Metabolic arsenal of giant viruses: Host hijack or self-use?
Figures
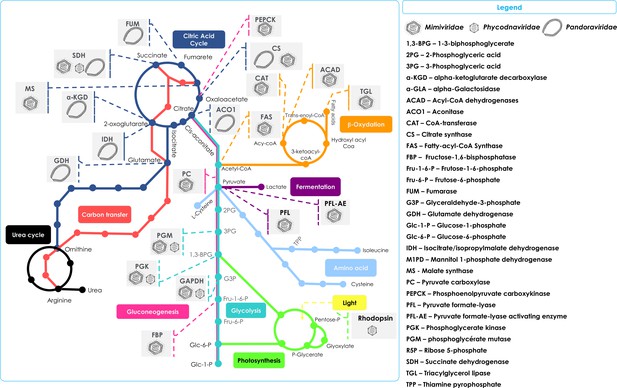
Schematic representation of the metabolic enzymes and pathways detected in NCLDVs.
Schematic of the TCA cycle (dark blue) feeding into the Urea cycle (black); Carbon transfer (red); Gluconeogenesis (scarlet); Glycolysis (marine); Photosynthesis (green); Amino acid metabolism (blue); Fermentation (purple); and lipid β-oxidation (orange). Also shown in the Legend are the identified cellular enzymes and putative substrates which have been identified as being encoded in specific NCLDV genomes, here represented by Mimiviridae, Phycodnaviridae, and Pandoraviridae.
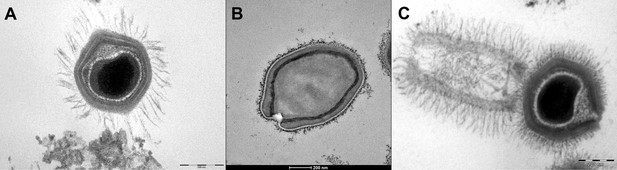
Transmission electron microscopy images of Mimivirus (A), Pandoravirus massiliensis (B) and Tupanvirus (C).
(A) Mimivirus particle is composed of an external layer of dense fibers surrounding an icosahedral capsid and an internal membrane sac enveloping the virus genomic material. (B) Pandoravirus massiliensis virion is ovoid-shaped with an ostiole-like apex, measuring 1.0 μm in length and 0.5 μm in diameter. (C) Tupanvirus exhibits an icosahedral capsid similar to those of Mimivirus measuring ~450 nm. However, Tupanvirus virion harbors a large cylindrical tail (550 nm extension;~450 nm diameter, including fibrils) attached to the base of the capsid. Electron micrographs were acquired on a Tecnai G2 transmission electron microscope (Scale bar, 200 nm).
Tables
List of metabolic enzymes detected in NCLDVs.
The enzymes were grouped according to the metabolic pathway to which they belong and associated with the giant virus and/or the family in which they were identified.
| Pathway | Enzyme | Function | KEGG* | Detected in | Family | Reference(s) |
|---|---|---|---|---|---|---|
| Amino acid catabolism | Glutamate dehydrogenase | Reversible conversion of glutamate to α-ketoglutarate and ammonia | R00243 | Pandoravirus and others uncharacterized viruses† | Mimiviridae, Pandoraviridae and Phycodnaviridae | Moniruzzaman et al., 2020; Aherfi et al., 2018 |
| Glutamine synthetase | Condensation of glutamate and ammonia to form glutamine: | R00253 | Uncharacterized viruses† | Mimiviridae | Ha et al., 2021 | |
| Glutaminase | Hydrolysis of glutamine into glutamate | R00256 | Uncharacterized viruses† | Mimiviridae | Moniruzzaman et al., 2020; Ha et al., 2021 | |
| Lipide catabolism and β-Oxydation | Triacylglycerol lipase | Degrades triacylglycerol into glycerol and fatty acids | R01369 | Prymnesium kappa virus RF01 | Mimiviridae | Blanc-Mathieu et al., 2021 |
| Fatty-acyl-CoA Synthase | Conversion of a acetyl-CoA and seven malonyl-CoA molecules to produce a Palmitoyl-CoA | R05190 | Prymnesium kappa virus RF01 | Mimiviridae | Blanc-Mathieu et al., 2021 | |
| CoA-transferase | Conversion acyl-CoA and acetate into fatty acid anion and acetyl-CoA. | R00393 | Prymnesium kappa virus RF01 | Mimiviridae | Blanc-Mathieu et al., 2021 | |
| Acyl-CoA dehydrogenase | Desaturation of the acyl-CoA esters | R00392 | Prymnesium kappa virus RF01 and others uncharacterized viruses† | Mimiviridae | Blanc-Mathieu et al., 2021 | |
| Citric Acid Cycle | Succinate dehydrogenase | Conversion of succinate into fumarate | R02164 | Prymnesium kappa virus RF01, Pandoravirus massiliensis and others uncharacterized viruses† | Mimiviridae, Pandoraviridae and Phycodnaviridae | Moniruzzaman et al., 2020; Aherfi et al., 2021; Blanc-Mathieu et al., 2021; Ha et al., 2021; |
| Citrate synthase | Claisen condensation between acetyl CoA and oxaloacetate to yield, after hydrolysis of the thioester bond, citrate and CoA | R00351 | Pandoravirus massiliensis and others uncharacterized viruses† | Pandoraviridae and Mimiviridae | Aherfi et al., 2021; Moniruzzaman et al., 2020 | |
