Sonic hedgehog-dependent recruitment of GABAergic interneurons into the developing visual thalamus
Figures
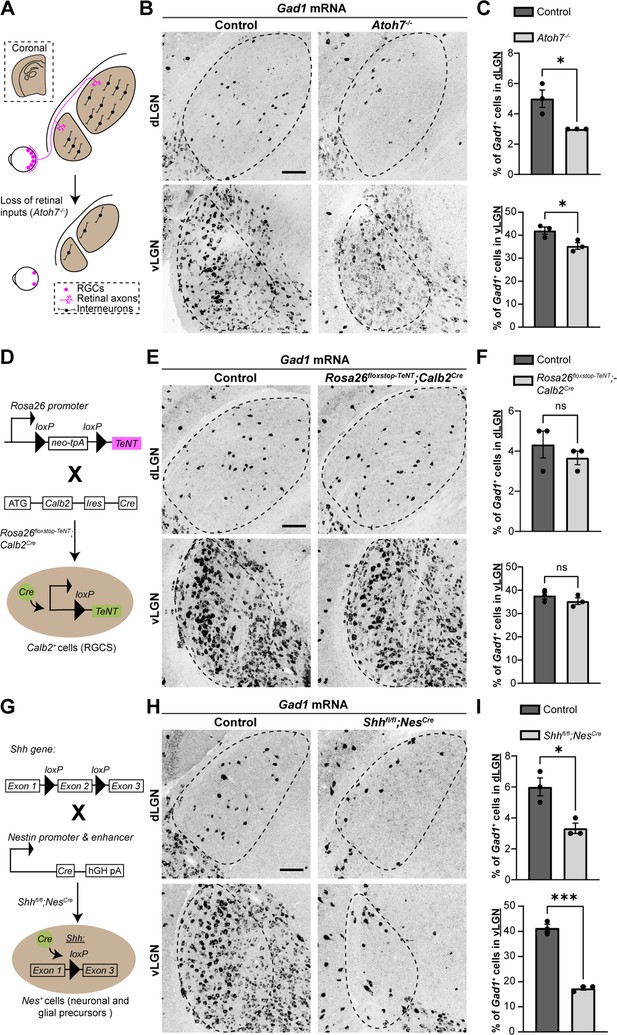
Sonic hedgehog (SHH), but not retinal activity, is required for the recruitment of interneurons into visual thalamus.
(A) Schematic representation of loss of retinal inputs and interneurons in visual thalamus of Atoh7−/− mice. (B) In situ hybridization (ISH) shows a reduction in Gad1+ cells in dorsal lateral geniculate nucleus (dLGN) and ventral lateral geniculate nucleus (vLGN) of >P150 Atoh7−/− mice compared with controls. (C) Quantification of percentage of Gad1+ cells in dLGN and vLGN of >P150 controls and Atoh7−/− mice. Each data point represents one biological replicate and bars depict means ± standard error of the mean (SEM). Asterisks (*) indicate p < 0.05 by Student’s t-test (n = 3 mice for each group). (D) Schematic representation of Calb2Cre-inducible expression of tetanus toxin (TeNT) in retinal ganglion cells (RGCs). Rosa26floxstop-TeNT mice with the construct containing a loxP-flanked neomycin (Neo) cassette and TeNT coding sequence under Rosa26 locus, were crossed with Calb2Cre mice that harbor a Cre recombinase and internal ribosome entry site (IRES) in the Calb2 locus. (E) ISH for Gad1 in dLGN and vLGN of P120 control and Rosa26floxstop-TeNT;Calb2Cre mice. (F) Quantification revealed no significant difference in Gad1+ cells in visual thalamus of control and Rosa26floxstop-TeNT;Calb2Cre mice. Each data point represents one biological replicate and bars depict means ± SEM. ns indicates no significant differences by Student’s t-test (n = 3 mice for each group). (G) Schematic representation of strategy to delete SHH from neural cells in the developing brain. This was achieved by crossing Shhfl/fl mice, which have two loxP sites flanking exon 2 of the Shh gene, with NesCre transgenic mice that contain a Cre recombinase and human growth hormone polyadenylation signal (hGH pA) under the control of Nestin promoter and enhancer. (H) ISH revealed a dramatic reduction in Gad1+ cells in dLGN and vLGN of P18 Shhfl/flNesCre mice compared with controls. (I) Quantification of percentage of Gad1+ cells in dLGN and vLGN of P18 control and Shhfl/flNesCre mice. Each data point represents one biological replicate and bars depict means ± SEM. Asterisks represent significant differences (***p < 0.001; *p < 0.05) by Student’s t-test (n = 3 mice for each group). Scale bars in C, E, H: 100 μm.
