Nanobodies combined with DNA-PAINT super-resolution reveal a staggered titin nanoarchitecture in flight muscles
Figures
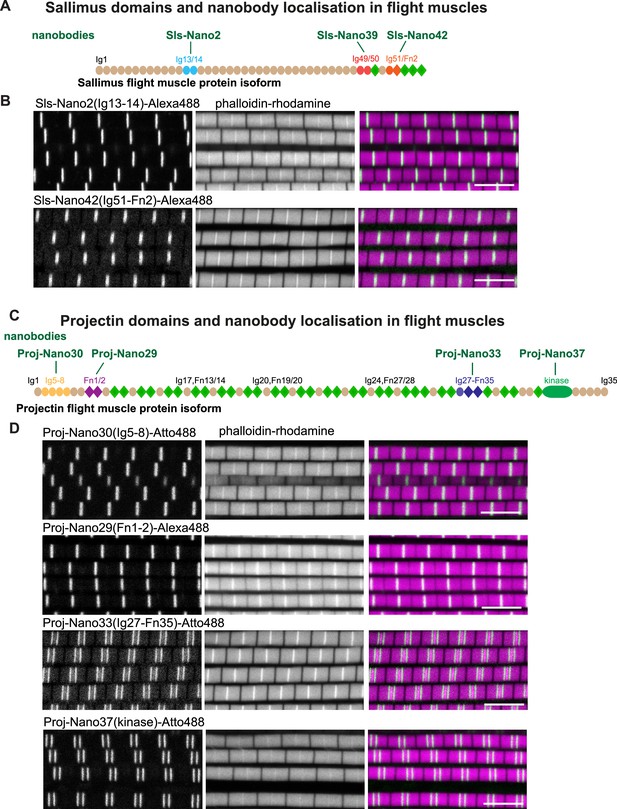
Drosophila titin domain organisation and nanobodies.
(A, C) Sallimus (A) and Projectin (C) flight muscle protein isoforms with the domains recognised by the used nanobodies highlighted in different colours. (B, D) Single confocal sections of flight muscle sarcomeres from adult hemi-thoraces stained for actin with phalloidin (magenta) and the indicated anti-Sls or anti-Projectin nanobodies directly coupled to Alexa488 or Atto488 (green). The Z-disc is revealed by the prominent actin signal. Scale bars 5 µm.
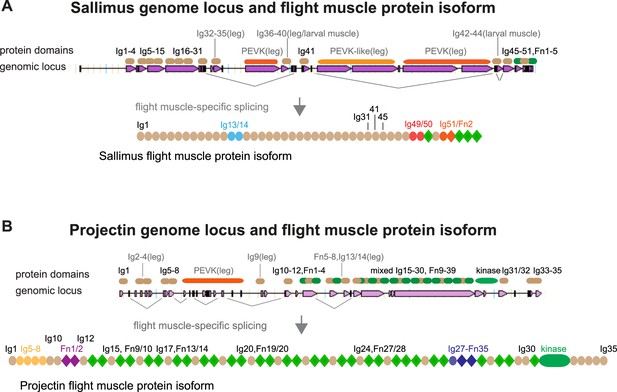
Flight muscle titin isoforms.
(A, B) Genomic organisation of sallimus (A) and projectin/bent (B) genomic loci taken from FlyBase. Pink rectangles indicate exons, lines indicate introns. Above the genomic locus is the rough position of respective protein domains indicated. The domains named in grey are largely specific to leg or larval muscles and not expressed in flight muscles. This is also indicated by the prominent flight muscle-specific splice junctions indicated in grey below the genomic loci. Note that the spring-like PEVK domains in sallimus or projectin are spliced out in flight muscles. This results in the respective Sallimus and Projection domain structure of the flight muscle isoforms.