| Aconitase | Catalyzes the stereospecific isomerization of citrate to isocitrate via cis-aconitate in a non-redox reaction | R01324 | Pandoravirus massiliensis and others uncharacterized viruses† | Pandoraviridae and Mimiviridae | Moniruzzaman et al., 2020; Rodrigues et al., 2019; Aherfi et al., 2021 | |
| Isocitrate/isopropyl malate dehydrogenase | Oxidative decarboxylation of isocitrate, resulting in alpha-ketoglutarate and carbon dioxide. | R00267 / R01652 | Pandoravirus massiliensis and others uncharacterized viruses† | Pandoraviridae and Mimiviridae | Aherfi et al., 2021; Moniruzzaman et al., 2020 | |
| Malate synthase | Conversion of enzyme are acetyl-CoA, H2O, and glyoxylate into (S)-malate and CoA. | R00472 | Uncharacterized viruses† | Mimiviridae | Ha et al., 2021 | |
| Alpha-ketoglutarate decarboxylase | Conversion of α-ketoglutarate to succinyl-CoA and produces NADH directly providing electrons for the respiratory chain | R00272 | Pandoravirus massiliensis | Pandoraviridae | Aherfi et al., 2021 | |
| Fumarase | Conversion of fumarate to L-malate | R01082 | Pandoravirus massiliensis | Pandoraviridae | Aherfi et al., 2021 | |
| Fermentation | Pyruvate formate-lyase | Catalyzes the reaction of pyruvate +CoA acetyl-CoA +formate | R00212 | Tetraselmis virus | Phycodnaviridae | Müller et al., 2012; Schvarcz and Steward, 2018; Sun et al., 2020 |
| Formate-lyase activating enzyme | Converts pyruvate and CoA into acetyl CoA and formate | R04710 | Tetraselmis virus | Phycodnaviridae | Müller et al., 2012; Schvarcz and Steward, 2018 | |
| Gluconeogenesis | Fructose bisphosphatase | Converts fructose-1,6-bisphosphate to fructose 6-phosphate | R00762 | Uncharacterized viruses† | Mimiviridae | Moniruzzaman et al., 2020; Ha et al., 2021 |
| Phosphoenolpyruvate carboxykinase | Converts oxaloacetate into phosphoenolpyruvate and carbon dioxide. | R00341 | Uncharacterized viruses† | Mimiviridae | Moniruzzaman et al., 2020; Ha et al., 2021 | |
| Pyruvate carboxylase | Catalyzes the conversion of pyruvate to oxaloacetate | R00344 | Uncharacterized viruses† | Mimiviridae | Moniruzzaman et al., 2020 | |
| Glycolysis | Glyceraldehyde-3-phosphate dehydrogenase | Conversion of pyruvate to oxaloacetate | R01061 | Uncharacterized viruses† | Mimiviridae and Phycodnaviridae | Moniruzzaman et al., 2020 |
| Phosphoglycerate mutase | Transfers the phosphate from 3-phosphoglyceric acid (3 PG) to the second carbon to form 2-phosphoglyceric acid (2 PG) | R01518 | Uncharacterized viruses† | Mimiviridae and Phycodnaviridae | Moniruzzaman et al., 2020; Ha et al., 2021 | |
| Phosphoglycerate kinase | Catalyzes the formation of ATP from ADP and 1,3-diphosphoglycerate | R01512 | Uncharacterized viruses† | Mimiviridae and Phycodnaviridae | Moniruzzaman et al., 2020 | |
| Photosynthesis | Rhodopsin | Generating a proton motive force across the cell membrane (light dependent) | R02903 | Organic Lake Phycodnavirus 2 and Phaeocystis globosa virus | Phycodnaviridae | Needham et al., 2019; Yutin and Koonin, 2012; Schulz et al., 2020 |
| Mannitol metabolism | Mannitol 1-phosphate dehydrogenase | Converts D-mannitol 1-phosphate and NAD +into fructose 6-phosphate, NADH and H+. | R00758 | Tetraselmis virus | Phycodnaviridae | Schvarcz and Steward, 2018 |
| Saccharide degradation | Alpha-galactosidase | Catalyzes the removal of terminal α-galactose groups from substrates such as glycoproteins and glycolipids | R01101 | Tetraselmis virus | Phycodnaviridae | Schvarcz and Steward, 2018 |
-
*
KEGG codes for the biochemical reactions described (https://www.genome.jp/kegg/reaction/).