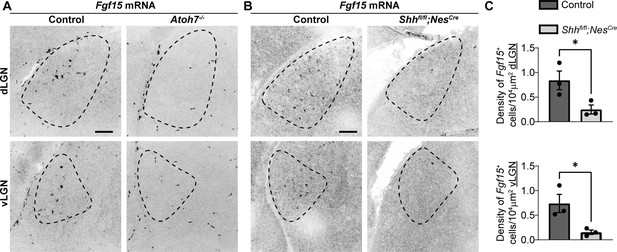
Thalamic Fgf15 expression is dependent on retinal inputs and sonic hedgehog (SHH).
(A) In situ hybridization (ISH) revealed a dramatic reduction in Fgf15 expression in dorsal lateral geniculate nucleus (dLGN) and ventral lateral geniculate nucleus (vLGN) of P2 Atoh7−/− mutants compared with controls. (B) ISH revealed a dramatic reduction in Fgf15 expression in dLGN and vLGN of P3 Shhfl/flNesCre mutants compared with controls. (C) Quantification for density of Fgf15+ cells in dLGN and vLGN of P3 control and Shhfl/flNesCre mutant mice. Each data point represents one biological replicate and bars depict means ± standard error of the mean (SEM). Asterisks (*) indicate *p < 0.05 by Student’s t-test. Scale bars in A, B: 100 μm.
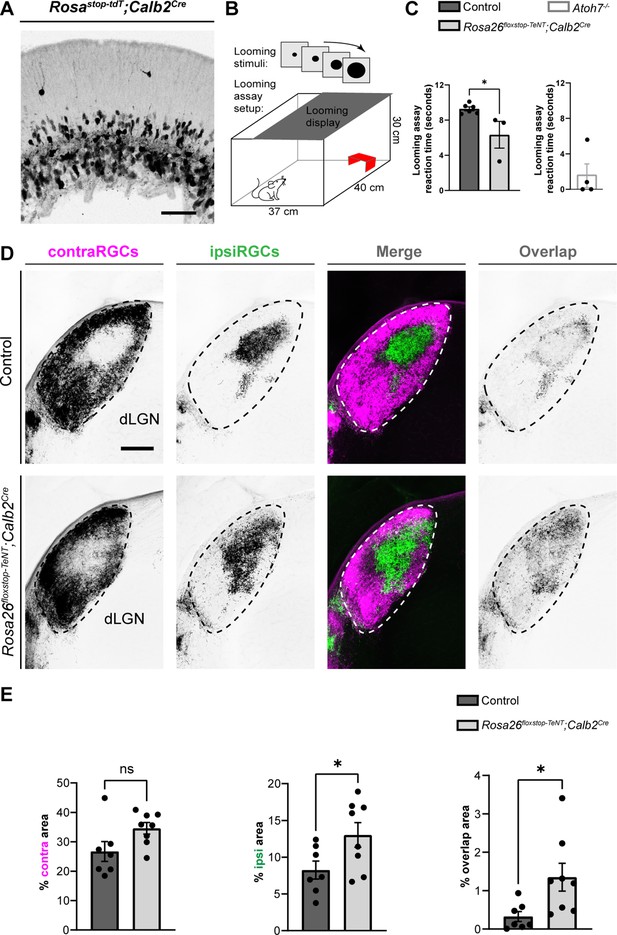
Anatomical and functional characterization of Rosa26floxstop-TeNT;Calb2Cre mice.
(A) Expression of tdT in the GCL of P0 Rosastop-tdT;Calb2Cre mouse retina. (B) Schematic representation of the behavioral response to a looming visual stimulus. (C) Adult Rosa26floxstop-TeNT;Calb2Cre (age-matched and littermate mutants to controls) and Atoh7−/− mutants (not age-matched and not littermate mutants to controls) had reduced reaction times to a looming visual stimulus compared with controls. Each data point represents one biological replicate and bars depict means ± standard error of the mean (SEM). Asterisks represent significant differences (***p < 0.001; *p < 0.05) by Student’s t-test. (D) Cholera Toxin Subunit B (CTB)-labeled eye-specific retinal projections to dorsal lateral geniculate nucleus (dLGN) in P16 control and P24 Rosa26floxstop-TeNT;Calb2Cre mutant mice. (E) Quantification of the percentage of dLGN area covered by contraRGCs projections, ipsiRGCs projections, or overlapping eye-specific projections in controls and Rosa26floxstop-TeNT;Calb2Cre mutants. Each data point represents one biological replicate and bars depict means ± SEM. Asterisks (*) indicate *p < 0.05 by Student’s t-test (n = 7 mice for control group and n = 8 mice for mutant group). Scale bars in A: 50 μm and D: 200 μm.
