory et al., 2021); (ii) spike-raster (each tick demarcates the time of an action potential) from simultaneously recorded putative single-units after spike-sorting of extracellular signals from medial prefrontal cortex (mPFC; blue), ventral striatum (v. Striatum, red), and orbital frontal cortex (OFC, green) (color corresponds to b.ii) (Chung et al., 2019); (iii) high-gamma band activity from electrodes over the speech sensorimotor cortex (SMC), with dorsal-ventral distance from Sylvian fissure color coded red-to-black (color corresponds to b.iii) (Bouchard et al., 2013); (iv) simulated intracellular membrane potentials from different cell-types from large-scale biophysical simulation of the hippocampus (BC, Basket Cell); HC, Hilar Interneuron (with axon associated with the) Perforant Path; HCC, Hilar Interneuron (with axon associated with the) Commissural/Associational Path; IS, Interneuron-Specific Interneuron; MCPP, medial Perforant Path; NGFC, neurogliaform cell; MC, mossy cell; GC, granule cell](Raikov and Soltesz, unpublished data). (d) Neurodata Without Borders (NWB) provides a robust, extensible, and maintainable software ecosystem for standardized description, storage, and sharing of the diversity of experimental subjects, behaviors, experimental designs, data acquisition systems, and measures of neural activity exemplified in a – c.
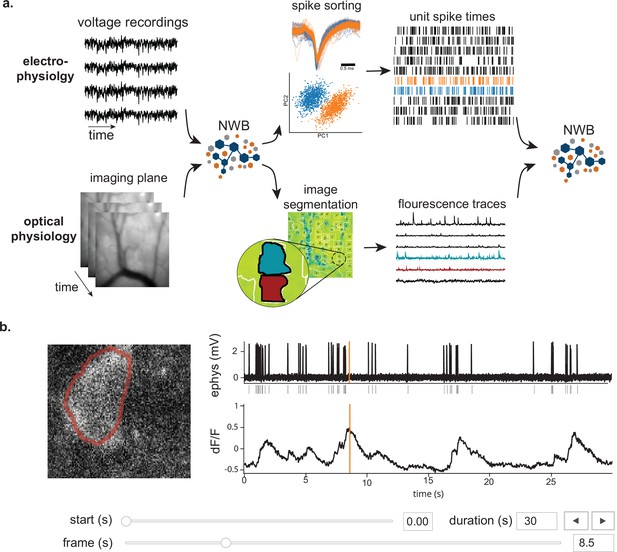
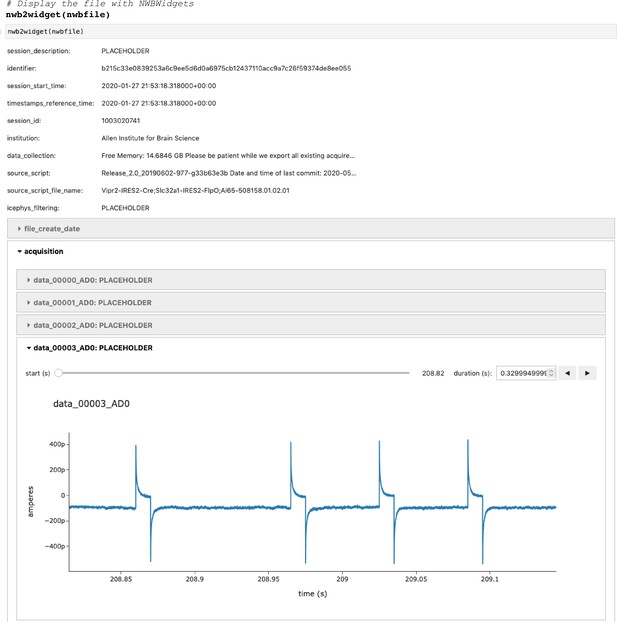
NWB enables unified description and storage of multimodal raw and processed data.