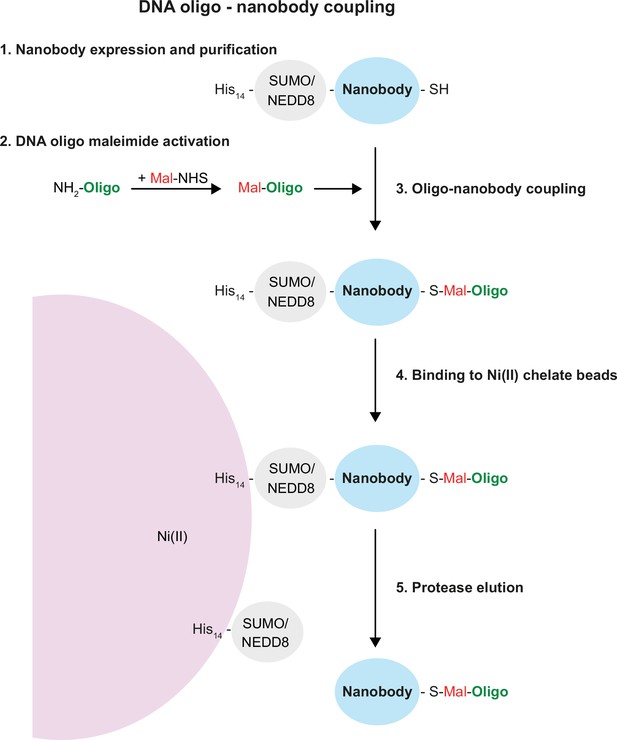
Single-strand DNA oligonucleotide-nanobody coupling.
Schematic representation of the five steps from nanobody purification, oligo activation and coupling, purification, and elution of the oligo-coupled nanobody. See ‘Results’ and ‘Methods’ sections for details.
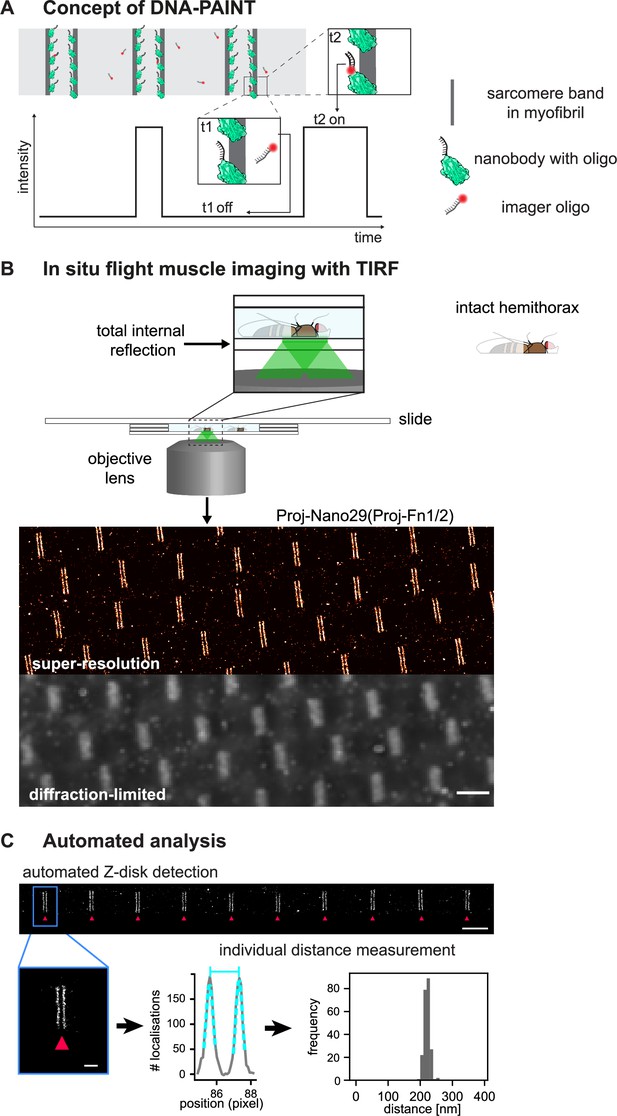
Drosophila flight muscle DNA-PAINT imaging and automated extraction of sarcomeric protein domain positions.
