Actin-regulated Siglec-1 nanoclustering influences HIV-1 capture and virus-containing compartment formation in dendritic cells
Figures
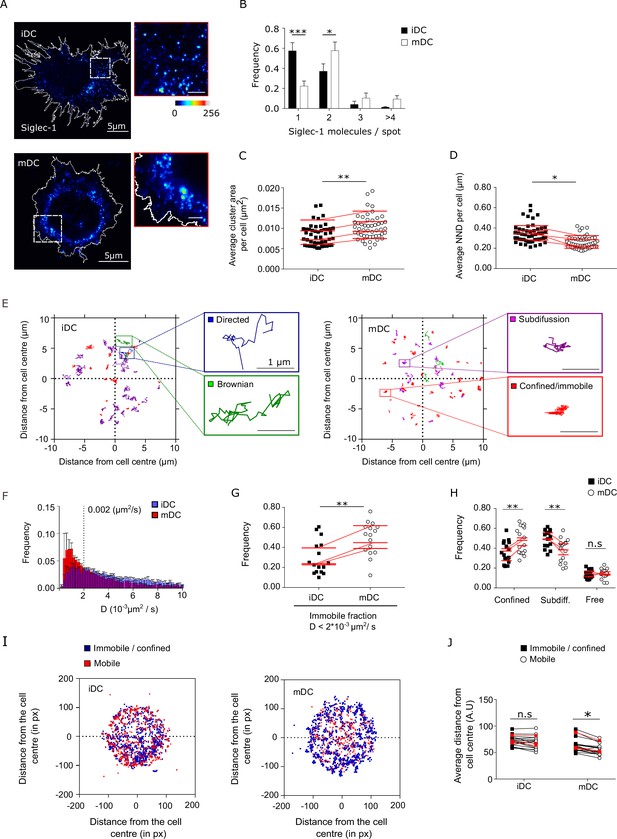
Dendritic cell (DC) activation induces the formation of Siglec-1 nanoclusters with decreased mobility.
(A) Representative STED images of Siglec-1 in immature DCs (iDCs) and mature DCs (mDCs). The pseudo-color code denotes the intensity of individual Siglec-1-labeled spots, from monomer (dark blue) to nanocluster (red-to-white). The insets show enlarged regions highlighted by the white boxes on the main images (scale bar: 1 µm). Siglec-1 has been labeled using full Abs (see Materials and methods). Control experiments using single-chain Abs and different fixation methods are shown in Figure 1—figure supplement 1E–I, Figure 1—figure supplement 1—source data 1, and Figure 1—figure supplement 1—source data 2. (B) Frequency of the number of Siglec-1 molecules per spot, in iDC and mDC. Bars represent the mean ± standard error of the mean (SEM) of 3 different donors (minimum of 9 cells/donor and condition). (C) Average size of Siglec-1 spots and (D) proximity between individual spots, calculated by measuring the nearest neighbor distance (NND) between spots per cell, in iDC and mDC. Each symbol in (C, D) corresponds to an individual cell, red lines are the average value on iDCs and mDCs for each donor (4 donors, 9 cells/donor and cell type). (E) Representative Siglec-1 trajectories with the receptor labeled with single-chain Abs at sub-labeling conditions as recorded on an iDC (left) and an mDC (right). The magnified insets show examples of different types of motion as classified by the MSS analysis. (F) Frequency of the diffusion coefficients for individual Siglec-1 trajectories on iDCs and mDCs. The dash vertical line corresponds to the diffusion threshold to separate immobile from mobile particles (0.002 µm2/s). Each data set represents the mean ± SEM of 3 donors (minimum of 3 cells and 83 trajectories/cell). (G) Fraction of immobile trajectories (<0.002 μm2/s). Each symbol corresponds to an individual cell, red lines are the average value on iDCs and mDCs for each donor (3 donors). (H) Fraction of mobile trajectories (>0.002 μm2/s) classified as confined, sub-diffusive or free. Each symbol corresponds to an individual cell, red lines show the average value on iDCs and mDCs for 3 donors analyzed. (I) Plots showing the center of mass of individual Siglec-1 trajectories (average x,y position in all the frames of a given trajectory) in iDC and mDC. Blue dots correspond to immobile and confined trajectories, and red dots correspond to sub-diffusive and free trajectories. The graph shows all the trajectories analyzed for a minimum of 8 cells per condition from one donor. (J) Distance from the cell center of immobile/confined (black squares) and sub-diffusive/free (empty circles) trajectories in iDC and mDC. Black squares are paired to the empty circles within the same cell. Red symbols correspond to the average values of a minimum of 3 iDC and mDC cells per donor (3 donors). ns, p > R 0.05, * p < % 0.05, ** p < % 0.001; *** p < % 0.0001.
-
Figure 1—source data 1
Excel file containing the source data for Figure 1B–D, G, H.
- https://cdn.elifesciences.org/articles/78836/elife-78836-fig1-data1-v2.zip
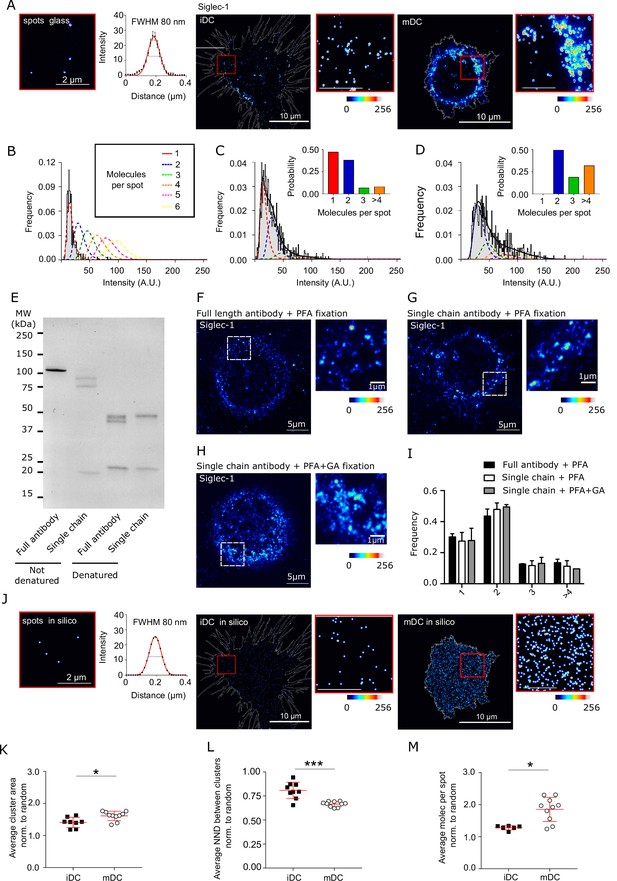
Different nanoscale distribution of Siglec-1 in immature versus mature DCs.
