NAD kinase promotes Staphylococcus aureus pathogenesis by supporting production of virulence factors and protective enzymes
Figures
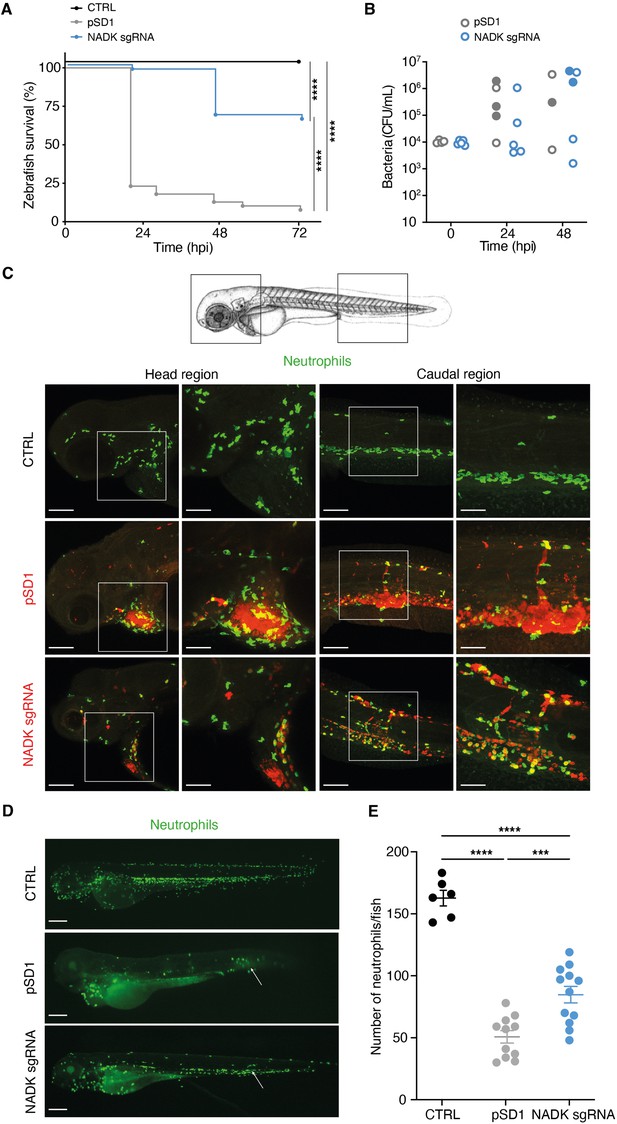
Nicotinamide adenine dinucleotide kinase (NADK) promotes S. aureus virulence in zebrafish.
(A) Survival of zebrafish larvae uninfected (CTRL) or intravenously injected at 60 hpf with 104 S. aureus containing the empty vector (pSD1) or the NADK knockdown strain (NADK sgRNA) between 0 and 72 hpi (n=48). (B) Bacterial burden in zebrafish larvae upon intravenous injection with 104 S. aureus containing the empty vector (pSD1) or the NADK knockdown strain (NADK sgRNA). For each strain, CFU was determined in living larvae (open circles) or dead larvae (filled circles) 0, 24, and 48 hpi. (C) Representative fluorescence confocal images of transgenic mpx:GFP zebrafish larvae uninfected (CTRL), or intravenously injected with 104 S. aureus containing the empty vector (pSD1) or the NADK knockdown strain (NADK sgRNA) at 12 hpi. Maximum intensity Z-projection images (2 μm serial optical sections) of bacteria (red) and neutrophils (green). Scale bars, 25 μm. Insets are shown at higher magnification on the right panels for head and caudal regions. Scale bars, 10 μm. Neutrophils containing NADK knockdown bacteria can be seen in the caudal inset. (D) Representative fluorescence confocal images of transgenic mpx:GFP zebrafish larvae uninfected (CTRL), or intravenously injected with 104 S. aureus containing the empty vector (pSD1) or the NADK knockdown strain (NADK sgRNA) at 24 hpi, showing neutrophils (green). White arrows indicate the injection site. Scale bars, 500 μm (E) Number of neutrophils in uninfected zebrafish larvae (CTRL) or in zebrafish larvae intravenously injected with 104 S. aureus containing the empty vector (pSD1) or the NADK knockdown strain (NADK sgRNA) at 24 hpi. Comparison of data was performed using one-way analysis of variance (***p<0.001, ****p<0.0001).