(A) Concept of DNA-PAINT imaging of sarcomeres labelled with an oligo-conjugated nanobody. Binding of the imager oligo to one nanobody results in a strong, detectable intensity burst (t2, blink). (B) Schematic of a mounted intact Drosophila hemi-thorax in a DNA-PAINT imaging chamber enabling total internal reflection (TIRF) illumination. Comparison of the diffraction-limited and the super-resolved result illustrated in one hemi-thorax labelled with Proj-Nano29. Note that the super-resolved image can readily resolve the two bands flanking each Z-disc. Scale bar 2 µm. (C) Automated image analysis for individual Z-discs detection (see Figure 3—figure supplement 2 and ‘Methods’ for details). Individual bands are detected automatically and their centre position is obtained using a Gaussian fit (bottom centre). The distance between the centre of bands for tens of sarcomeres from a single hemi-thorax is then reported in a histogram (bottom right). Scale bar 2 µm (top) and 0.5 µm (bottom).
-
Figure 3—source data 1
Distances between Proj-Nano29 bands in shown example.
- https://cdn.elifesciences.org/articles/79344/elife-79344-fig3-data1-v3.xlsx
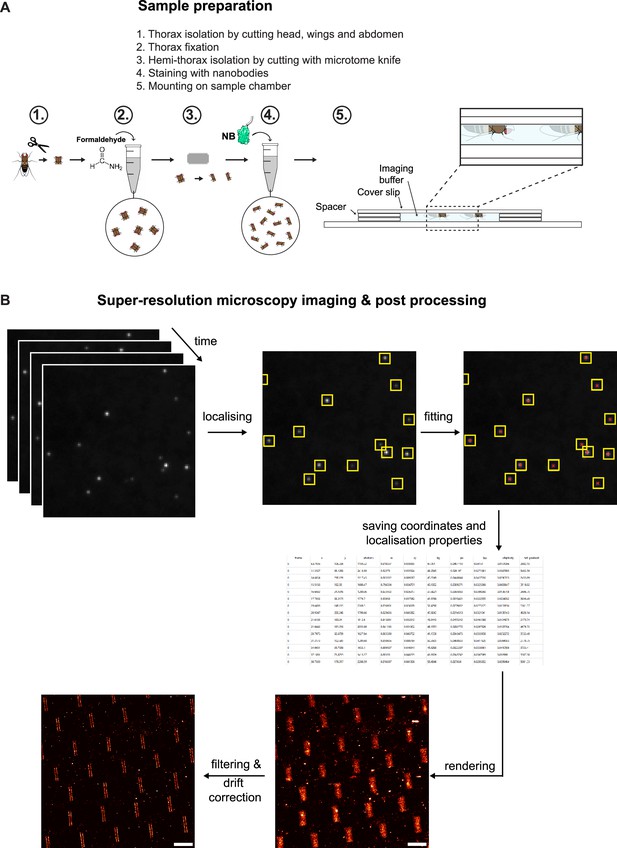
Sample preparation and data processing.
(A) Schemes illustrating the different steps of sample preparation. Thorax isolation, followed by PFA fixation and hemithorax preparation. Fixed hemi-thoraces were stained with nanobodies and were mounted in a sample chamber in imager solution positioning the flight muscles close to the coverslip to enable total internal reflection (TIRF) imaging. (B) DNA-PAINT post-processing workflow. The single-molecule ‘blinking’ events were identified using a threshold detection, and the maximum is determined via a Gaussian fit. The coordinates in time and space and other localisation properties were saved in an hdf5 file. The hdf5 was then drift-corrected and rendered using Picasso render.