(A) STED images of Siglec-1 antibodies non-specifically adhered to glass and labeled with Fab-Atto488 secondary antibodies with a corresponding Gaussian fit of the intensity (left); and representative STED images of Siglec-1 on immature dendritic cells (iDCs) and mature dendritic cells (mDCs) together with enlarged views. Scale bars in the enlarged views are 1μm (B) Histogram of the spot intensities for Siglec-1 antibodies adhered on glass. The black line (and shaded region) shows the lognormal fit of the experimental data, and the dashed lines correspond to the calculated intensity distribution functions for different numbers of molecules per spot. Intensity distributions of Siglec-1 spots from the representative images of iDC (C) and mDC (D). Black lines are the fit of experimental intensity distributions from which we extract the probability of having 1 up to >4 molecules/spot (see bar graphs in the insets). (E) Representative Coomassie-stained NuPAGE gel showing the generation of single-chain antibodies from a mouse monoclonal [Hsn 7D2] anti-Siglec-1 antibody. Full (lanes 1 and 3) and single-chain antibodies (lanes 2 and 4) that were either left untreated (not denatured; lanes 1 and 2) or reduced with beta-mercaptoethanol (denatured; lanes 3 and 4) were loaded in the gel, as denoted. Representative STED images of Siglec-1 in mDCs fixed using 4% paraformaldehyde (PFA) (F, G) or 1% PFA + 0.2% glutaraldehyde (GA) (H) and labeled using the full length anti-Siglec-1 antibody (F) or the single-chain Siglec-1 antibody (G, H). The pseudo-color code denotes the intensity of individual Siglec-1-labeled spots, from monomer (dark blue) to nanocluster (red-to-white). The insets show enlarged regions highlighted by the white dashed boxes on the main images (scale bars are 5 μm in the main images and 1 μm in the zoomed-in panels). (I) Frequency of the number of Siglec-1 molecules per spot in mDCs corresponding to panels (F–H). Bars represent the mean ± standard error of the mean (SEM) of at least 2 different donors (minimum of 3 cells/donor and condition). (J) In silico spots generated with an Full-width-at-half-maximum (FWHM) as extracted from the experimental data shown in panel A for spots on glass (left), Siglec-1 staining in iDC (middle), and mDC (right). To generate the in silico images we quantified the total number of Siglec-1 receptors by taking into account the number of molecules per spot and total number of spots. The resulting number of molecules was randomly distributed within the cell area as spots that were convoluted with the FWHM of experimental data (60–80 nm). Average area of Siglec-1 spots per cell (K), average nearest neighbor distance (NND) between Siglec-1 spots per cell (L), and average number of molecules per spot per cell (M), in iDC and mDC. For each cell analyzed the values are normalized to the respective in silico values of Siglec-1 random distributions (mean + standard deviation [SD] of one representative experiment; N = 3). * p< % 0.05, *** < p % 0.0001.
-
Figure 1—figure supplement 1—source data 1
Zip folder containing the uncropped gel shown in Figure 1—figure supplement 1E with the relevant bands clearly labeled.
- https://cdn.elifesciences.org/articles/78836/elife-78836-fig1-figsupp1-data1-v2.zip
-
Figure 1—figure supplement 1—source data 2
Excel file containing the source data for panels I, K–M.
- https://cdn.elifesciences.org/articles/78836/elife-78836-fig1-figsupp1-data2-v2.zip
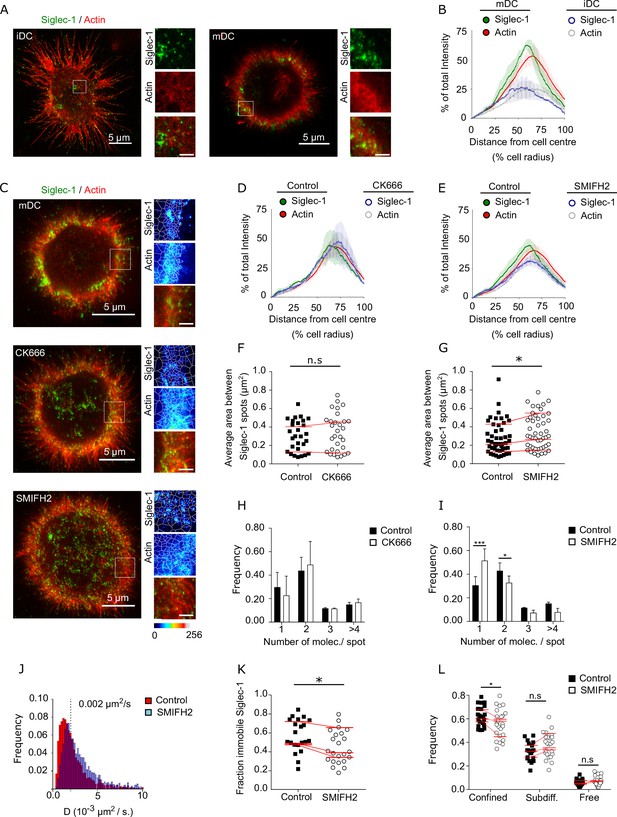
Formin-dependent actin polymerization regulates Siglec-1 nanoscale organization and mobility.