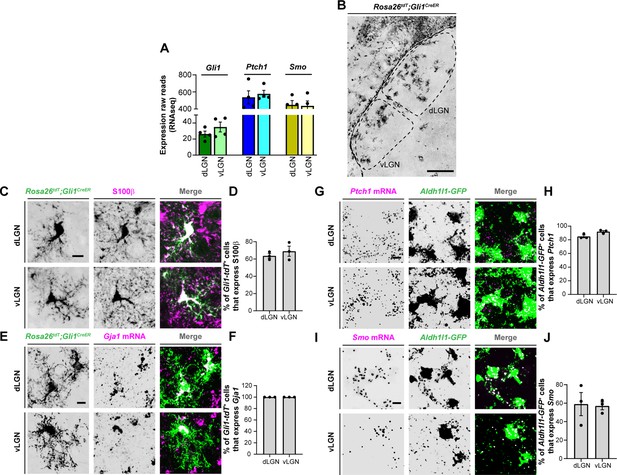
Expression of sonic hedgehog (SHH) signaling molecules by astrocytes in visual thalamus.
(A) Raw transcript reads of mRNAs for downstream SHH signaling components in P3 dorsal lateral geniculate nucleus (dLGN) and ventral lateral geniculate nucleus (vLGN) by RNAseq. Each data point represents a different biological replicate and bars depict means ± standard error of the mean (SEM). (B) Presence of Gli1-tdT+ cells in dLGN and vLGN of P7 Rosa26tdT;Gli1CreER mice. (C) IHC for S100ß in P7 Rosa26tdT;Gli1CreER mice revealed S100ß expression in Gli1-tdT+ cells in visual thalamus. (E) Quantification of percentage of Gli1-tdT+ cells that express S100ß in P7 visual thalamus. Each data point represents one biological replicate and bars depict means ± SEM (n = 3 mice for each region). (D) In situ hybridization (ISH) for Gja1 in P7 Rosa26tdT;Gli1CreER mice revealed expression of Gja1 mRNA by Gli1-tdT+ cells in visual thalamus. (F) Quantification shows 100% of Gli1-tdT+ cells express the astrocytic marker Gja1 in P7 visual thalamus. Each data point represents one biological replicate and bars depict means ± SEM (n = 3 mice for each region). (G) RNAscope-based ISH detected Ptch1 mRNA in dLGN and vLGN of P3 Aldh1l1-GFP mice. This revealed expression of Ptch1 mRNA in the cell bodies as well as in the processes of Aldh1l1-GFP+astrocytes. (H) Quantification of percentage of Aldh1l1-GFP+astrocytes that express Ptch1 mRNA in P3 visual thalamus. Each data point represents one biological replicate and bars depict means ± SEM (n = 3 mice for each region). (I) RNAscope-based ISH detected Smo mRNA in dLGN and vLGN of P3 Aldh1l1-GFP mice. This revealed expression of Smo mRNA in the cell bodies as well as in the processes of Aldh1l1-GFP+ astrocytes.(J) Quantification of percentage of Aldh1l1-GFP+astrocytes that express Smo mRNA in P3 visual thalamus. Each data point represents one biological replicate and bars depict means ± SEM (n = 3 mice for each region). Scale bars in B: 200 μm and in C, E, G, I: 10 μm.
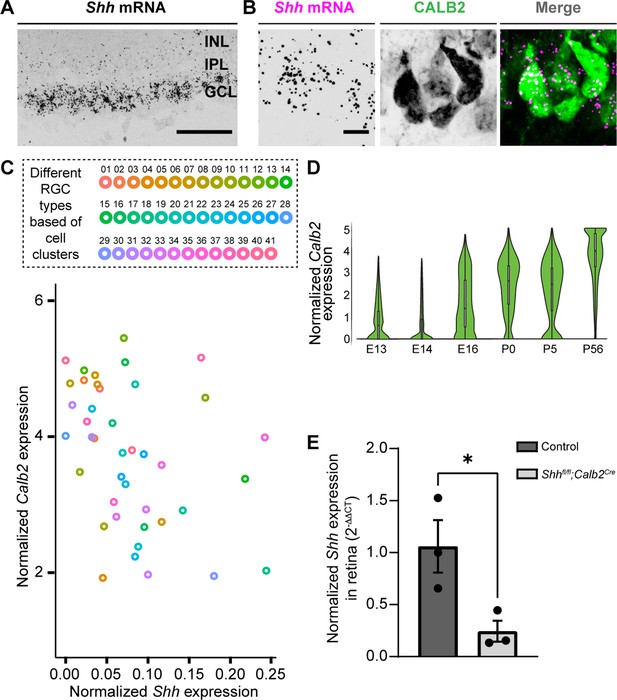
Deletion of sonic hedgehog (SHH) from retinal ganglion cells (RGCs) in the perinatal retina.