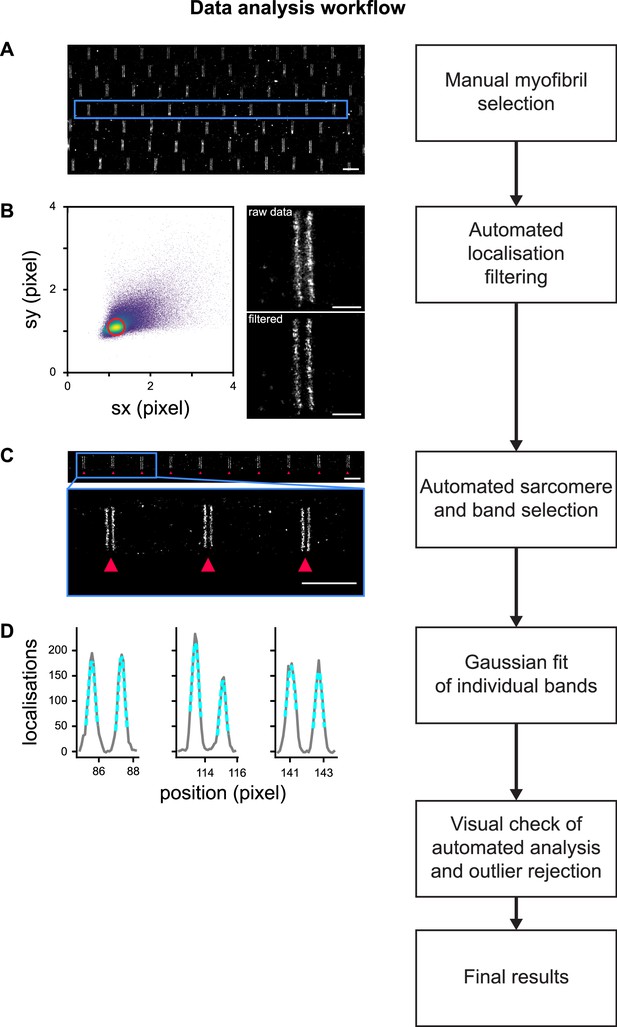
Data analysis workflow.
(A) Individual myofibrils were manually selected using the rectangular selection tool from Picasso (Schnitzbauer et al., 2017). Scale bar 2 µm. (B) To limit localisation events arising from multiple emitters that create artefacts (Lelek et al., 2021), localisations are filtered based on the standard deviation of their Gaussian fits. Localisations kept are within a disc in the standard deviation space (sx, sy), centred on the maximum of the distribution and of radius 0.2 pixel. Scale bars 0.5 µm. (C) Individual Z-discs were automatically detected (see ‘Methods’ for details). Scale bars 2 µm. (D) Individual bands generated by the accumulation of protein epitopes were detected automatically and their centre position was obtained using a Gaussian fit. After a visual check of the automated detection result, the results were compiled for further analysis.
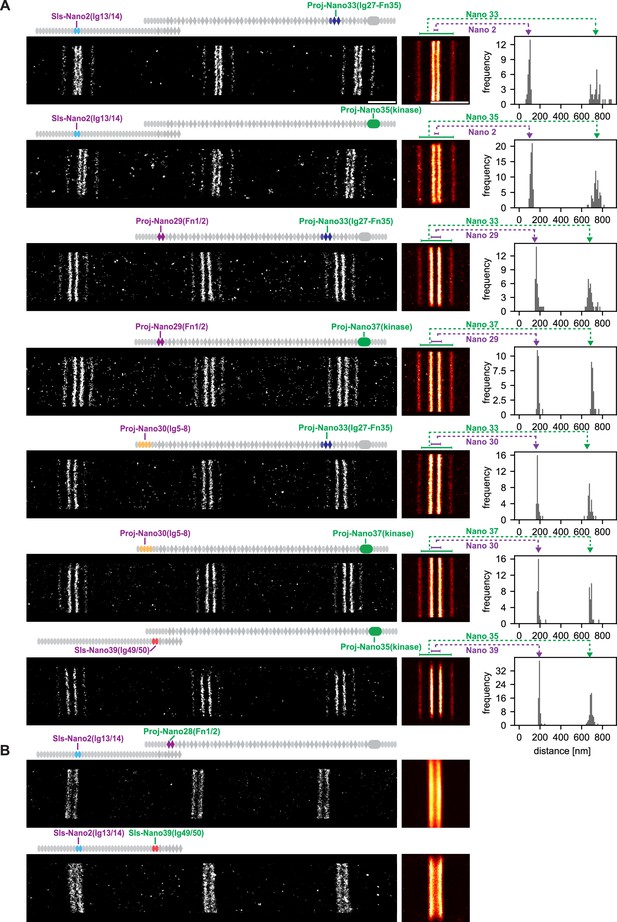
Single-colour DNA-PAINT imaging of Sallimus (Sls) and Projectin domains.