(A) Representative dual color STED images of immature dendritic cell (iDC; left) and mature dendritic cell (mDC; right) stained with Siglec-1 and actin (labeled with SiR-actin). Insets show magnified areas with pseudo-color-coded intensities of Siglec-1 (upper), actin (middle), and merged image (lower) (scale bar: 1 µm). (B) Siglec-1 and actin intensity from the cell center toward the edge expressed as percentage of total intensity for each marker in iDC and mDC. Lines represent the mean ± standard error of the mean (SEM) of 3 different donors (minimum of 9 cells/donor and condition). (C) Representative STED images of mDC stained for Siglec-1 and actin, together with enlarged insets, in control conditions and after 1-hr treatment with CK666 (100 µM) or SMIFH2 (25 µM). The white lines in the insets delimit the Voronoi areas between adjacent Siglec-1 spots; the smaller the areas, the closer single Siglec-1 spots reside from each other (scale bars insets: 1 µm). Siglec-1 and actin intensity from the cell center toward the edge expressed as percentage of total intensity for each marker in control mDC and treated with CK666 (D) or SMIFH2 (E). Lines represent the mean ± SEM of 2 (D) and 4 (E) different donors (minimum 10 cells/donor and condition). (F) Average Voronoi areas between contiguous Siglec-1 spots per cell in control and CK666-treated cells (2 donors, at least 10 cells/donor). Red lines connect the average of all the cells in each donor. (G) Similar to (F) for control and SMIFH2-treated cells (3 donors, at least 10 cells/donor). (H) Frequency histogram of the number of Siglec-1 molecules per spot in control and CK666-treated mDC. Mean ± SEM of a minimum of 10 cells/donor from 2 donors. (I) Similar to (H) for control and SMIFH2-treated cells (3 donors, at least 10 cells/donor). (J) Frequency of Siglec-1 diffusion coefficients in control mDC (n = 8 cells, 4424 trajectories) and cells treated with SMIFH (n = 8 cells, 5002 trajectories) from one donor. (K) Fraction of immobile trajectories in control and SMIFH2-treated mDCs. A total of 21 (control) and 24 (SMIFH2) cells were quantified from 3 donors (minimum 6 cells and 1261 trajectories/donor and condition). Red lines connect the average of each individual donor. (L) Fraction of mobile trajectories classified as confined, sub-diffusive or free in control and SMIFH2-treated mDC. Each symbol corresponds to an individual cell, red lines show the average value of all cells from 3 donors (minimum 6 cells/donor and condition). ns, p > R 0.05, * p < % 0.05, *** p < % 0.0001.
-
Figure 2—source data 1
Excel file containing the source data for Figure 2B, D–L.
- https://cdn.elifesciences.org/articles/78836/elife-78836-fig2-data1-v2.zip
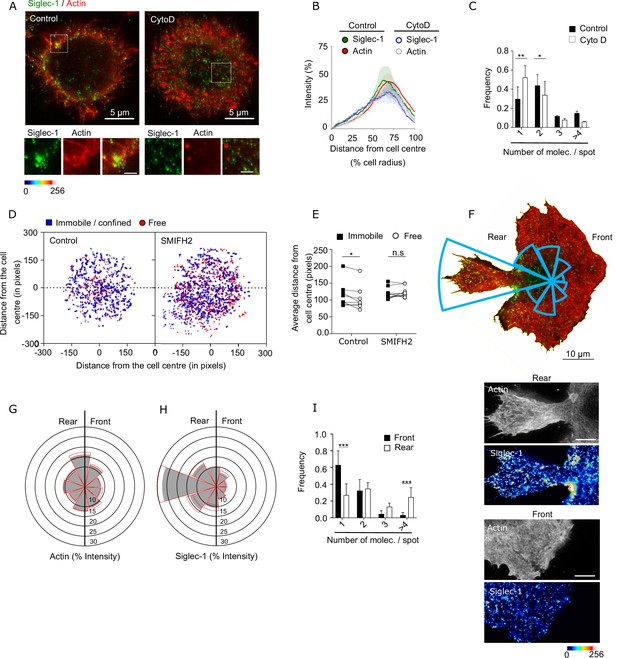
Formin-dependent actin polymerization regulates Siglec-1 nanoscale organization and mobility.
(A) Representative dual color STED images of mature dendritic cell (mDC) stained with Siglec-1 and SiR-actin in control conditions and after 1-hr treatment with Cytochalasin D (Cyto D, 2 µg/ml). The bottom panels correspond to magnified images of the dashed squares (scale bars: 2 µm). (B) Siglec-1 and actin intensity from the cell center toward the edge expressed as percentage of total intensity for each marker in immature dendritic cell (iDC) control mDC and cells treated with Cyto D. Lines represent the mean + standard error of the mean (SEM) of two different donors in which a minimum of 10 cells/donor and condition were analyzed. (C) Frequency histogram of the number of Siglec-1 molecules per spot in control mDCs and cells treated with Cyto D. Mean + SEM of a minimum of 9 cells/donor from two donors. (D) Plots showing the center mass of individual Siglec-1 trajectories (average x,y position in all the frames of a trajectory) in control mDC and cells treated with SMIFH2, colored-coded according to their mobility. The graph shows the centers of all the trajectories analyzed for a minimum of 8 cells per condition from one representative experiment (N = 3). (E) Distance from the cell center of immobile/confined and sub-diffusive/free trajectories in control mDC and cells treated with SMIFH2. Black squares show the average distance from the cell center of all immobile/confined trajectories in one cell, and are paired to the average values of all sub-diffusive/free trajectories within the same cell (empty circles). (F) Representative STED image of a mDC fixed and stained for Siglec-1 and SiR-actin after injection between a glass coverslip and a layer of agarose with CCL19 at 2.5 µg/ml. The rose plot histogram shows the distribution of Siglec-1 staining from rear to front in 40° bins. On the bottom magnified insets of Siglec-1 and actin at the rear and the front of the cell (scale bar: 2 µm). (G, H) Rose plot histograms of actin and Siglec-1 intensity distributions on the front and rear end of mDCs subjected to a CCL19 homogeneous gradient in an under-agarose assay. Graphs show the mean + SEM (red lines) of two donors with a minimum of 10 cells/donor. (I) Frequency histogram of the number of Siglec-1 molecules per spot at the rear and the front of mDCs treated as in G. Mean + SEM of a minimum of 10 cells/donor from two donors. ns, p > R 0.05, * p < % 0.05, ** p < % 0.001; *** p< % 0.0001.
-
Figure 2—figure supplement 1—source data 1
Excel file containing the source data for panels B, C, E, I.
- https://cdn.elifesciences.org/articles/78836/elife-78836-fig2-figsupp1-data1-v2.zip
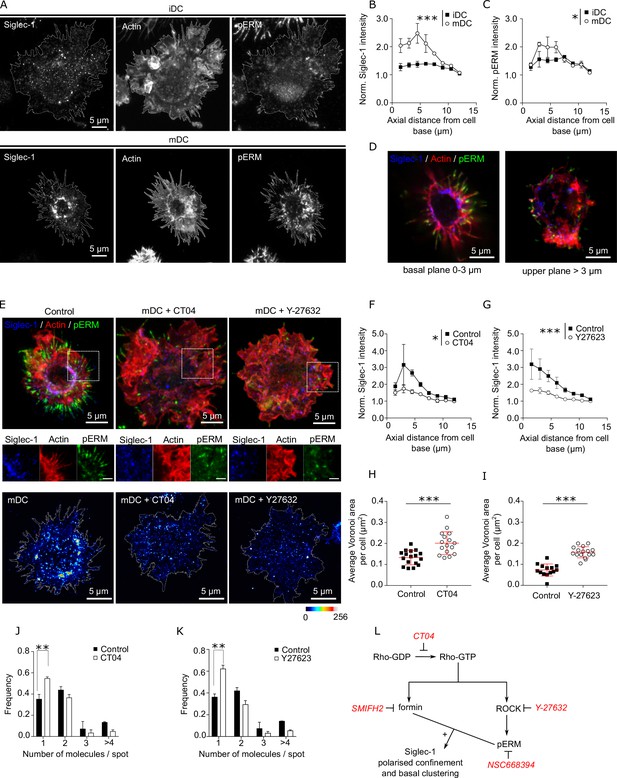
Siglec-1 confinement and basal nanoclustering occur in polarized regions of the plasma membrane characterized by RhoA activity.