(A) RNAscope-based in situ hybridization (ISH) revealed dense Shh mRNA in the GCL of P3 retina (inner nuclear layer, INL; inner plexiform layer, IPL; ganglion cell layer, GCL). (B) RNAscope-based ISH revealed Shh mRNA in CALB2+ cells in the GCL of P3 retina. (C) Single-cell RNAseq data (from Rheaume et al., 2018) analyzed to show Calb2 and Shh mRNA expression by different subtypes of RGCs in P5 mouse retina. (D) Single-cell RNAseq data (from Shekhar et al., 2022) analysis for developmental expression of Calb2 by RGCs. (E) Real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR) showed a reduction in Shh mRNA in retina of P3 Shhfl/fl;Calb2Cre mice compared to controls. Each data point represents one biological replicate and bars depict means ± standard error of the mean (SEM). Asterisks (*) indicate p < 0.05 by Student’s t-test (n = 3 mice for each group). Scale bars in A: 100 μm and in B: 10 μm.
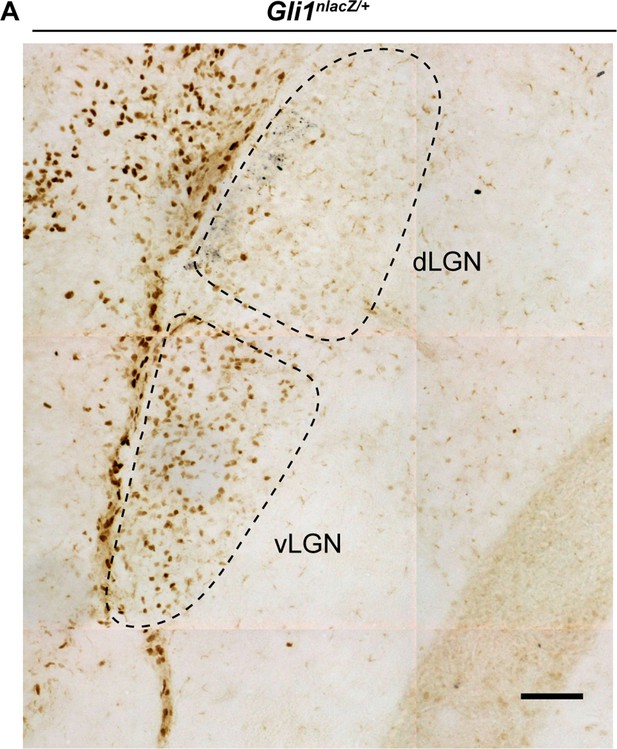
Presence of active sonic hedgehog (SHH) signaling in developing visual thalamus.
(A) Bright-field immunohistochemistry for βGal in dorsal lateral geniculate nucleus (dLGN) and ventral lateral geniculate nucleus (vLGN) of P3 Gli1nlacZ/+ mice. Scale bar in A: 100 μm.
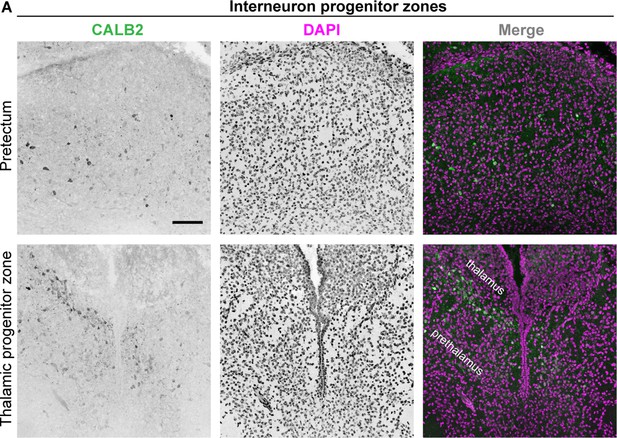
Expression of CALB2 in developing LGN and interneuron progenitor zones.
(A) IHC for CALB2 shows sparse expression in P3 tectal and thalamic progenitor zones of thalamic GABAergic interneurons. Scale bar in A: 100 μm.