(A) Left: representative DNA-PAINT images of myofibrils stained with two different anti-Sls or anti-Projectin nanobodies labelling two epitopes and imaged with the same fluorescent imager oligo. The different Sls or Projectin nanobody combinations are indicated above each image. Middle: pseudo-coloured sum image centred around Z-discs resulting from one hemi-thorax. Right: histogram of distances between bands centred around Z-discs with the respective nanobody combinations indicated in green or magenta. The frequency (y-axis) refers to the number of Z-discs present in the respective length bins. Note that four bands can be readily distinguished for all shown nanobody combinations. The number of Z-discs scored in the shown samples are from top to bottom: 34, 59, 35, 25, 28, 28, and 63 (see Figure 4—source data 1 for the individual measurements). (B) Similar representations as in (A). However, the positions of neighbouring Sls or Projectin epitopes cannot be resolved in a single colour. Scale bar 1 µm.
-
Figure 4—source data 1
Band distances and imaging conditions os samples shown in Figure 4.
- https://cdn.elifesciences.org/articles/79344/elife-79344-fig4-data1-v3.xlsx
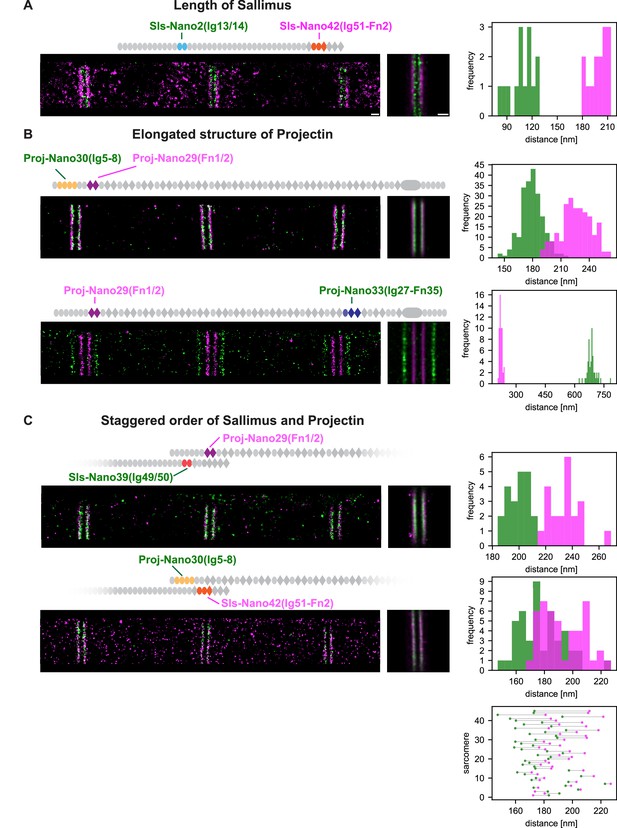
Dual-colour DNA-PAINT imaging reveals staggered order of Sallimus (Sls) and Projectin.