(A) Representative maximum intensity projections of 3D confocal images of immature dendritic cell (iDC; upper row) and mature dendritic cell (mDC; lower row) immunostained with anti-Siglec-1, SiR-actin, and pERM. Plots of the axial distribution from the cell base of Siglec-1 (B) and pERM (C) intensities. Values in each plane show the mean intensity in steps of 1.5 µm from the cell base, normalized by the minimum mean intensity within the whole stack. Each symbol represents the mean ± standard error of the mean (SEM) from two donors (at least 10 cells per condition). (D) Confocal projections of the basal (0–5 µm) and the apical (>5 µm) parts of the mDC example shown in (A). (E) Top panels: confocal maximum intensity projections images of control mDC and cells treated for 3 hr with CT04 (2 µg/ml), or for 1 hr with Y-27632 (30 µM), fixed and immunostained with anti-Siglec-1, SiR-actin, and pERM. Middle panels: enlarged images from the highlighted regions of the top images (scale bar: 2 µm). Bottom panels: STED images of mDCs treated as described above and immunostained with anti-Siglec-1. Siglec-1 intensity as function of axial distance from the cell base in control mDC and cells treated with CT04 (F) and Y-27623 (G). Results show the mean ± standard error of the mean (SEM) of two donors, with at least 7 (F) and 9 (G) cells/donor and condition. Average Voronoi areas between contiguous Siglec-1 spots in control mDC, and cells treated with CT04 (H) and Y-27623 (I). Each dot shows the average Voronoi area per cell; results show the average ± SD of one representative experiment (n = 2) with a minimum of 16 (H) and 14 (I) cells. Number of Siglec-1 molecules per spot in control mDCs and cells treated with CT04 (J) and Y-27623 (K). Mean ± SEM of 2 donors, with a minimum of 10 (J) and 9 (K) cells/donor. (L) Scheme of the different pathways downstream of Rho activation targeted by different inhibitors. Statistics in the legends of panels B, C, F, G correspond to the significance of a two-way analysis of variance (ANOVA) test depending on maturation status (iDC vs. mDC in A, B) or treatment control vs. CT04 or Y-27623 in F, G. * p < % 0.05, ** p < % 0.001; *** p < % 0.0001.
-
Figure 3—source data 1
Excel file containing the source data for Figure 3B, C, F–K.
- https://cdn.elifesciences.org/articles/78836/elife-78836-fig3-data1-v2.zip
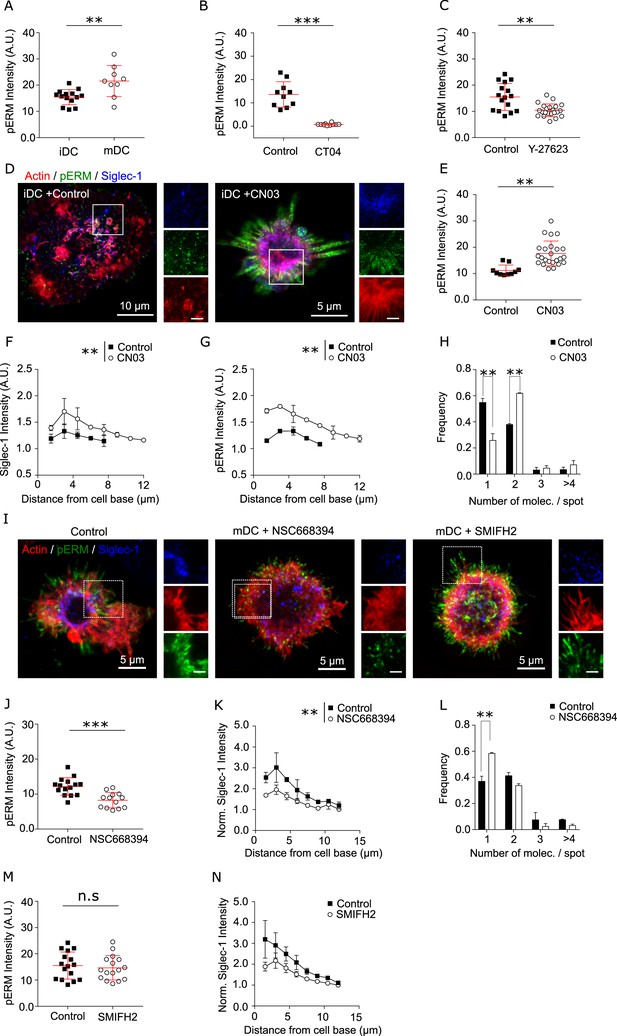
Siglec-1 confinement and basal nanoclustering occurs in polarized regions of the plasma membrane characterized by RhoA activity.
pERM mean intensity values in control immature dendritic cells (iDCs) and control mature dendritic cells (mDCs) (A) or mDCs treated with CT04 (B) or Y-27623 (C). Results show the mean + standard deviation (SD) of one representative experiment (N = 2) in which a minimum of 9 (A), 7 (B), and 10 (C) cells/donor and condition were analyzed. (D) STED images of the basal plane for control iDCs or cells treated for 3 hr with CN04 (2 µg/ml) and immunostained with anti-Siglec-1, SiR-actin, and pERM. Insets correspond to enlarged images of the regions highlighted in the main images (scale bars: 2 µm). (E) pERM mean intensity values in control iDCs and cells treated with CN03. Results show the mean + SD of one representative experiment (N = 2) with a minimum of 9 cells/donor and condition. (F) Plots of the axial distribution of Siglec-1 intensity in control iDCs and cells treated with CN03 obtained from 3D confocal images (10–15 stacks). Values in each plane show the mean intensity in steps of 1.5 µm from the cell base, normalized by the minimum mean intensity within the whole stack. Each symbol represents the mean + standard error of the mean (SEM) of two donors with at least 9 cells per condition. (G) Same as in (F) but for pERM intensity. (H) Frequency histogram of the number of Siglec-1 molecules per spot in control iDCs and cells treated with CN03. Mean + SEM of two donors, at least 9 cells/donor and condition. (I) Confocal maximum intensity projection images of control mDCs and cells treated for 3 hr with NSC668394 (250 µM), or for 1 hr with SMIFH2 (25 µM), immunostained with anti-Siglec-1, SiR-actin, and pERM. Insets correspond to enlarged images of the regions highlighted in the main images. Scale bars: 2 µm. (J) pERM mean intensity values in control mDC and cells treated with NSC668394. Mean + SD of a minimum of 8 cells per condition from one representative experiment (N = 2). (K) Same as in (F) but in control mDCs and cells treated with NSC668394. Each symbol represents the mean + SEM of two donors, with at least 9 cells per condition. (L) Frequency histogram of the number of Siglec-1 molecules per spot in control mDCs and cells treated with NSC668394. Mean + SEM of two donors, at least 9 cells/donor and condition. (M) pERM mean intensity values in control and SMIFH2-treated mDC. Data show the mean + SD of one representative experiment (N = 2) with a minimum of 10 cells per condition. (N) Same as in (K) but in control mDC and cells treated with SMIFH2. Each symbol represents the mean + SEM of two donors, with a minimum of 10 cells per condition. * p < % 0.05, ** p < % 0.001; *** p < % 0.0001.