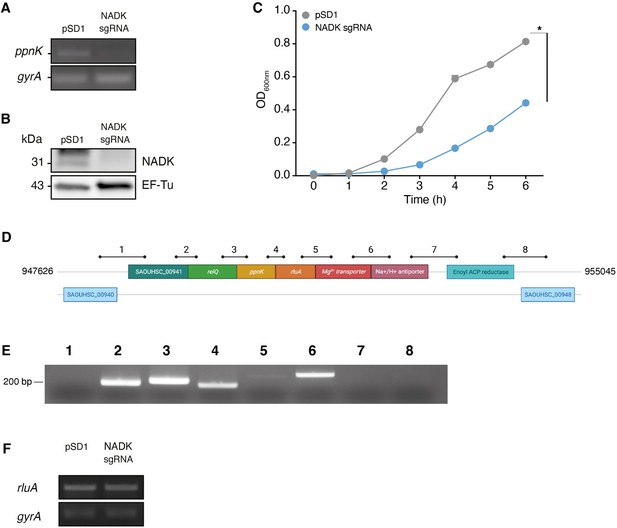
Nicotinamide adenine dinucleotide kinase (NADK) is important for S. aureus growth.
(A) Total RNAs of S. aureus containing the empty vector (pSD1) or the NADK knockdown strain (NADK sgRNA) were analyzed by RT-PCR using oligonucleotides specific to the target gene ppnK or the control gene gyrA. (B) Bacterial protein extracts of S. aureus containing the empty vector (pSD1) or the NADK knockdown strain (NADK sgRNA) were analyzed by immunoblotting using antibodies against the target protein NADK or against the control protein EF-Tu. (C) Bacterial growth of S. aureus containing the empty vector (pSD1) or the NADK knockdown strain (NADK sgRNA) monitored for 6 hr at OD600nm in BHI broth at 37°C upon ATc induction. Data shown are representative of three independent experiments. The bar indicates the standard error of the means of biological replicates. Comparison of data was performed using a t-test (*p<0.05). (D) S. aureus ppnK gene is part of the relQ operon. Genomic region surrounding the ppnK gene of S. aureus Xen36 between nucleotides 947,626 and 955,045. Bars and numbers represent the regions amplified by RT-PCR shown in B. (E) Ethidium bromide staining of DNA in an agarose gel following RT-PCR amplification using total RNA from S. aureus Xen36 and primers located in regions indicated in D. (F) Ethidium bromide staining of DNA in an agarose gel following RT-PCR amplification using total RNA from S. aureus Xen36/pSD1 or Xen36/NADK sgRNA strains and oligonucleotides specific to the gene rluA or the control gene gyrA, respectively.
-
Figure 1—figure supplement 1—source data 1
NADK transcript levels (A).
- https://cdn.elifesciences.org/articles/79941/elife-79941-fig1-figsupp1-data1-v1.pdf
-
Figure 1—figure supplement 1—source data 2
NADK protein levels (B).
- https://cdn.elifesciences.org/articles/79941/elife-79941-fig1-figsupp1-data2-v1.pdf
-
Figure 1—figure supplement 1—source data 3
NADK operon transcript levels (E).
- https://cdn.elifesciences.org/articles/79941/elife-79941-fig1-figsupp1-data3-v1.pdf
-
Figure 1—figure supplement 1—source data 4
RluA transcript levels (F).
- https://cdn.elifesciences.org/articles/79941/elife-79941-fig1-figsupp1-data4-v1.pdf
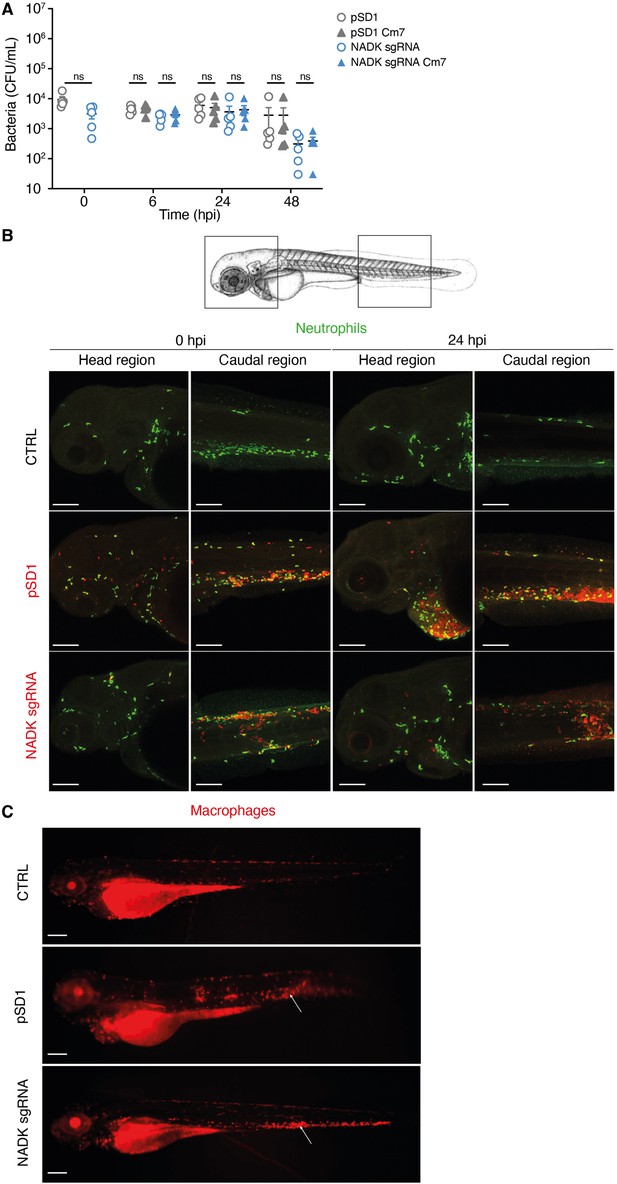
Nicotinamide adenine dinucleotide kinase (NADK) contributes to S. aureus virulence in zebrafish.