(a) Example pipelines for extracellular electrophysiology and optical physiology demonstrate how NWB facilitates data processing. For extracellular electrophysiology (top), raw acquired data is written to the NWB file. The NWB ecosystem provides interfaces to a variety of spike sorters that extract unit spike times from the raw electrophysiology data. The spike sorting results are then stored into the same NWB file (bottom). Separate experimental data acquired from an optical technique is converted and written to the NWB file. Several modern software tools can then be used to process and segment this data, identifying regions that correspond to individual neurons, and outputting the fluorescence trace of each putative neuron. The fluorescence traces are written to the same NWB file. NWB handles the time alignment of multiple modalities, and can store multiple modalities simultaneously, as shown here. The NWB file also contains essential metadata about the experimental preparation. (b) NWBWidgets provides visualizations for the data within NWB files with interactive views of the data across temporally aligned data types. Here, we show an example dashboard for simultaneously recorded electrophysiology and imaging data. This interactive dashboard shows on the left the acquired image and the outline of a segmented neuron (red) and on the right a juxtaposition of extracellular electrophysiology, extracted spike times, and simultaneous fluorescence for the segmented region. The orange line on the ephys and dF/F plots indicate the frame that is shown to the left. The controls shown at the bottom allow a user to change the window of view and the frame of reference within that window.
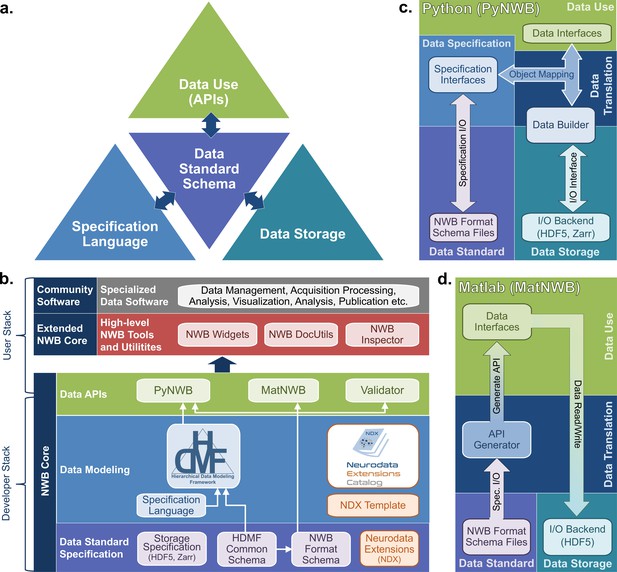
The NWB software architecture modularizes and integrates all components of a data language.
(a) Illustration of the main components of the NWB software stack consisting of: (i) the specification language (light blue) to describe data standards, (ii) the data standard schema (lilac), which uses the specification language to formally define the data standard, (iii) the data storage (blue gray) for translating the data primitives (e.g., groups and datasets) described by the schema to/from disk, and (iv) the APIs (green) to enable users to easily read and write data using the standard. Additional data translation components (dark blue arrows) defined in the software then insulate and separate these four main components to enable the individual components to evolve while minimizing impacts on the other components. For example, by insulating the schema from the storage we can extend the standard schema without having to modify the data storage and conversely also integrate new storage backends without having to modify the standard schema. (b) Software stack for defining and extending the NWB data standard and creating and using NWB data files. The software stack covers all aspects of data standardization: (i) data specification, (ii) data modeling, (iii) data storage, (iv) data APIs, (v) data translation, and (vi) tools. Depending on their role, different stakeholders typically interact with different subsets of the software ecosystem. End users typically interact with the data APIs (green) and higher-level tools (red, gray) while tool developers typically interact with the data APIs and data modeling layers (green, blue). Working groups and developers of extensions then typically interact with the data modeling and data standard specification components. Finally, core NWB developers typically interact with the entire developer stack, from foundational documents (lilac) to data APIs (green). (c) Software architecture of the PyNWB Python API. PyNWB provides interfaces for interacting with the specification language and schema, data builders, storage backends, and data interfaces. Additional software components (arrows) insulate and formalize the transitions between the various components. The object-mapping-based data translation describes: (i) the integration of data interfaces (which describe the data) with the specification (which describes the data model) to generate data builders (which describe the data for storage) and (ii) vice versa, the integration of data builders with the specification to create data interfaces. The object mapping insulates the end-users from specifics of the standard specification, builders, and storage, hence, providing stable, easy-to-use interfaces for data use that are agnostic of the data storage and schema. The I/O interface then provides an abstract interface for translating data builders to storage which specific I/O backends must implement. Finally, the specification I/O then describes the translation of schema files to/from storage, insulating the specification interfaces from schema storage details. Most of the data modeling, data translation, and data storage components are general and implemented in HDMF. This approach facilitates the application of the general data modeling capabilities we developed to other science applications and allows PyNWB itself to focus on the definition of data interfaces and functionality that are specific to NWB. (d) Software architecture of the MatNWB Matlab API. MatNWB generates front-end data interfaces for all NWB types directly from the NWB format schema. This allows MatNWB to easily support updates and extensions to the schema while enabling development of higher-level convenience functions.