-
Figure 3—figure supplement 1—source data 1
Excel file containing the source data for panels B, C, E–H, J–N.
- https://cdn.elifesciences.org/articles/78836/elife-78836-fig3-figsupp1-data1-v2.zip
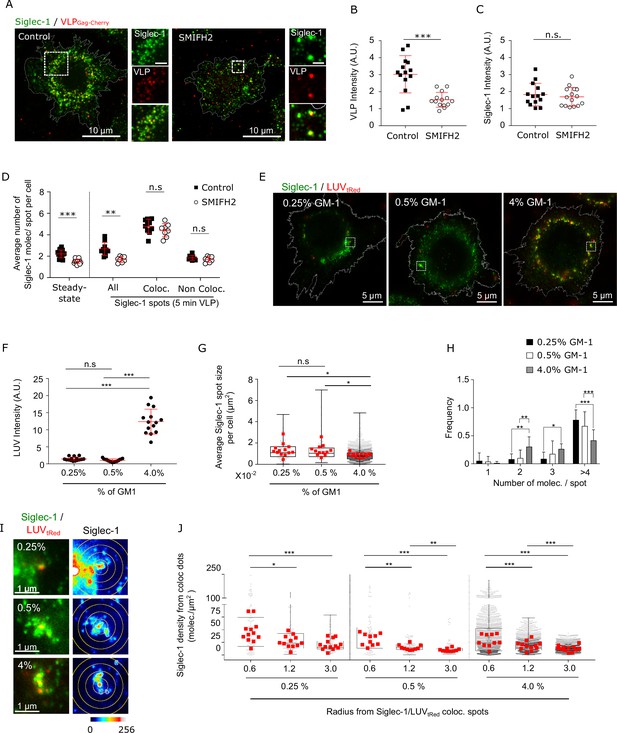
Siglec-1 basal nanoclustering increases the capture of HIV-1-VLPs and the avidity to gangliosides carrying sialic acid ligands.
(A) Confocal images of control mature dendritic cell (mDC), and cells treated with SMIFH2, pulsed for 5 min with VLP-Gag-Cherry and immunostained with anti-Siglec-1. Insets on the right correspond to enlarged views of the regions highlighted in the main images (scale bars: 2 µm). (B) VLP Intensity in control and SMIFH2-treated cells, after 5 min of VLP capture. Mean ± standard deviation (SD) of one representative experiment (n = 2) with 14 cells per condition. (C) Levels of Siglec-1 in control and SMIFH2-treated cells, after 5 min of VLP capture. Mean ± SD of one representative experiment (n = 2), 14 cells per condition. (D) Average number of Siglec-1 molecules per spot per cell in control and SMIFH2-treated mDC, at steady state and after 5 min of VLP capture. Siglec-1 data after 5 min VLP exposure are further separated according to whether Siglec-1 spots colocalize (or not) with VLP particles. Data correspond to the mean + SD of one representative experiment (n = 3 for the absence of VLP and n = 2 for the 5 min VLP) in which a minimum of 8 cells per experiment and condition were analyzed. (E) Representative dual color STED images of mDC immunostained with anti-Siglec-1 after 5-min pulse with tRed-LUVs carrying different GM-1 concentrations. Squares are shown as magnified regions in panel (I). (F) LUV intensity in mDC after 5 min of LUV capture. Mean ± SD of a minimum of 14 cells per condition. (G) Sizes of Siglec-1 spots colocalizing with LUVs. Small dots represent each spot analyzed and red squares correspond to the average size of all colocalizing spots in a cell (at least 12 cells per condition). (H) Number of Siglec-1 molecules per spot colocalizing with LUVs. Results correspond to the mean ± SD of a minimum of 12 cells per condition. (I) Magnified STED images of Siglec-1 colocalizing with LUVs carrying different GM-1 concentrations. Circles with different radii on the Siglec-1 images are drawn starting from the center of each colocalizing spot. (J) Siglec-1 density (i.e., number of Siglec-1 molecules per µm2) at different radius from Siglec-1 spots colocalizing with 0.25–4% GM-1 LUVs (each small dot is an individual colocalizing spot, red squares are the mean values in each cell analyzed, 12 cells per condition). ns, p > R 0.05, * p < % 0.05, ** p < % 0.001; *** p < % 0.0001.
-
Figure 4—source data 1
Excel file containing the source data for Figure 4B–D, F–H, J.
- https://cdn.elifesciences.org/articles/78836/elife-78836-fig4-data1-v2.zip
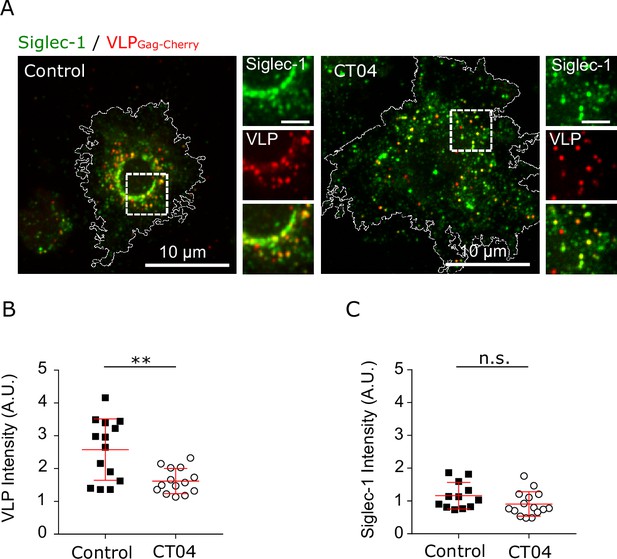
Siglec-1 basal nanoclustering increases the capture of HIV-1-VLPs and the avidity to gangliosides carrying sialic acid ligands.