(A) The number of pSD1 (gray) or NADK sgRNA (blue) S. aureus was monitored in zebrafish larvae after intravenous injection with 5.103 bacteria. For each strain, CFU was determined 0, 6, 24, and 48 hpi by plating fish lysates on BHI agar (open circles) and BHI agar supplemented with chloramphenicol (triangles). (B) Representative fluorescence confocal images of transgenic mpx:GFP zebrafish larvae fixed and labeled with specific antibodies to detect GFP or S. aureus, uninfected (CTRL), or intravenously injected with 104 S. aureus containing the empty vector (pSD1) or the NADK knockdown strain (NADK sgRNA) at 0 or 24 hpi. Maximum intensity Z-projection images (2 μm serial optical sections) of bacteria (red) and neutrophils (green) in the caudal and head regions. Scale bars, 25 μm. (C) Representative fluorescence confocal images of transgenic mfap4::mCherryF zebrafish larvae fixed and labeled with specific antibodies to detect mCherry or S. aureus, uninfected (CTRL), or intravenously injected with 104 S. aureus containing the empty vector (pSD1) or the NADK knockdown strain (NADK sgRNA) at 24 hpi, showing macrophages (red). White arrows indicate the injection site. Scale bars, 500 μm.
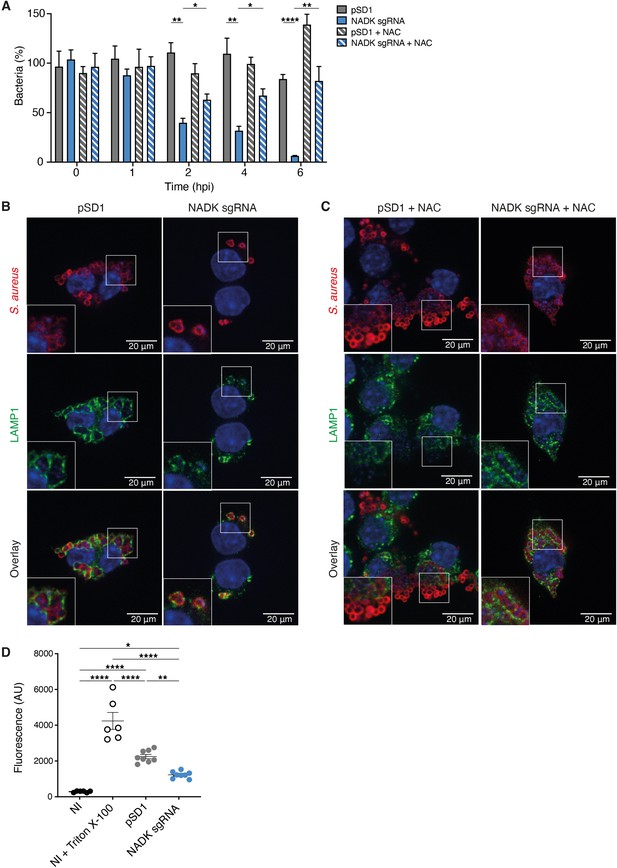
Nicotinamide adenine dinucleotide kinase (NADK) promotes S. aureus survival and cytotoxicity in macrophages.