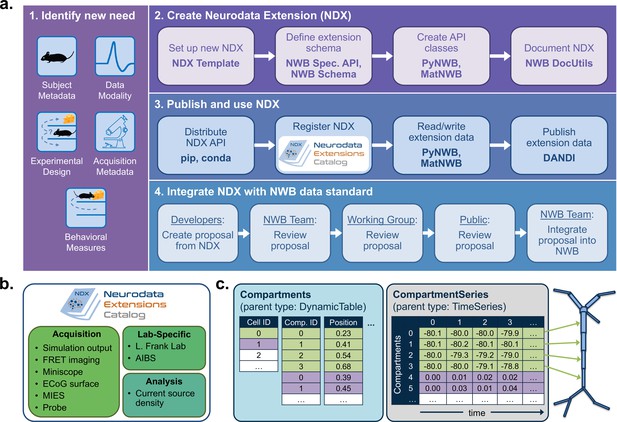
NWB enables creation and sharing of extensions to incorporate new use cases.
(a) Schematic of the process of creating a new neurodata extension (NDX), sharing it, and integrating it with the core NWB data standard. Users first identify the need for a new data type, such as additional subject metadata or data from a new data modality. Users can then use the NDX Template, NWB Specification API, PyNWB/MatNWB data APIs, and NWB DocUtils tools to set up a new NDX, define the extension schema, define and test custom API classes for interacting with extension data, and generate Sphinx-based documentation in common formats, for example, HTML or PDF. After the NDX is completed, users can publish the NDX on PyPI and conda-forge for distribution via the pip and conda tools, and share extensions via the NDX Catalog, a central, searchable catalog. Users can easily read/write extension data using PyNWB/MatNWB and publish extension data in DANDI and other archives. Finally, extensions are used to facilitate enhancement, maintenance, and governance of the NWB data standard. Users may propose the integration of an extension published in the NDX Catalog with the core standard. The proposal undergoes three phases of review: an initial review by the NWB technology team, an evaluation by a dedicated working group, and an open, public review by the broader community. Once approved, the proposal is integrated with NWB and included in an upcoming version release. (b) Sampling of extensions currently registered in the NDX catalog. Users can search extensions based on keywords and textual descriptions of extensions. The catalog manages basic metadata about extensions, enabling users to discover and access extensions, comment and make suggestions, contribute to the source code, and collaborate on a proposal for integration into the core standard. While some extensions have broad applicability, others represent data and metadata for a specific lab or experiment. (c) Example extension for storing simulation output data using the SONATA framework. The new Compartments type extends the base DynamicTable type and contains metadata about each cell and compartment within each cell, such as position and label. The CompartmentSeries type extends the base TimeSeries type and contains a link to the Compartments type to associate each row of its data array with a compartment from the Compartments table.
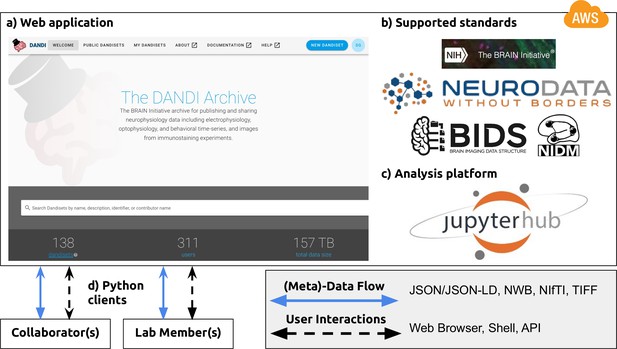
NWB is foundational for the DANDI data repository to enable collaborative data sharing.