(A) Left: representative DNA-PAINT image of a myofibril stained with two nanobodies labelling Sls-Ig13/14 (Sls-Nano2) and Sls-Ig51/Fn2 epitopes (Sls-Nano42). Middle: sum image centred around Z-discs resulting from one hemi-thorax. Right: histogram of distances between bands centred around Z-discs (Sls-Ig13/14 in green, Sls-Ig51/Fn2 in magenta). The frequency (y-axis) refers to the number of Z-discs present in the respective length bins (number of Z-discs scored: 14, see Figure 5—source data 1 for the respective measurements). (B). Top: representative DNA-PAINT image of a myofibril stained with two nanobodies labelling Proj-Ig5-8 (Proj-Nano30) and Proj-Fn1/2 (Proj-Nano29) epitopes, sum image, and histograms of distances between bands (Proj-Ig5-8 in green, Proj-Fn1-2 in magenta, number of Z-discs scored: 219). Bottom: representative myofibril stained for Proj-Fn1/2 (Proj-Nano29) and Proj-Ig27-Fn35 (Proj-Nano33) epitopes, sum image, and histogram of distances between bands centred around Z-discs (Proj-Fn1/2 in magenta, Proj-Ig27-Fn35 in green, number of Z-discs scored: 52) (C). Top: representative DNA-PAINT image of a myofibril stained with two nanobodies labelling SlsIg49/50 (Sls-Nano39) and Proj-Fn1/2 (Proj-Nano29) epitopes, sum image, and histogram of distances between bands centred around Z-discs (Sls-Ig49/50 in green, Proj-Fn1/2 in magenta, number of Z-discs scored: 21). Bottom: same as top for Sls-Ig51/Fn2 (Sls-Nano42) and Proj-Ig5-8 (Proj-Nano30) epitopes, sum image, histogram of distances, and plot showing the epitope positions from the Z-discs in the individual sarcomeres analysed (bottom right, Sls-Ig51/Fn2 in magenta, Proj-Ig5-8 in green, number of Z-discs scored: 45). Note that in 42 of 45 cases the Proj-Ig5-8 (green) is closer to the Z-disc than Sls-Ig51/Fn2 (magenta). Scale bar 250 nm.
-
Figure 5—source data 1
Band distances and imaging conditions for all samples shown in Figure 5.
- https://cdn.elifesciences.org/articles/79344/elife-79344-fig5-data1-v3.xlsx
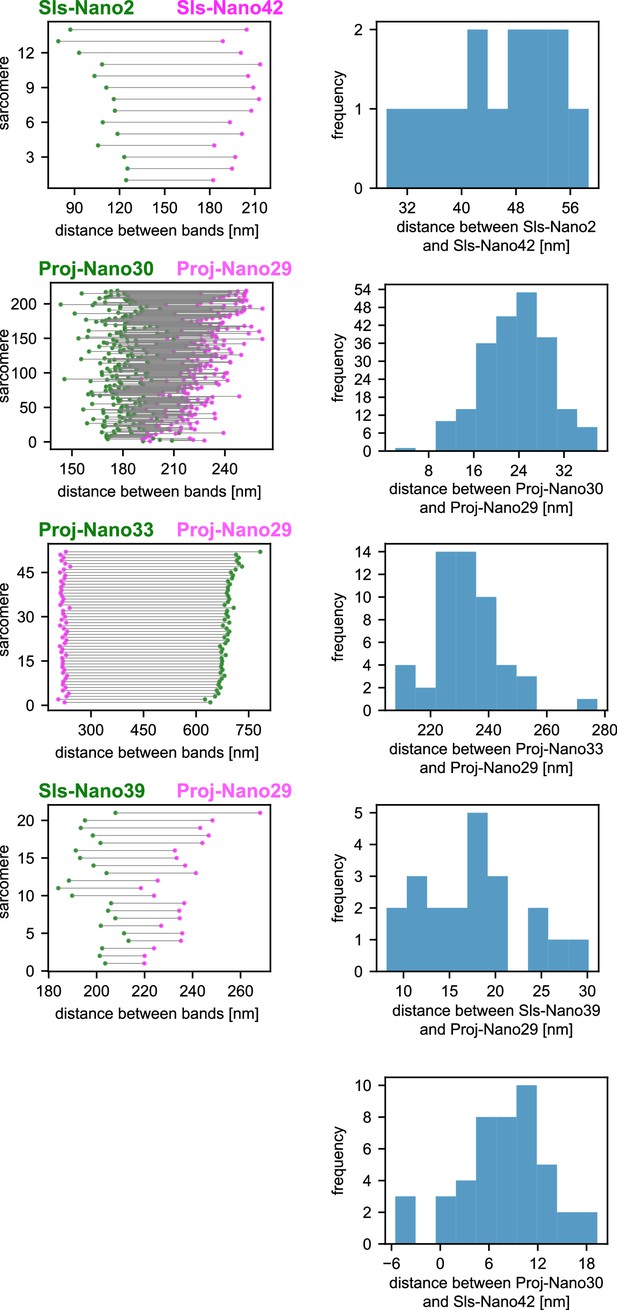
Dual-colour DNA-PAINT sarcomere quantifications.