(A) Percentage of growth of the S. aureus strains containing the empty vector (pSD1) or the ppnK knockdown strain (NADK sgRNA) at time 0, and 1, 2, 4, and 6 hpi of RAW264.7 macrophages left untreated or treated with N-acetylcysteine (NAC). Bars indicate standard errors of the means of biological replicates (n=4). Comparison of data was performed using two-ways analysis of variance (*p<0.05, **p<0.01, ****p<0.0001). (B–C) Representative images of RAW264.7 macrophages infected with S. aureus/pSD1 or S. aureus/NADK sgRNA strains and analyzed 6 hpi by confocal fluorescence microscopy using antibodies to label S. aureus (Cy5, red) and LAMP-1 (FITC, green). Nuclei were labeled with DAPI (blue). (B) shows untreated macrophages. (C) shows macrophages treated with NAC. Insets are shown at higher magnification. Images are representative of three independent experiments. (D) Cell death of RAW264.7 macrophages uninfected (NI), uninfected and treated with Triton X-100 (lysis control), or 6 hpi with S. aureus strains containing the empty vector (pSD1) or the ppnK knockdown strain (NADK sgRNA). Bars indicate standard errors of the means of biological replicates. Comparison of data was performed using two-ways analysis of variance (*p<0.05, **p<0.01, ****p<0.0001). Data are representative of at least three independent experiments.
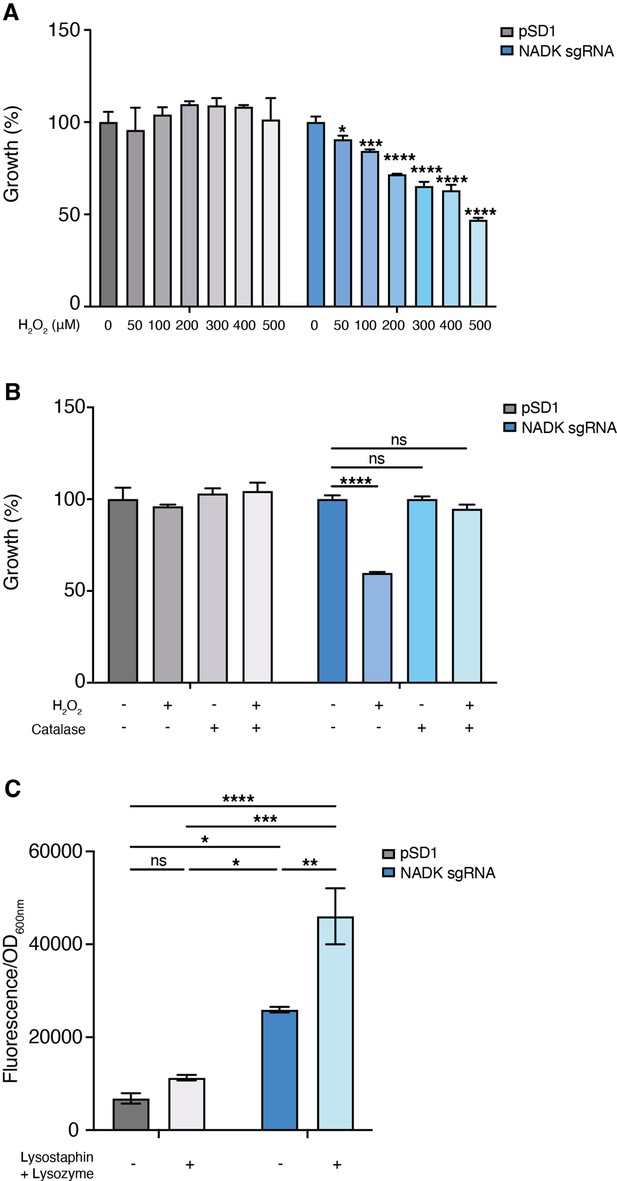
Nicotinamide adenine dinucleotide kinase (NADK) protects S. aureus from antimicrobial defense compounds.
Bacterial growth was monitored at OD600nm in BHI broth at 37°C. (A) Percentage of growth of the S. aureus strain containing the empty vector (pSD1) and the ppnK knockdown strain (NADK sgRNA) exposed for 6 hr to increasing concentrations of H2O2 relative to the untreated condition. (B) Percentage of growth of the S. aureus strain containing the empty vector (pSD1) and the ppnK knockdown strain (NADK sgRNA) exposed for 6 hr to H2O2 and catalase alone or in combination, relative to the untreated condition. Data shown are representative of three independent experiments. Bars indicate the standard error of the means of biological replicates. Comparison of data was performed using one-way analysis of variance (ns: nonsignificant, *p<0.05, ***p<0.001, ****p<0.0001). (C) Bacterial cell death of the S. aureus strain containing the empty vector (pSD1) and the ppnK knockdown strain (NADK sgRNA) untreated or exposed for 30 min to 0.05 mg/mL lysostaphin and 5 mg/mL lysozyme. Bacterial permeability was assessed using the CellTox Green cytotoxicity assay. Bars indicate standard errors of the means of biological replicates. Comparison of data was performed using two-ways analysis of variance (*p<0.05, **p<0.01, ***p<0.001, ****p<0.0001). Data are representative of at least three independent experiments.