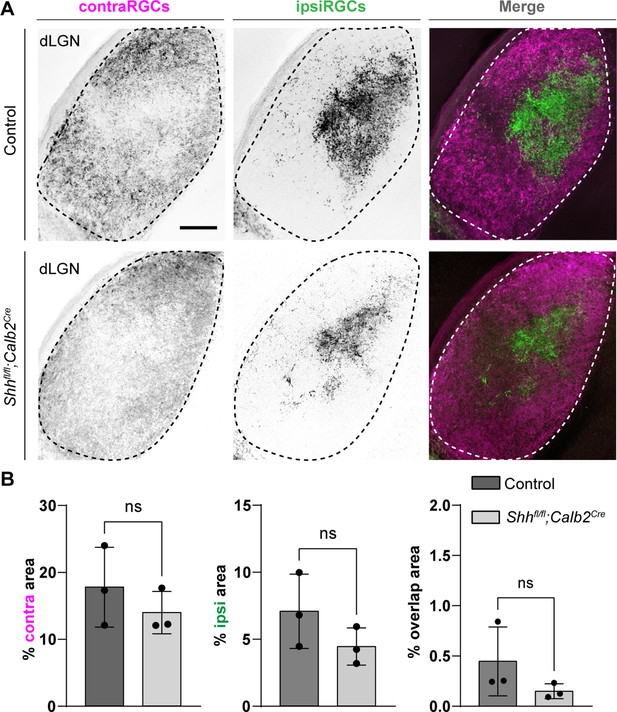
Retinal innervations innervate visual thalamus in the absence of retinal ganglion cell (RGC)-derived sonic hedgehog (SHH).
(A) Cholera Toxin Subunit B (CTB)-labeled eye-specific retinal projections to dorsal lateral geniculate nucleus (dLGN) in P25 control and P25 Shhfl/fl;Calb2Cre mutant mice. (B) Quantification for the percentage of dLGN area covered by contraRGCs projections, ipsiRGCs projections, or overlapping eye-specific projections in controls and Shhfl/fl;Calb2Cre mutants. Each data point represents one biological replicate and bars depict means ± standard error of the mean (SEM). ns indicates no significant differences by Student’s t-test (n = 3 mice for each group). Scale bar in A: 100 μm.
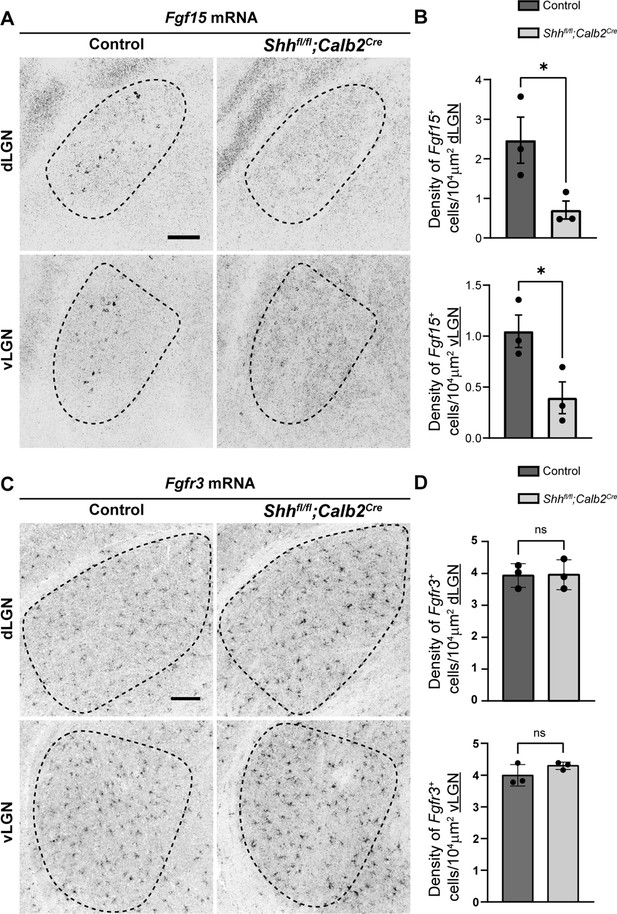
Absence of retinal ganglion cell (RGC)-derived sonic hedgehog (SHH) does not impact astrocyte distribution but decreases Fgf15 expression in the visual thalamus.