Distance quantifications of the dual-colour DNA-PAINT data shown in Figure 5. Plots on the left show the epitope distances from the Z-discs in each sarcomere analysed with green and magenta colours indicating the respective nanobodies used. Plots on the right display histograms plotting the distances in between the individual two nanobody epitopes in each sarcomere. These are half the values from the plots on the left, as the organisations are symmetric around the Z-discs. The frequency (y-axis) refers to the number of sarcomeres present in the respective length bins.
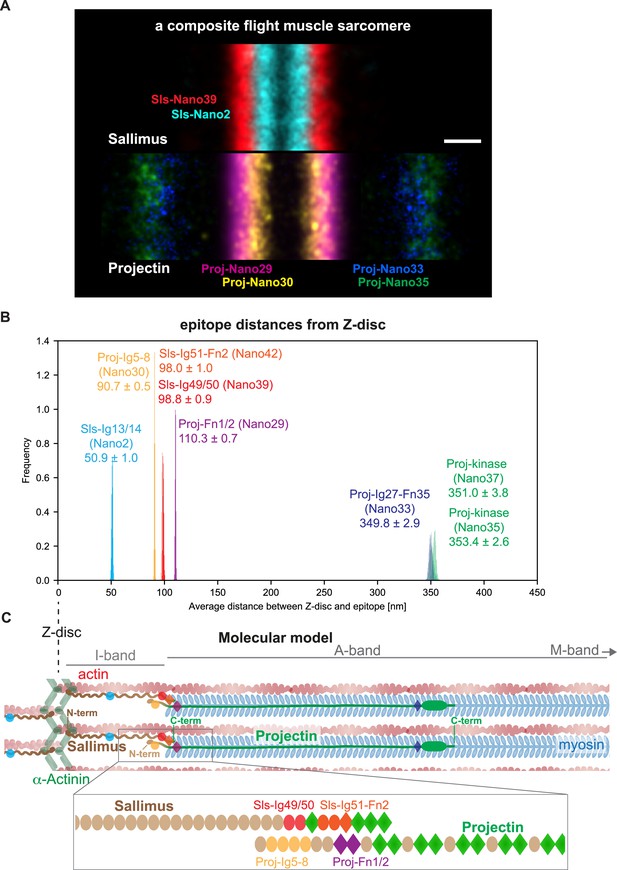
Summary and model.
(A) A sarcomere displayed as a composite of two summed anti-Sallimus nanobody bands (top) and four summed anti-Projectin nanobody bands (bottom), each originating from one individual hemi-thorax imaged. Note the overlay of the positions of both proteins. Scale bar is 100 nm. (B) Distribution of the average distances from the Z-disc for all Sls and Projectin epitopes measured using bootstrapping (see ‘Methods’). (C) Cartoon model of the relative arrangement of Sls and Projectin within the flight muscle sarcomere. The positions of the measured Sls and Projectin domains are highlighted in colours. The zoomed regions illustrate the suggested staggered architecture of the C-terminal Sls and the N-terminal Projectin protein parts.