-
†
Enzymes detected in NCLDVs from metagenome-assembled genome analysis.
List of enzymes with other biological roles detected in NCLDVs.
The enzymes were grouped according to the biological process to which they belong and associated with the giant virus and/or the family in which they were identified.
| Biological process | Enzyme | Function | Detected in | Family |
|---|---|---|---|---|
| Oxidative stress regulation | Superoxide dismutase | Catalyzes the dismutation of the superoxide radical into ordinary molecular oxygen and hydrogen | Emiliania huxleyi virus, Megavirus chiliensis, and others uncharacterized viruses* | Mimiviridae and Phycodnaviridae |
| Glutathione peroxidase | Reduces free hydrogen peroxide to water. | Emiliania huxleyi virus and others uncharacterized viruses* | Mimiviridae and Phycodnaviridae | |
| Ion’s transport and assimilation | Ammonium transporter | Mediates the transport of ammonium ions | Ostreococcus virus 6 | Phycodnaviridae |
| Phosphate transporter | Mediates the transport of phosphate ions | Uncharacterized viruses | Mimiviridae and Phycodnaviridae | |
| Sulfur transporter | Mediates the transport of sulfur ions | Uncharacterized viruses | Mimiviridae and Phycodnaviridae | |
| Magnesium transporter | Mediates the transport of magnesium ions | Uncharacterized viruses | Mimiviridae and Phycodnaviridae | |
| Iron transporter | Mediates the transport of iron ions | Uncharacterized viruses | Mimiviridae and Phycodnaviridae | |
| Ferritin | Iron storage protein | Uncharacterized viruses | Mimiviridae and Phycodnaviridae | |
| Ferric reductases | Oxidation of NADPH and transference the electron to reduce metals like iron and copper | Uncharacterized viruses | Mimiviridae and Phycodnaviridae | |
| Multicopper oxidases | Oxidation of different substrates by accepting electrons at a mononuclear copper centre and transferring them to a trinuclear copper centre. | Uncharacterized viruses | Mimiviridae and Phycodnaviridae | |
| Biosynthesis of glycosphingolipids | Serine palmitoyltransferase | Catalyzes the decarboxylative condensation of L-serine and palmitoyl coenzyme A to 3-ketodihydrosphingosine. | Coccolitho virus | Phycodnaviridae |
| Polysaccharide biosynthesis | Hyaluronan synthase | Produces the glycosaminoglycan hyaluronan from UDP-α-N-acetyl-D-glucosamine and UDP-α-D-glucuronate | Chlorovirus CVK2 | Phycodnaviridae |
| Chitin synthase | Produces Uridine diphosphate (UDP) and [[[1,4-(N-acetyl-beta-D-glucosaminyl)]n+1]] from UDP-GlcNAc and [[[1,4-(N-acetyl-beta-D-glucosaminyl)]n]] | Chlorovirus CVK2 | Phycodnaviridae | |
| Sugar metabolism | GDP-D-mannose 4,6 dehydratase | Conversion of GDP-(d)-mannose to GDP-4-keto, 6-deoxy-(d)-mannose | Paramecium bursaria Chlorella virus 1 | Phycodnaviridae |
| GDP-4-keto-6-deoxy-D-mannose epimerase/reductase | Converts GDP-4-keto-6-deoxy-d-mannose into GDP-l-fucose | Paramecium bursaria Chlorella virus 1 | Phycodnaviridae | |
| Polysaccharides degradation | Chitinase | Chitin degradation by cleaves the disaccharide to its monomer subunits | Chlorella virus PBCV-1 | Phycodnaviridae |
| 1–3-beta glucanase | Successive hydrolysis at the nonreducing end of the glucan, resulting in the formation of oligosaccharides and glucose | Chlorella virus PBCV-1 | Phycodnaviridae | |
| Pectate lyase | Randomly cleaves α–1,4-polygalacturonic acid via a β-elimination reaction | Aureococcus anophagefferens virus | Phycodnaviridae |
-
*
Enzyme’s genes were detected in NCLDVs from metagenome-assembled genome analysis.