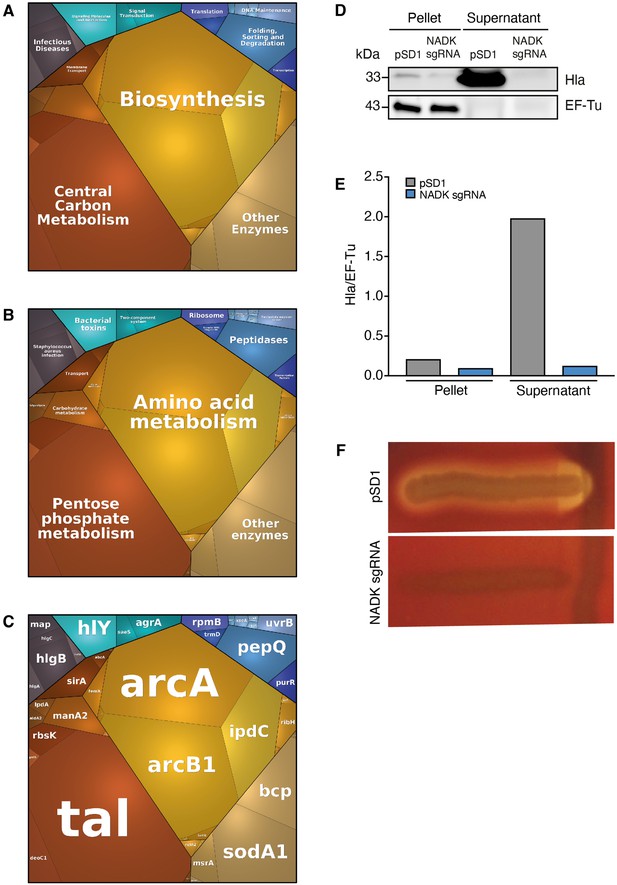
Nicotinamide adenine dinucleotide kinase (NADK) promotes expression of S. aureus virulence determinants.
The S. aureus strain containing the empty vector (pSD1) and the ppnK knockdown strain (NADK sgRNA) were grown in BHI broth at 37°C. (A–C) Whole bacterial cell lysates were analyzed by LC-MS/MS. Voronoi treemaps were generated to visualize proteins more abundant in S. aureus/pSD1 than in the knockdown strain at three hierarchical levels according to KEGG database: top level (A), second level (B), third level (C). Colors represent functional categories. Category size based on LFQ intensity corresponds to differences in protein abundance. (D) Whole bacterial cell lysates and culture supernatants were analyzed by immunoblotting using antibodies against alpha-hemolysin and EF-Tu. (E) Quantification of the Hla immunoblots normalized to corresponding EF-Tu protein levels in the pellet. (F) A Christie-Atkins-Munch-Peterson test was performed by streaking the S. aureus strain containing the empty vector (pSD1) and the ppnK knockdown strain (NADK sgRNA) perpendicularly to the S. aureus RN4220 strain that produces only beta-hemolysin (vertical streak) on sheep blood agar. Clearing zones indicate hemolysis.
-
Figure 4—source data 1
Alpha-hemolysin protein levels (D).
- https://cdn.elifesciences.org/articles/79941/elife-79941-fig4-data1-v1.pdf
-
Figure 4—source data 2
Hemolysis levels (F).