-
Figure 6—source data 1
Data summary and all band distances generated in the manuscript.
- https://cdn.elifesciences.org/articles/79344/elife-79344-fig6-data1-v3.xlsx
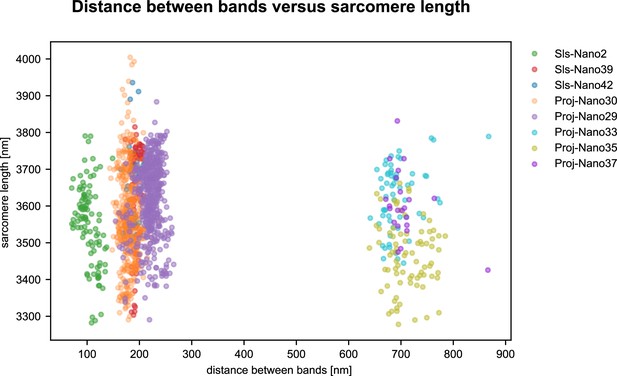
Distance between bands versus sarcomere length.
Scatter plot of distance between bands of nanobodies used in this study versus the individual sarcomere length. Each dot corresponds to one sarcomere. The sarcomere length reported for nanobodies centred on a given Z-disc was obtained by measuring half the distance between Z-discs located directly on the left and on the right. Note that the distances between bands do not correlate with the slight variations in sarcomere length. A total of 1442 distances were quantified (see Figure 6—source data 1). Each point is obtained from three consecutive Z-discs. The first and third Z-disc result in the sarcomere length (middle of bands in the Z-disc defined the Z-disc centr, which are then used as references to measure the length of two sarcomeres – first to second and second to third – which is then divided by two to obtain the sarcomere length). The distance between bands is obtained from the second sarcomere.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Drosophila melanogaster) | Luminy | Leonte et al., 2021 | ||
| Gene (D. melanogaster) | sls | http://flybase.org/reports/FBgn0086906 | FBgn0086906 | |
| Gene (D. melanogaster) | bt (Projectin) | http://flybase.org/reports/FBgn0005666 | FBgn0005666 | |
| Other | Sls-Ig13/14 (Nano2) | Loreau et al., 2023 | Nanobody – use at about 50 nM | |
| Other | Sls-Ig49/50 (Nano39) | Loreau et al., 2023 | Nanobody – use at about 50 nM | |
| Other | Sls-Ig51-Fn2 (Nano42) | Loreau et al., 2023 | Nanobody – use at about 50 nM | |
| Other | Sls-Ig13/14 (Nano2) | Loreau et al., 2023 | Nanobody – use at about 50 nM | |
| Other | Proj-Ig5-8 (Nano30) | Loreau et al., 2023 | Nanobody – use at about 50 nM | |
| Other | Proj-Fn1/2 (Nano29) | Loreau et al., 2023 | Nanobody – use at about 50 nM | |
| Other | Proj-Ig27-Fn35 (Nano33) | Loreau et al., 2023 | Nanobody – use at about 50 nM | |
| Other | Proj-kinase (Nano37) | Loreau et al., 2023 | Nanobody – use at about 50 nM | |
| Other | Proj-kinase (Nano35) | Loreau et al., 2023 | Nanobody – use at about 50 nM | |
| Chemical compound, drug | Rhodamine-phalloidin | Invitrogen, Cat#R415 | 1 in 500 | |
| Chemical compound, drug | P1 imager – Atto643 | Metabion | TAGATGTAT – Atto643 | |
| Chemical compound, drug | P3 imager – Cy3b | Metabion | TAATGAAGA – Cy3B | |
| Chemical compound, drug | PS3 imager – Atto643 | Metabion | TCCTCCC – Atto643 | |
| Software, algorithm | PAINT data band extraction | https://github.com/PierreMangeol/titin_PAINT; Mangeol, 2022 |