The DANDI project makes data and software for cellular neurophysiology FAIR. DANDI stores electrical and optical cellular neurophysiology recordings and associated MRI and/or optical imaging data. NWB is foundational for the DANDI data repository to enable collaborative data sharing. (a) DANDI provides a Web application allowing scientists to share, collaborate, and process data from cellular neurophysiology experiments. The dashboard provides a summary of Dandisets and allows users to view details of each dataset. (b) DANDI works with US BRAIN Initiative awardees and the neurophysiology community to curate data using community data standards such as NWB, BIDS, and NIDM. DANDI is supported by the US BRAIN Initiative and the Amazon Web Services (AWS) Public Dataset Program. (c) DANDI provides a JupyterHub interface to visualize the data and interact with the archive directly through a browser, without the need to download any data locally. (d) Using Python clients and/or a Web browser, researchers can submit and retrieve standardized data and metadata from the archive. The data and metadata use standard formats such as HDF5, JSON, JSON-LD, NWB, NIfTI, and TIFF.
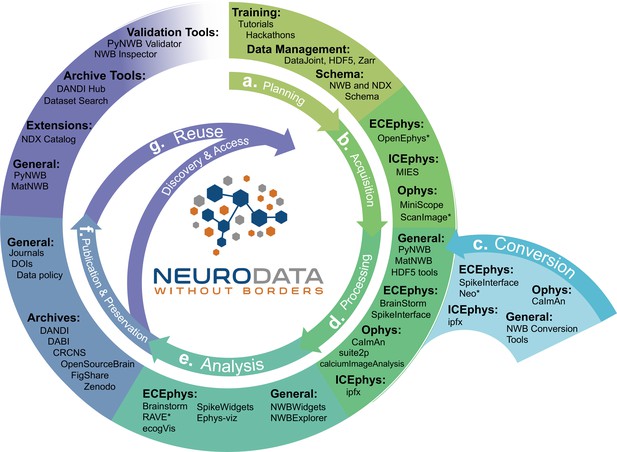
NWB is integrated with state-of-the-art analysis tools throughout the data life cycle.
NWB technologies are at the heart of the neurodata lifecycle and applications. Data standards are a critical conduit that facilitate the flow of data throughout the data lifecycle and integration of data and software across all phases (a. to g.) of the data lifecycle. (a) NWB supports experimental planning through integration with data management, best practices, and by allowing users to clearly define what metadata to collect. (b–c) NWB supports storage of unprocessed acquired electrical and optical physiology signals, facilitating integration already during data acquisition. NWB is already supported by several acquisition systems (b) as well as a growing set of tools for conversion (c) of existing data to NWB. (d) Despite its young age, NWB is already supported by a large set of neurophysiology processing software and tools. Being able to access and evaluate multiple processing methods, e.g., different spike sorting algorithms and ROI segmentation methods, is important to enable high-quality data analysis. Through integration with multiple different tools, NWB provides access to broad range of spike sorters, including, MountainSort, KiloSort, WaveClust, and others, and ophys segmentation methods, e.g., CELLMax, CNMF, CNMF-E, and EXTRACT. (e) For scientific analysis, numerous general tools for exploration and visualization of NWB files (e.g. NWBWidgets and NWBExplorer) as well as application-specific tools for advanced analytics (e.g. Brainstorm) are accessible to the NWB community. (f–g) NWB is supported by a growing set of data archives (e.g. DANDI) for publication and preservation of research data. Data archives in conjunction with NWB APIs, validation tools, and the NDX Catalog play a central role in facilitating data reuse and discovery.