- https://cdn.elifesciences.org/articles/79941/elife-79941-fig4-data2-v1.pdf
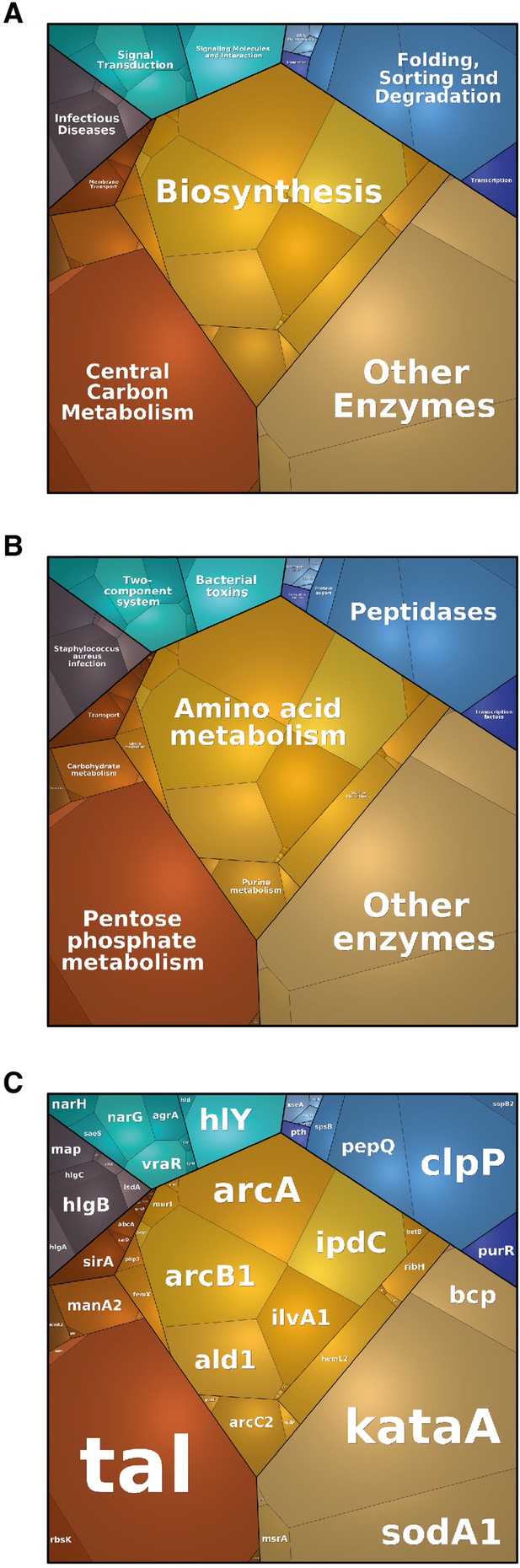
Nicotinamide adenine dinucleotide kinase (NADK) promotes expression of S. aureus virulence determinants.
The S. aureus strain containing the empty vector (pSD1) and the ppnK knockdown strain (NADK sgRNA) were grown at 37°C in BHI broth supplemented with H2O2. (A–C) Whole bacterial cell lysates were analyzed by LC-MS/MS. Voronoi treemaps were generated to visualize proteins more abundant in S. aureus/pSD1 than in the knockdown strain at three hierarchical levels according to KEGG database: top level (A), second level (B), third level (C). Colors represent functional categories. Category size based on LFQ intensity corresponds to differences in protein abundance.
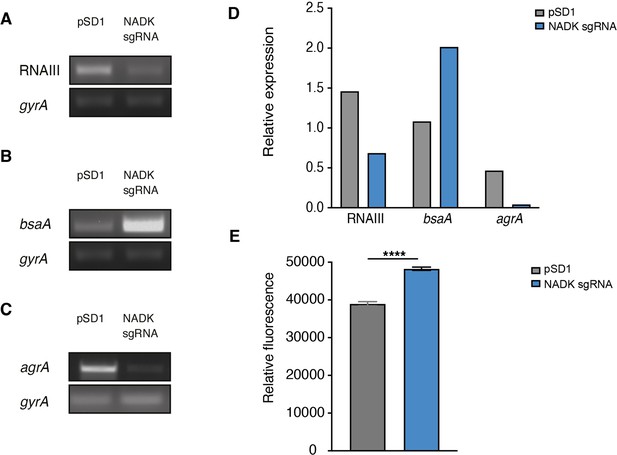
Nicotinamide adenine dinucleotide kinase (NADK) is required for AgrA expression and S. aureus redox control.
The S. aureus strain containing the empty vector (pSD1) and the ppnK knockdown strain (NADK sgRNA) were grown aerobically in BHI broth at 37°C for 4 hr. (A–C) RNA was extracted and subjected to RT-PCR using oligonucleotides specific to gyrA, RNAIII, bsaA, and agrA genes, respectively. (D) Relative gene expression was normalized to gyrA transcript levels. (E) S. aureus ROS levels were quantified using the DCFH2-DA assay. Comparison of data was performed using t-test (****p<0.0001). Data are representative of at least three independent experiments.
-
Figure 5—source data 1
RNAIII transcript levels (A).
- https://cdn.elifesciences.org/articles/79941/elife-79941-fig5-data1-v1.pdf
-
Figure 5—source data 2
BsaA transcript levels (B).
- https://cdn.elifesciences.org/articles/79941/elife-79941-fig5-data2-v1.pdf
-
Figure 5—source data 3
AgrA transcript levels (C).