(A) In situ hybridization (ISH) revealed reduced Fgf15 expression in dorsal lateral geniculate nucleus (dLGN) and ventral lateral geniculate nucleus (vLGN) of P3 Shhfl/fl;Calb2Cre mutants compared to controls. (B) Quantification for density of Fgf15+ cells in dLGN and vLGN of P3 control and Shhfl/fl;Calb2Cre mutant mice. Each data point represents one biological replicate and bars depict means ± standard error of the mean (SEM). Asterisks (*) indicate *p < 0.05 by Student’s t-test (n = 3 mice for each group). (C) ISH revealed no change in Fgfr3+ astrocytes in dLGN and vLGN of P7 Shhfl/fl;Calb2Cre mutants compared to controls. (D) Quantification for density of Fgfr3+ cells in dLGN and vLGN of P7 control and Shhfl/fl;Calb2Cre mutant mice. Each data point represents one biological replicate and bars depict means ± SEM. ns indicates no significant differences by Student’s t-test (n = 3 mice for each group). Scale bars in A, C: 100 μm.
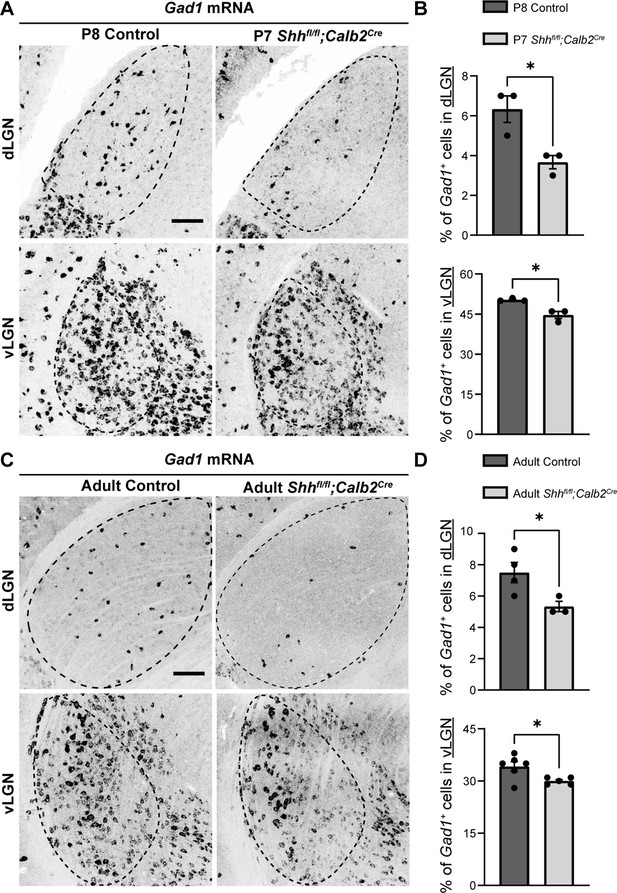
Retinal ganglion cell (RGC)-derived sonic hedgehog (SHH) is required for the recruitment of Gad1+ + into visual thalamus.
(A) In situ hybridization (ISH) revealed a reduction in Gad1+ cells in dorsal lateral geniculate nucleus (dLGN) and ventral lateral geniculate nucleus (vLGN) of P7 Shhfl/fl;Calb2Cre mutants compared with controls. (B) Quantification of percentage of Gad1+ cells in dLGN and vLGN of P7 Shhfl/fl;Calb2Cre and control mice. Each data point represents one biological replicate and bars depict means ± standard error of the mean (SEM). Asterisks (*) indicate *p < 0.05 by Student’s t-test (n = 3 mice for each group). (C) ISH revealed a reduction in Gad1+ cells in dLGN and vLGN of >P90 Shhfl/fl;Calb2Cre mutants compared with controls. (D) Quantification of percentage of Gad1+ cells in dLGN and vLGN of adult Shhfl/fl;Calb2Cre and control mice. Each data point represents one biological replicate and bars depict means ± SEM. Asterisks (*) indicate *p < 0.05 by Student’s t-test (n = 4 mice for control dLGN group, n = 3 mice for mutant dLGN group, n = 6 mice for control vLGN group, and n = 5 mice for mutant vLGN group). Scale bars in A, C: 100 μm.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | anti-GFP (rabbit polyclonal) | Thermo Fisher Scientific | Cat#A-11122; RRID:AB_221569 | 1:250 |
| Antibody | anti-S100 (rabbit polyclonal) | Dako | Cat# Z0311; RRID:AB_10013383 | 1:200 |
| Antibody | anti-Calretinin (rabbit polyclonal) | Swant | Cat#7697; RRID: AB_2619710 | 1:1000 |
| Antibody | anti-RFP (rabbit polyclonal) | Thermo Fisher Scientific | Cat#600-401-379-RTU; RRID:AB_2209751 | 1:500 |
| Antibody | Anti-Digoxigenin-POD (sheep polyclonal) | Millipore Sigma | Cat#11207733910; RRID:AB_514500 | 1:1000 |
| Antibody | Anti-Fluorescein-POD (sheep polyclonal) | Millipore Sigma | Cat#11426346910; RRID:AB_840257 | 1:1000 |
| Biological sample (Mus musculus) | Rosa26tdT;Gli1CreER brains | A.D.R. Garcia, Drexel University | JAX #007913, #007914; RRID: IMSR_JAX:007913, IMSR_JAX:007914 | |