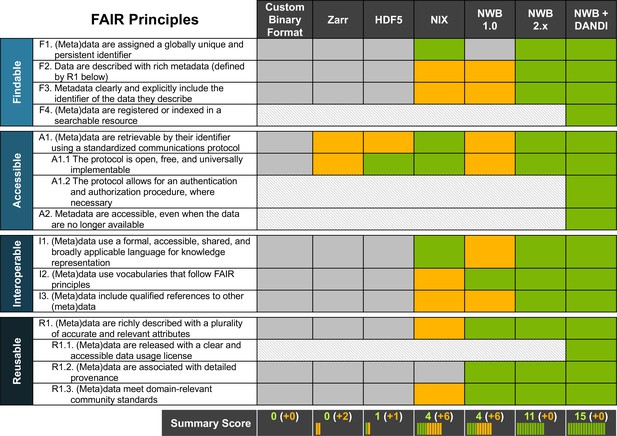
NWB together with DANDI provides an accessible approach for FAIR sharing of neurophysiology data.
The table above assesses various approaches for sharing neurophysiology data with regard to their compliance with FAIR data principles. Here, cells shown in gray/green indicate non-compliance and compliance, respectively. Cells shown in yellow indicate partial compliance, either due to incomplete implementation or optional support, leaving achieving compliance ultimately to the end user. The larger, shaded blocks indicate areas that are typically not covered by data standards directly but are the role of other resources in a FAIR data ecosystem, e.g., the DANDI data archive.
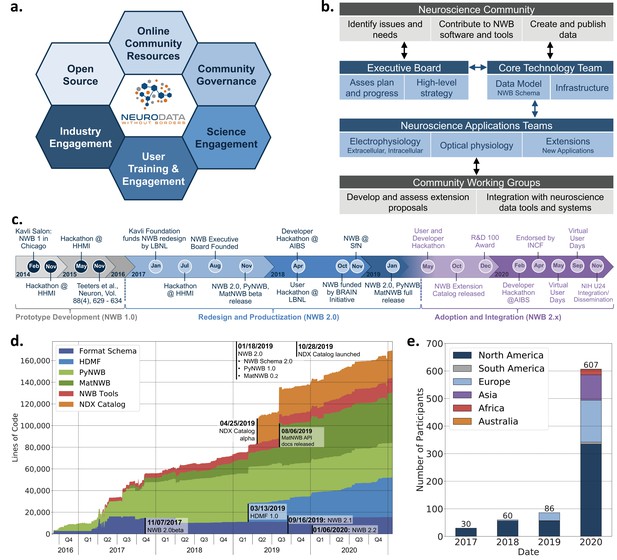
Coordinated community engagement, governance, and development of NWB.
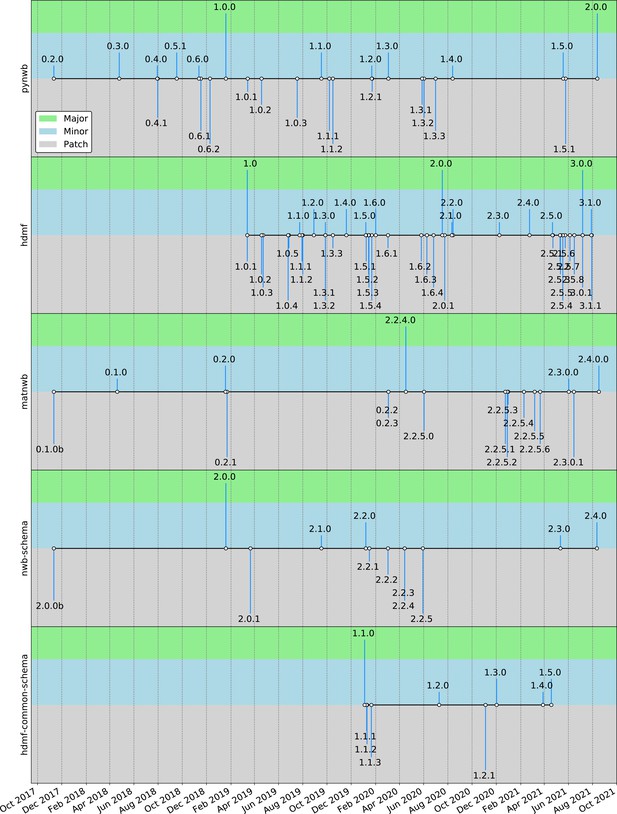
(a) NWB is open source with all software and documents available online via GitHub and the nwb.org website. NWB provides a broad range of online community resources, e.g., Slack, online Help Desk, GitHub, mailing list, or Twitter, to facilitate interaction with the community and provides a broad set of online documentation and tutorials. NWB uses an open governance model that is transparent to the community. Broad engagements with industry partners (e.g. DataJoint, Kitware, MathWorks, MBFBioscience, Vidrio, CatalystNeuro, etc.) and targeted science engagements with neuroscience labs and tool developers help sustain and grow the NWB ecosystem. Broad user training and engagement activities, e.g., via