- https://cdn.elifesciences.org/articles/79941/elife-79941-fig5-data3-v1.pdf
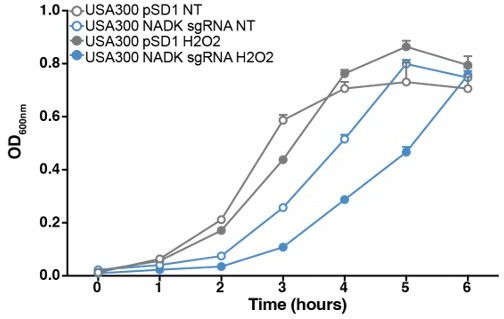
NADK protects S. aureus USA300 from hydrogen peroxide toxicity.
Growth of the USA300 strain containing the empty plasmid (USA300 pSD1, grey) and the NADK knockdown strain (USA300 sgRNA, blue) was monitored at OD600nm in BHI broth at 37°C (NT, open circles) or in BHI exposed to hydrogen peroxide (H2O2, closed circles). Bars indicate the standard errors of the means of biological replicates (n=3).
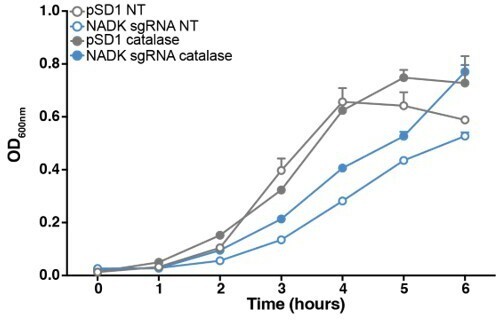
Reactive oxygen species contribute to S. aureus growth defect upon NADK knockdown.
Aerobic growth of the Xen36 strain containing the empty plasmid (pSD1, grey) and the NADK knockdown strain (NADK sgRNA, blue) was monitored at OD600nm in BHI broth at 37°C (NT, open circles) or in BHI supplemented with 10 ug/mL catalase (closed circles). Bars indicate the standard errors of the means of biological replicates (n=3).
Tables
Relative abundance of a subset of protective proteins differentially expressed by S. aureus pSD1 and NADK sgRNA strains.
| Uniprot | Protein | Description | Log2R* | P† |
|---|---|---|---|---|
| Q2FV54 Q2G000 Q2G261 Q2FZZ3 Q2G0D9 Q2G280 P0A086 Q2FVL7 Q2G0E0 Q2FYU7 | OatA Trx2 SodM - GraS - MsrA2 - GraR KatA | O-acetyl transferase Thioredoxin 2 Superoxide dismutase [Mn/Fe] Thioredoxin domain-containing protein Sensor histidine kinase Peroxiredoxin Methionine sulfoxide reductase Thioredoxin domain-containing protein Response regulator protein Catalase | 3.07 1.79 1.68 1.59 1.31 1.26 1.10 1.02 1.00 0.59 | 1.78E-05 2.75E-05 2.11E-06 7.22E-04 4.00E-04 6.69E-07 2.25E-05 1.94E-03 2.23E-03 1.00E-05 |
-
*
Log2R=Log2[pSD1]/[NADK sgRNA].
-
†
Adjusted p-value.
Relative abundance of a subset of virulence-related proteins differentially expressed by S. aureus pSD1 and NADK sgRNA strains.
| Uniprot | Protein | Description | Log2R* | P† |
|---|---|---|---|---|
| Q2FWM4 P0C818 Q2FVK2 P0C7Y1 Q2FUU5 Q2FVK1 Q2G1 × 0 Q2FWN9 Q2FVK3 Q2FWP0 Q2G2R8 | AgrA PsmA4 HlgC PsmA1 Lip1 HlgB Hly LukL2 HlgA LukL1 SspP | Accessory gene regulator protein Phenol-soluble modulin alpha 4 Gamma-hemolysin component C Phenol-soluble modulin alpha 1 Lipase Gamma-hemolysin component B Alpha-hemolysin Leukocidin-like protein 2 Gamma-hemolysin component A Leukocidin-like protein 1 Staphopain A | + + + + + 7.88 6.99 3.88 3.84 3.09 2.89 | NA NA NA NA NA 3.85E-10 3.85E-10 4.14E-08 1.47E-08 2.25E-06 4.08E-07 |
-
*
Log2R=Log2[pSD1]/[NADK sgRNA]; +: protein detected in pSD1 strain and not detected from NADK sgRNA strain.
-
†
Adjusted p-value: NA: not applicable.