| Peptide, recombinant protein | Fluorescein RNA Labeling Mix | Roche | Cat#11685619910 | |
| Peptide, recombinant protein | DIG RNA Labeling Mix | Roche | Cat#11277073910 | |
| Peptide, recombinant protein | SuperScript II Reverse Transcriptase | Thermo Fisher Scientific | Cat#18064022 | |
| Peptide, recombinant protein | Cholera Toxin Subunit B (CTB, Recombinant), Alexa Fluor 488 Conjugate | Thermo Fisher Scientific | CAT#C22841 | |
| Peptide, recombinant protein | Tamoxifen | Sigma | CAT#T5648-1G | |
| Peptide, recombinant protein | CTB (Recombinant), Alexa Fluor 555 Conjugate | Thermo Fisher Scientific | CAT#C34776 | |
| Commercial assay, kit | SuperScript II Reverse Transcriptase First Strand cDNA Synthesis kit | Invitrogen | Cat#18064014 | |
| Commercial assay, kit | pGEM-T Easy Vector Systems | Promega | Cat#A1360 | |
| Commercial assay, kit | MAXIscript in vitro Transcription Kit | Ambion | Cat#AM1312 | |
| Commercial assay, kit | Tyramide Signal Amplification system | PerkinElmer | Cat#NEL753001KT | |
| Commercial assay, kit | iTaq Universal SYBR Green Supermix | Bio-Rad | Cat#1725124 | |
| Commercial assay, kit | Bio-Rad Total RNA Extraction from Fibrous and Fatty Tissue kit | Bio-Rad | Cat#7326870 | |
| Commercial assay, kit | RNAscope Multiplex Fluorescent Reagent Kit V2 | Advanced Cell Diagnostics (ACD) | Cat#323100 | |
| Other | RNAseq datasets for the developing mouse dLGN and vLGN | DOI: https://doi.org/10.7554/eLife.33498.006 | Monavarfeshani et al., 2018 | |
| Other | Single-cell RNAseq dataset for RGC subtypes | DOI: https://doi.org/10.1038/s41467-018-05134-3 | Accession # GSE115404 | Rheaume et al., 2018 |
| Other | Single-cell RNAseq dataset for RGCs at various ages | DOI: https://doi.org/10.7554/eLife.73809 | Accession # GSE185671 | Shekhar et al., 2022 |
| Strain, strain background (Mus musculus) | C57BL/6J mice | The Jackson Laboratory | JAX#000664; RRID:IMSR_JAX:000664 | |
| Strain, strain background (Mus musculus) | Calb2Cre | The Jackson Laboratory | JAX#010774; RRID:IMSR_JAX:010774 | |
| Strain, strain background (Mus musculus) | Shhfl/fl | The Jackson Laboratory | JAX#004293; RRID:IMSR_JAX:004293 | |
| Strain, strain background (Mus musculus) | NesCre | The Jackson Laboratory | JAX#003771; RRID:IMSR_JAX:003771 | |
| Strain, strain background (Mus musculus) | Aldh1l1-GFP | S. Robel, Virginia Tech | Stock#011015-UCD; RRID: MMRRC_011015-UCD | |
| Strain, strain background (Mus musculus) | Rosa26floxstop-TeNT | A. Maximov, The Scripps Research Institute | MGI:3839913 | Zhang et al., 2008 |
| Strain, strain background (Mus musculus) | Rosa26tdT(Ai14) | The Jackson Laboratory | JAX#007914; RRID: IMSR_JAX:007914 | |
| Strain, strain background (Mus musculus) | Gli1CreER | Ahn and Joyner, 2005 | JAX#007913; RRID: IMSR_JAX:007913 | |
| Strain, strain background (Mus musculus) | Rosa26tdT | The Jackson Laboratory | JAX#007909; RRID:IMSR_JAX:007909 | |
| Strain, strain background (Mus musculus) | Atoh7−/− | S.W. Wang, University of Texas MD Anderson Cancer Center | Stock# 042298-UCD; RRID:MMRRC_042298-UCD | |