(A) Confocal images of control mature dendritic cell (mDC), and cells treated with CT04, pulsed for 5 min with VLP-Gag-Cherry and immunostained with anti-Siglec-1. Insets on the right correspond to enlarged views of the regions highlighted in the main images (scale bar: 2 µm). (B) VLP Intensity in control and CT04-treated cells, after 5 min of VLP capture. Mean + standard deviation (SD) of one representative experiment (N = 2) with 14 cells per condition. (C) Levels of Siglec-1 in control and CT04-treated cells, after 5 min of VLP capture. Mean + SD of one representative experiment (N = 2) with 14 cells per condition. ns, p > R 0.05, ** p < % 0.001.
-
Figure 4—figure supplement 1—source data 1
Excel file containing the source data for panels B, C.
- https://cdn.elifesciences.org/articles/78836/elife-78836-fig4-figsupp1-data1-v2.zip
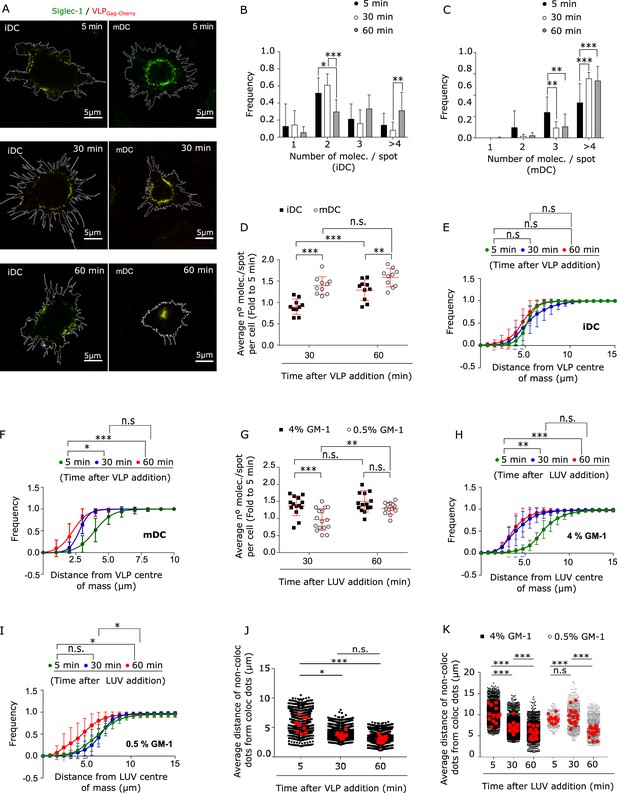
Interaction with HIV-1 particles induces Siglec-1 changes at the nano- and meso-scale toward the formation of the virus-containing compartment (VCC).
(A) Representative STED images of immature dendritic cell (iDC; left) and mature dendritic cell (mDC; right) immunostained with anti-Siglec-1 after different times of VLP-Gag-Cherry capture. Frequency histograms of Siglec-1 molecules/spot colocalizing with VLP-Gag-Cherry in iDC (B) and mDC (C) at different times of VLP capture. Mean ± standard deviation (SD) of a minimum of 10 cells per incubation time in iDC and mDC. (D) Fold increase in the number of Siglec-1 molecules per spot with respect to the average value at 5 min. Mean ± SD of a minimum of 10 cells per time condition in iDC and mDC. Cumulative frequency plots of VLP particles colocalizing with Siglec-1 as a function of the distance from the VLP intensity center-of-mass, in iDC (E) and mDC (F) at different times of VLP capture. Dots show the mean ± SD of a minimum of 9 cells per condition and lines show the sigmoidal fit to the data. (G) Same as in D, but in mDC incubated for different times with LUVs at 0.5% and 4% GM-1 concentrations. Mean ± SD of a minimum of 13 cells per condition. (H–I) Same as in (E, F) but in mDCs incubated for different times with 4% (H) and 0.5% (I) of GM-1. Dots show the mean ± SD of a minimum of 7 cells per time and condition. (J) Box plots showing the distance of non-colocalizing spots to colocalizing Siglec-1/VLP spots in mDC for different incubation times. Small dots show the average distance of the whole population of non-colocalizing spots from each Siglec-1/VLP colocalizing spot, and red squares denote the average of all colocalizing spots in an individual cell (at least 9 cells per time). (K) Same as in (J) but in mDC incubated with 4% and 0.5% GM-1 LUVs (minimum of 11 cells per time and condition). Statistical analysis in the legends of panels (E, F, H, I) corresponds to a one-way analysis of variance (ANOVA) comparing the distance from the center-of-mass of VLPs (E, F) or LUVs (H, I) at which we recover 50% of the total intensity of VLP-Gag-Cherry or LUVs-tRed. ns, p > R 0.05, * p < % 0.05, ** p < % 0.001; *** p < % 0.0001.
-
Figure 5—source data 1
Excel file containing the source data for Figure 5B–K.
- https://cdn.elifesciences.org/articles/78836/elife-78836-fig5-data1-v2.zip
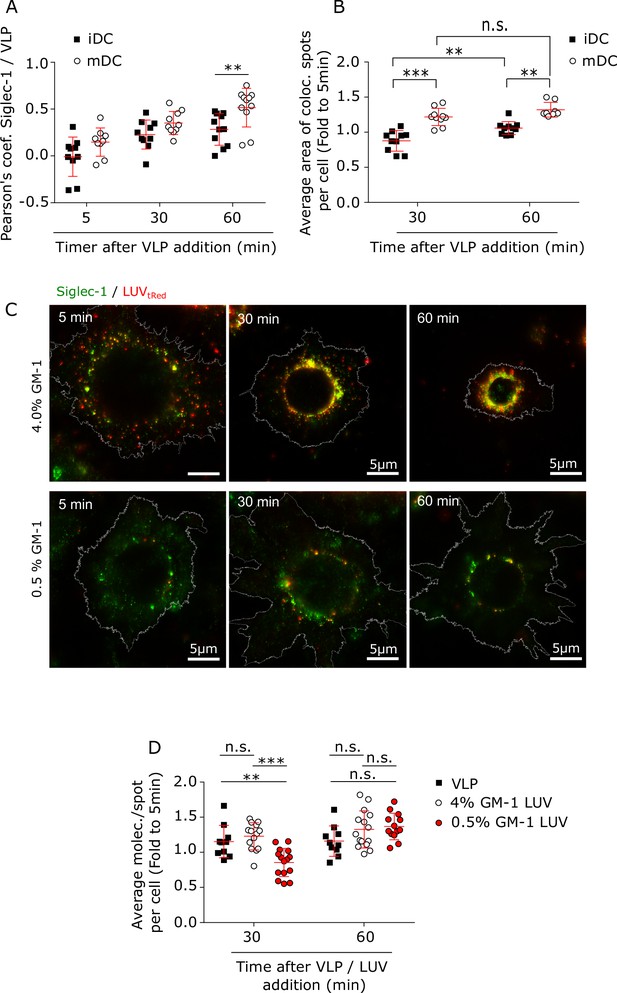
Interaction with HIV-1 particles induces Siglec-1 changes at the nano- and meso-scale towards the formation of the VCC.