Strains and plasmids used in this study.
| Strain/plasmid | Relevant characteristics | Reference |
|---|---|---|
| Strains | ||
| E. coli strain | ||
| TOP10 | F-mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu) 7,697 galU galK λ- rpsL(StrR) endA1 nupG | Invitrogen |
| S. aureus strains | ||
| Xen36 | Strain derived from a clinical isolate from a bacteremic patient | PerkinElmer, ATCC49525 |
| Xen36/pSD1 | Xen36 strain carrying plasmid pSD1 | Gelin et al., 2020 |
| Xen36/NADK sgRNA | Xen36 strain carrying plasmid pSD1 ppnK | Gelin et al., 2020 |
| RN4220 | Restriction-deficient strain derived from NCTC 8325–4 | Kreiswirth et al., 1983 |
| Plasmids | ||
| pSD1 | dCas9 ATc-inducible and sgRNA constitutive expression plasmid | Zhao et al., 2017 |
| pSD1 ppnK | pSD1 plasmid carrying sgRNA targeting S. aureus ppnK | Gelin et al., 2020 |
Primers used in this study.
| Primer | Sequence (5’–3’) |
|---|---|
| RT-ppnK-F | GTGACTCCAAGTCTAATGCC |
| RT-ppnK-R | ATTTTTCAACTTCATGAGGTAACC |
| RT-gyrA-F | GTGTTATCGTTGCTCGTG |
| RT-gyrA-R | CGGTGTCATACCTTGTTC |
| RT-RNAIII-F | AGTTTCCTTGGACTCAGTGCT |
| RT-RNAIII-R | AGGGGCTCACGACCATACTT |
| RT-bsaA-F | GCGAAGAAGCAGCTCAAAAC |
| RT-bsaA-R | CCTTCGCGATCCACTAAAAA |
| RT-agrA-F | GCCCTCGCAACTGATAATCC |
| RT-agrA-R | CACCGATGCATAGCAGTGTTC |
| RT-rluA-F | CCGCGAGAAATACCGAGTGT |
| RT-rluA-R | TCTCCCACAATTGGATGCCC |
| RT-operon-F1 | TGACTTGCTTAAAAAGCACACTG |
| RT-operon-R1 | ACGAGCATTTGTCCTACTTCAGA |
| RT-operon-F2 | AACCGTTGAAGAAACATTCGACA |
| RT-operon-R2 | GACGCTTGTTCACCAACTTCA |
| RT-operon-F3 | CATCGTTTGGAAAGAGCGGC |
| RT-operon-R3 | GGCATTAGACTTGGAGTCACCT |
| RT-operon-F4 | ACGTGTGCACGATTCTTTCAT |
| RT-operon-R4 | ATGGCGCTCACTGTCTTCT |
| RT-operon-F5 | AGTTCATTTGCATACGGGACG |
| RT-operon-R5 | ACGCTCTTTTTCATCTGTGTTCA |
| RT-operon-F6 | TAACTTGTGCGATGACGGTGG |
| RT-operon-R6 | TTCCAATCACAATCCCCATCAA |
| RT-operon-F7 | ACGTTGATGAATTGAAGCAAGAG |
| RT-operon-R7 | ACTTTAGCGACACCAAAAGCA |
| RT-operon-F8 | TCAAGTGGCGTTACAGGTGA |
| RT-operon-R8 | TTCAAATACCGCCAACGCAT |
Additional files
-
Supplementary file 1
Proteins more abundant in the S. aureus pSD1 strain than in the NADK sgRNA strain after growth in BHI broth.
- https://cdn.elifesciences.org/articles/79941/elife-79941-supp1-v1.xlsx
-
Supplementary file 2
Relative abundance of a subset of protective proteins differentially expressed by S. aureus pSD1 and NADK sgRNA strains exposed to hydrogen peroxide.
- https://cdn.elifesciences.org/articles/79941/elife-79941-supp2-v1.docx
-
Supplementary file 3
Proteins more abundant in the S. aureus pSD1 strain than in the NADK sgRNA strain after growth in BHI broth supplemented with hydrogen peroxide.
- https://cdn.elifesciences.org/articles/79941/elife-79941-supp3-v1.xlsx
-
Supplementary file 4
Relative abundance of a subset of protective proteins differentially expressed by S. aureus pSD1 and NADK sgRNA strains upon zebrafish infection.
- https://cdn.elifesciences.org/articles/79941/elife-79941-supp4-v1.docx
-
Supplementary file 5
Proteins more abundant in the S. aureus pSD1 strain than in the NADK sgRNA strain upon zebrafish infection.
- https://cdn.elifesciences.org/articles/79941/elife-79941-supp5-v1.xlsx
-
Supplementary file 6
Relative abundance of a subset of virulence-related proteins differentially expressed by S. aureus pSD1 and NADK sgRNA strains exposed to hydrogen peroxide.
- https://cdn.elifesciences.org/articles/79941/elife-79941-supp6-v1.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/79941/elife-79941-mdarchecklist1-v1.docx