hackathons, virtual training, tutorials at conferences, or online training resources, aim at facilitating adoption and growing the NWB community knowledge base. (b) Organizational structure of NWB showing the main bodies of the NWB team (blue boxes) and the community (gray boxes), their roles (light blue/gray boxes), and typical interactions (arrows). (c) The timeline of the NWB project to date can be roughly divided into three main phases. The initial NWB pilot project (2014–2015) resulted in the creation of the first NWB 1.0 prototype data standard. The NWB 2.0 effort then focused on facilitating use and long-term sustainability by redesigning and productizing the data standard and developing a sustainable software strategy and governance structure for NWB (2017–2019). The release of NWB 2.0 in Jan. 2019 marked the beginning of the third main phase of the project, focused on adoption and integration of NWB with neuroscience tools and labs, maintenance, and continued evolution and refinement of the data standard. (d) Overview of the growth of core NWB 2.x software in lines of code over time. (e) Number of participants at NWB outreach and training events over time. In the count we considered only the NWB hackathons and User Days (see c.), the 2019 and 2020 NWB tutorial at Cosyne, and 2019 training at the OpenSourceBrain workshop (i.e. not including attendees at presentations at conferences, e.g., SfN).
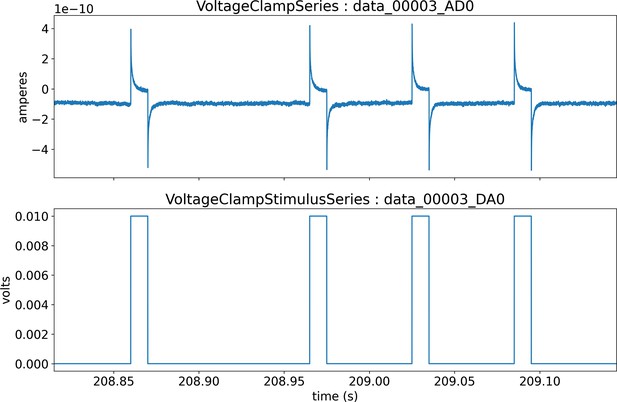
Visualization of the stimulus (bottom) and response (top) signals recorded via intracellular electrophysiology and stored in NWB.