(A) Colocalization between Siglec-1 and VLP-Gag-Cherry in immature dendritic cell (iDC) and mature dendritic cell (mDC) after different times of VLP capture as determined by the Pearson coefficient. Mean + standard deviation (SD) of a minimum of 9 cells per condition. (B) Size of Siglec-1 spots colocalizing with VLP-Gag-Cherry in iDC and mDC after different times of VLP capture. The average value of all colocalizing spots in each cell after 30 and 60 min of VLP capture is expressed as a fold increase to the average value after a 5-min pulse. Mean + SD of a minimum of 8 per condition. (C) Representative STED images of mDC fixed and immunostained with anti-Siglec-1 after different times of incubation with LUVs carrying 4% (upper panels) and 0.5% (lower panels) GM-1. (D) Number of Siglec-1 molecules per spot not colocalizing with either VLP, 0.5% or 4% GM-1 LUVs in mDC. Each symbol represents the average value of all non-colocalizing spots in each cell after 30 and 60 min of VLP/LUV addition expressed as a fold increase to the average at 5 min. Mean + SD of a minimum of 10 cells per time and condition. ns, p > R 0.05, ** p < % 0.001; *** p < % 0.0001.
-
Figure 5—figure supplement 1—source data 1
Excel file containing the source data for panels A, B, D.
- https://cdn.elifesciences.org/articles/78836/elife-78836-fig5-figsupp1-data1-v2.zip
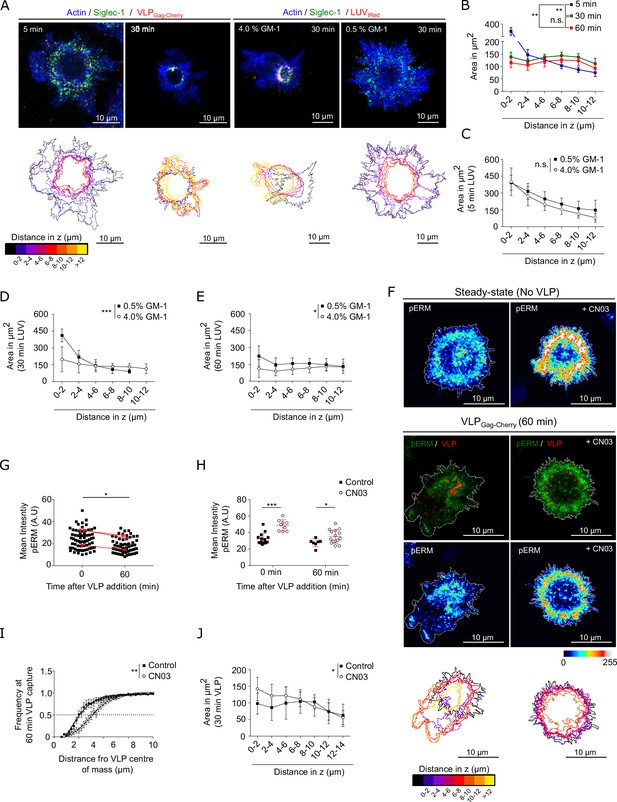
Binding of Siglec-1 to HIV-1 induces a global reorganization of the actin cytoskeleton which allows the final formation of a single virus-containing compartment (VCC) in mature dendritic cells (mDCs).
(A) Confocal maximum intensity projections of 3D confocal images on fixed mDCs immunostained with anti-Siglec-1, SiR-actin and incubated for different times with VLP-Gag-Cherry or tRedLUVs. A ring-shape region or compartment is clearly observed at 30 min of incubation with VLPs or 4% GM1 LUVs, with increased colocalization of Siglec-1, VLPs (or GM1), and actin occurring at axial distances of 4–6 μm above the basal membrane plane (see Figure 6—figure supplement 1A, B and Figure 6—figure supplement 1—source data 1for quantification). Color-coded outlines below each image show the cell perimeter in the axial plane obtained from 3D confocal stacks images (10–15 stacks of 1.5 µm per cell). (B) Cell area in the axial plane in mDC for different incubation times with VLPs. Data correspond to the mean ± standard error of the mean (SEM) of 4 (time points 5 and 30 min) and 3 donors (60 min), 7 cells/donor and condition. (C–E) Plots as in B for mDC pulsed with LUVs carrying 0.5% and 4% GM-1 for different incubation times (C, 5 min; D, 30 min; E, 60 min). Mean ± standard deviation (SD) of minimum of 7 cells per time and condition. (F) Confocal projections of mDC stained with anti-pERM in control conditions (left) or pre-treated for 4 hr with CN03 (right), in the absence (top panels) or the presence of VLP-Gag-Cherry for 60 min (bottom panels). Below scale-colored projections of the cell edges from the bottom to the top of control and CN03-treated mDC after 60 min of VLP capture. (G) Average pERM intensity in mDC before and after 60 min of VLP capture (3 donors, at least 7 cells/donor). Red dots are the mean values of each donor. (H) Average pERM intensity in control mDC and cells pre-treated with the Rho activator CN03 before and after 60 min of VLP capture. Mean ± SD of a minimum of 7 cells per condition. (I) Cumulative frequency plots of VLP intensity as a function of the distance from their center of mass in control mDC and cells pre-treated with CN03 after 60 min of VLP capture. Mean + SD of a minimum of 7 cells per condition. (J) Cell area in the axial plane in control and CN03-treated mDC after 60 min of incubation with VLPs. Mean ± SD of a minimum of 7 cells per condition. Statistics in the legends of panels B–E, J correspond to the significance of a two-way analysis of variance (ANOVA) test depending on time after VLP addition (A) percentage of GM-1 (C–E) or treatment (control mDC vs. CN03-treated cells). Statistics in the legends of panel I correspond to the significance of a Mann–Whitney comparing the distance from the center-of-mass of VLP at which we recover 50% of the total intensity of VLP-Gag-Cherry. ns, p > R 0.05, * p < % 0.05, ** p < % 0.001; *** p < % 0.0001.