| Strain, strain background (Mus musculus) | Gli1nlacZ/+ | The Jackson Laboratory | JAX#008211; RRID:IMSR_JAX:008211 | Bai et al., 2002 |
| Sequence-based reagent | Gad1 cloning primer F: TGTGCCCAAACTGGTCCT; R: TGGCCGATGATTCTGGTT | Integrated DNA Technologies | N/A | |
| Sequence-based reagent | Gja1 cloning primer F: CGTGAAGGGAAGAAGCGA; R: GCCTGCAAACTGCCAAGT | Integrated DNA Technologies | N/A | |
| Sequence-based reagent | Shh qPCR primer F: ACGTAGCCGAGAAGACCCTA; R: ACTTGTCTTTGCACCTCTGAGT | Integrated DNA Technologies | N/A | |
| Sequence-based reagent | Gapdh qPCR primer F: CGTCCCGTAGACAAAATGGT; R: TTGATGGCAACAATCTCCAC | Integrated DNA Technologies | N/A | |
| Sequence-based reagent | 18s qPCR primer F: GGACCAGAGCGAAAGCATTTG; R: GCCAGTCGGCATCGTTTATG | Integrated DNA Technologies | N/A | |
| Sequence-based reagent | Cre genotyping primer F: CGTACTGACGGTGGGAGAAT; R: TGCATGATCTCCGGTATTGA | Integrated DNA Technologies | N/A | |
| Sequence-based reagent | Shhfl/fl genotyping primer F: CAGAGAGCATTGTGGAATGG; R: CAGACCCTTCTGCTCATGG | Integrated DNA Technologies | N/A | |
| Sequence-based reagent | tdT genotyping primer F: ACCTGGTGGAGT TCAAGACCATCT; R: TTGATGACGGCCA TGTTGTTGTCC | Integrated DNA Technologies | N/A | |
| Sequence-based reagent | GFP genotyping primer F: AAGTTCATCTGCACCACCG; R: TCCTTGAAGAAGATGGTGCG | Integrated DNA Technologies | N/A | |
| Sequence-based reagent | TeNT genotyping primer FA: AAAGTCGCTCTGAGTTGTTAT; RA: GGAGCGGGAGAAATGGATATG; SA: CATCAAGGAAACCC TGGACTACTG | Integrated DNA Technologies | N/A | |
| Sequence-based reagent | Atoh7−/− genotyping primer (to see the wild-type band) F: ATGGCGCTCAGCTACATCAT; R: GGGTCTACCTGGAGCCTAGC | Integrated DNA Technologies | N/A | |
| Sequence-based reagent | Neo genotyping primer (to see the mutant Atoh7 band) F: GCCGGCCACAGTCGATGAATC; R: CATTGAACAAGATGGATTGCA | Integrated DNA Technologies | N/A | |
| Recombinant DNA reagent | Mouse Fgf15 cDNA | Horizon (Dharmacon) | Cat#MMM1013- 202768318, Clone ID: 5066286 | |
| Recombinant DNA reagent | RNA scope probe-Mm-Smo | ACD | Cat#318411 | |
| Recombinant DNA reagent | RNA scope probe-Mm-Ptch1-C2 | ACD | Cat#402811-C2 | |
| Recombinant DNA reagent | RNA scope probe-Mm-Shh-C3 | ACD | Cat#314361-C3 | |
| Recombinant DNA reagent | RNA scope 3-plex positive control probe-mm | ACD | Cat#320881 | |
| Recombinant DNA reagent | RNA scope 3-plex negative control probe-mm | ACD | Cat#320871 | |
| Software, algorithm | Prism | GraphPad | Version 8.0; RRID: SCR_002798 | |
| Software, algorithm | Adobe Photoshop | Adobe Inc | Version: 21.1.2 | |
| Software, algorithm | ZEN black edition | Carl Zeiss | Version: 14.0.12.201 | |
| Software, algorithm | Fiji ImageJ | NIH | Version: 1.52p | |
| Software, algorithm | RStudio | RStudio, Inc | Version: 1.2.5042 | |
| Other | Fgf15 riboprobe | This paper | N/A | Information in ‘Riboprobe production’ |
| Other | Gad1 riboprobe | This paper | N/A | Information in ‘Riboprobe production’ |
| Other | Gja1 riboprobe | This paper | N/A | Information in ‘Riboprobe production’ |