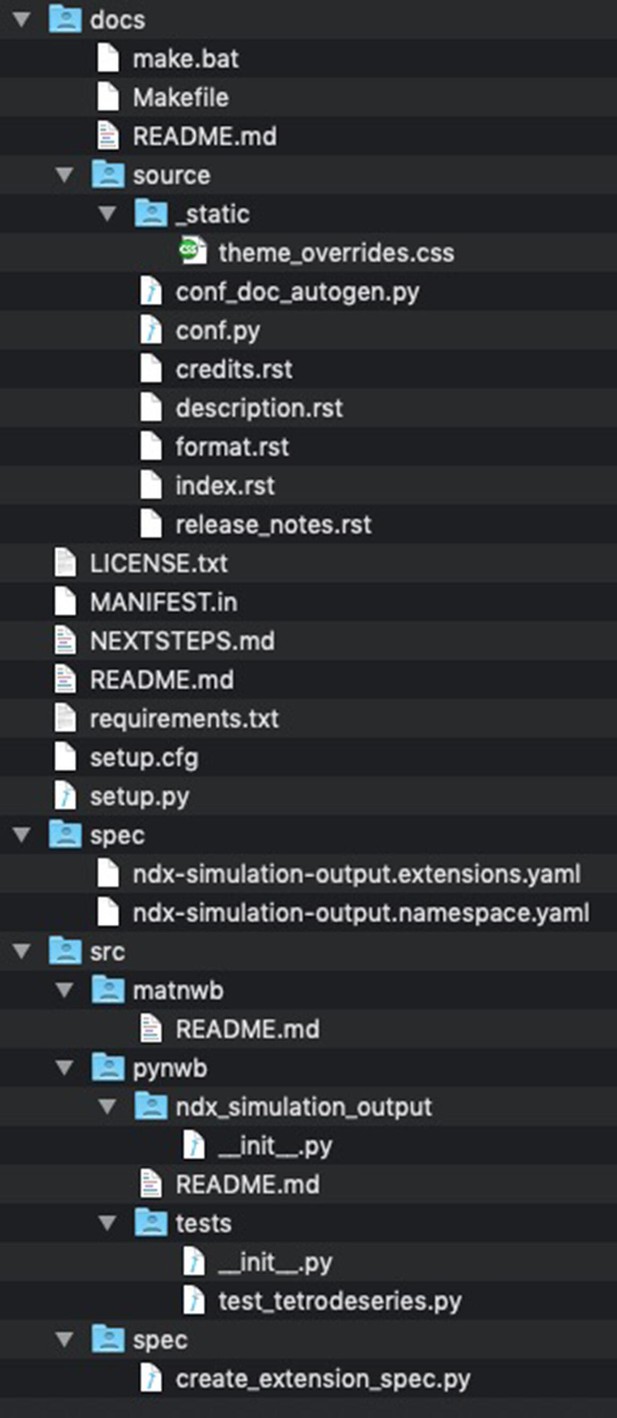
Files and folders generated by the cookiecutter ndx-template.
The main folder contains the license and readme file for extension along with files required for installing the extension (e.g., setup.py, setup.cfg, MANIFEST.in, and requirements.txt) as well a markdown file with instructions for next steps. The docs/ folder contains the Sphinx documentation setup for the extension. Without any additional changes required, the developer can with this setup automatically generate documentation in HTML, PDF, ePub and many other formats directly from the extension schema using the HDMF-DocUtils. Generating the documentation is as simple as executing “make html” in the docs/ folder. The spec/ folder contains the schema files for the extensions. The schema files are generated by the script in /src/spec/create_extension_spec.py (see Define the Extension Schema next), and are typically not modified manually by the developers. The /src folder then contains main source codes for the extension, including the: spec/ folder with the code to generated the extension schema matnwb/ folder with code for MatNWB pynwb/ folder with code for PyNWB.
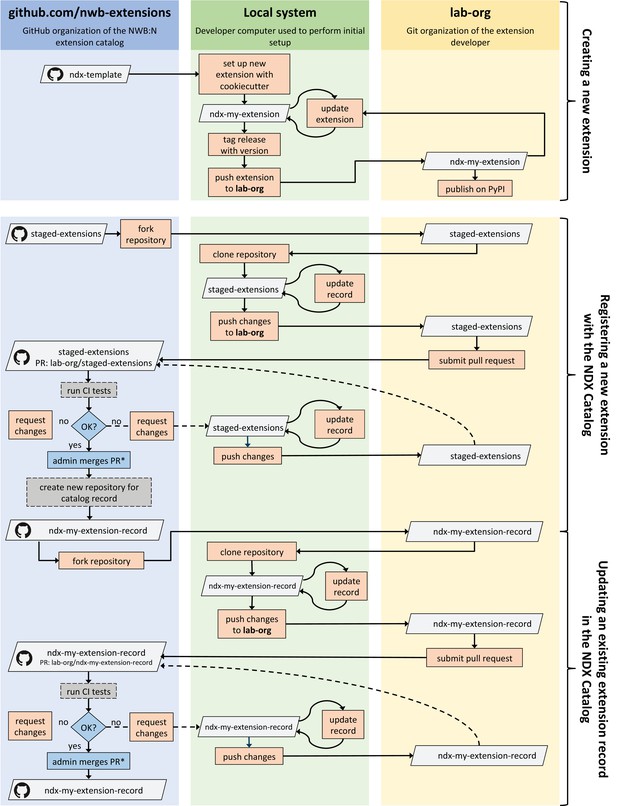
Illustration of the process for creating, publishing, and updating extensions via the Neurodata Extension Catalog (NDX Catalog), and (3) updating an extension/record.