-
Figure 6—source data 1
Excel file containing the source data for Figure 6B–E, G–J, and Figure 6G–J.
- https://cdn.elifesciences.org/articles/78836/elife-78836-fig6-data1-v2.zip
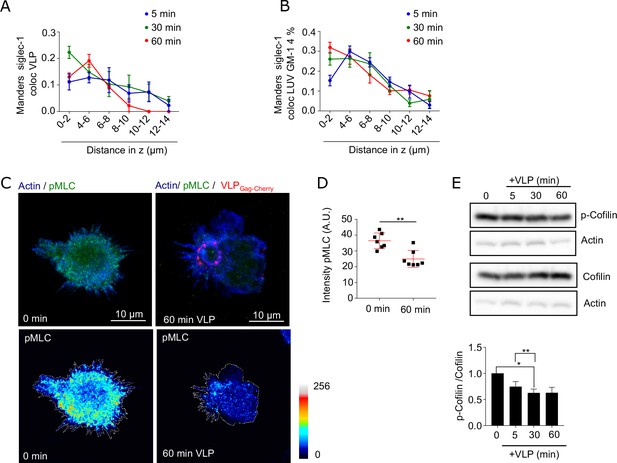
Binding of Siglec-1 to HIV-1 induces a global reorganization of the acti cytoskeleton which allows the final formation of a single VCC in mDCs.
(A) Manders coefficients in the axial plane of Siglec-1 colocalizing with VLP-Gag-Cherry (representative 3D confocal images in Figure 6A). Data in (A) show the mean + standard error of the mean (SEM) of 3 (time points of 5 and 30 min) and 2 donors (60 min) with a minimum of 9 cells/donor and time. (B) Same as in A but in cells incubated with 4.0% GM-1 LUVs. Data correspond to the mean + standard deviation (SD) of a minimum of 7 cells per time and condition. (C) Representative confocal images of mature dendritic cell (mDC) immunostained with anti-pMLC and SiR-actin at steady state and after 60 min of incubation with VLP-Gag-Cherry. Images in the bottom show projections of color-coded intensity values for pMLC. (D) pMLC intensity in mDC at steady state and after 60 min incubation with VLP-Gag-Cherry. Data correspond to the mean + SD from one representative experiment (N = 2) counting 7 cells per condition. (E) Representative immunoblots from cell extracts of mDC at steady state and incubated in the presence of VLP-Gag-Cherry for different times. Equal amounts of protein loaded in each line were revealed with anti-cofilin, p-cofilin (S19), and b-actin. Bars in the bottom graph show the mean + SEM. of three independent experiments quantifying the ratio between p-cofilin and cofilin levels normalized to time 0 min. * p < % 0.05, ** p < % 0.001.
-
Figure 6—figure supplement 1—source data 1
Excel file containing the source data for panels A, B, D, E.
- https://cdn.elifesciences.org/articles/78836/elife-78836-fig6-figsupp1-data1-v2.zip
-
Figure 6—figure supplement 1—source data 2
Zip folder containing the original files of the full raw unedited blots corresponding to Figure 6—figure supplement 1E.
- https://cdn.elifesciences.org/articles/78836/elife-78836-fig6-figsupp1-data2-v2.zip
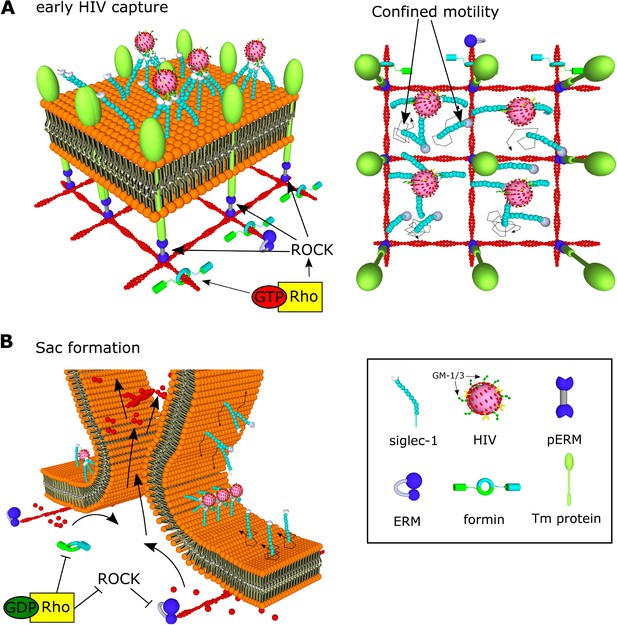
Model of HIV-1 capture by Siglec-1 and polarization toward the formation of the virus-containing compartment (VCC) in mature dendritic cell (mDC).
(A) In steady state, Siglec-1 receptors in mDCs are organized in small nanoclusters proximal to each other. This basal nanoclustering occurs by spatiotemporal confinement of Siglec-1 diffusion into plasma membrane regions dependent on Rho activity, its downstream effector ROCK, pERM, and formin-mediated actin polymerization (see in the middle panel a representation of the cross-section in the 3D reconstruction of a whole cell at early stages of HIV-1 capture). We propose that Siglec-1 mobility is restricted by transmembrane proteins that interact with the filamentous cortical actin cytoskeleton through activated ERM proteins in polarized regions of the mDC membrane (right). Siglec-1 basal nanoclustering is especially relevant to increase the avidity of Siglec-1 for the gangliosides in the membrane of the HIV-1, and therefore regulates the capture capacity of HIV-1 by mDC. (B) At late stages of HIV-1 capture, Siglec-1 clustering increases and most receptors accumulate in a polarized ring-shaped compartment that progressively constrains the membrane (see in the middle panel a representation of the cross-section in the 3D reconstruction of a whole cell at late stages of HIV-1 capture). These changes are accompanied by massive actin rearrangements that include extensive membrane ruffling above the plane in which Siglec-1 and HIV-1 accumulate and by a contraction of the membrane below the ring. This polarization requires a temporal drop in Rho activity. We propose that Rho inactivation would allow for the free diffusion of Siglec-1 through the membrane until being trapped by direct interaction with HIV-1 in the highly dense Siglec-1 regions in which initially most viruses are bound. In parallel, a drop in Rho activity would also lead to actin depolymerization and a decrease in membrane tension together with a retraction of the basal membrane. Both factors would favor the proximity between Siglec-1 bound to HIV-1 clusters and the progressive bending of the membrane until the formation of a single VCC.