Boxes shown in gray indicate Git repositories; boxes in orange describe user actions; and boxes in blue indicate actions by administrators of the NDX catalog.
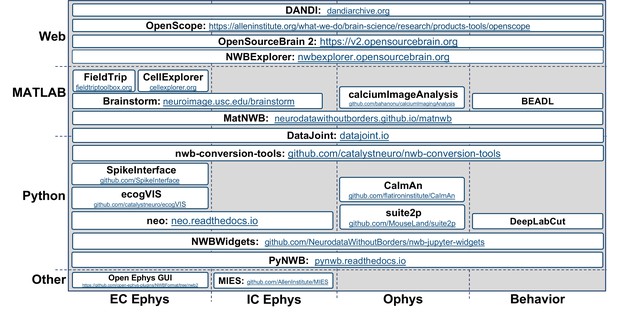
Overview of select data analysis, visualization, and management tools that support NWB.
Visualization showing select data analysis, visualization, and management tools that support NWB organized by their main application (x-axis) and programming environment (y-axis).
Tables
Compliance of NWB+DANDI with FAIR principles: Findability.
| Findable | ||||
|---|---|---|---|---|
| F1. (Meta)data are assigned a globally unique and persistent identifier | F2. Data are described with rich metadata (defined by R1 below) | F3. Metadata clearly and explicitly include the identifier of the data they describe | F4. (Meta)data are registered or indexed in a searchable resource | |
| Custom | No | No | No |
|
| Zarr | No |
| ||
| HDF5 | No | |||
| NIX |
|
| ||
| NWB 1.0 | No |
|
| |
| NWB 2 .x |
|
|
| |
| DANDI |
|
|
|
|
Compliance of NWB+DANDI with FAIR principles: Accessibility.
| Accessible | ||||
|---|---|---|---|---|
| A1. (Meta)data are retrievable by their identifier using a standardised communications protocol | A1.1 The protocol is open, free, and universally implementable | A1.2 The protocol allows for an authentication and authorisation procedure, where necessary | A2. Metadata are accessible, even when the data are no longer available | |
| Custom | No | No |
| |
| Zarr |
|
|
| |
| HDF5 |
| |||
| NIX |
|
| ||
| NWB 1.0 |
|
| ||
| NWB 2 .x |
|
| ||
| DANDI |
|
|
|
|
Compliance of NWB+DANDI with FAIR principles: Interoperability.
| Interoperable | |||
|---|---|---|---|
| I1. (Meta)data uses a formal, accessible, shared, and broadly applicable language for knowledge representation. | I2. (Meta)data use vocabularies that follow FAIR principles | I3. (Meta)data include qualified references to other (meta)data | |
| Custom | No | No | No |
| Zarr | No | No | No |
| HDF5 | No | No | No |
| NIX |
|
|
|
| NWB 1.0 |
|
|
|
| NWB 2 .x |
|
|
|
| DANDI |
|
|
|
-
*
Support for external resources has been released in HDMF >2.3 and is currently undergoing community review for integration with the NWB core data standard.
Compliance of NWB+DANDI with FAIR principles: Reusability.
| Reusable | ||||
|---|---|---|---|---|
| R1. (Meta)data are richly described with a plurality of accurate and relevant attributes | R1.1. (Meta)data are released with a clear and accessible data usage license | R1.2. (Meta)data are associated with detailed provenance | R1.3. (Meta)data meet domain-relevant community standards | |
| Custom | No |
| No | No |
| Zarr | No | No | No | |
| HDF5 | No | No | No | |
| NIX |
| No |
| |
| NWB 1.0 |
|
|
| |
| NWB 2 .x |
|
|
| |
| DANDI |
|
|
|
|