Phage resistance profiling identifies new genes required for biogenesis and modification of the corynebacterial cell envelope
Figures
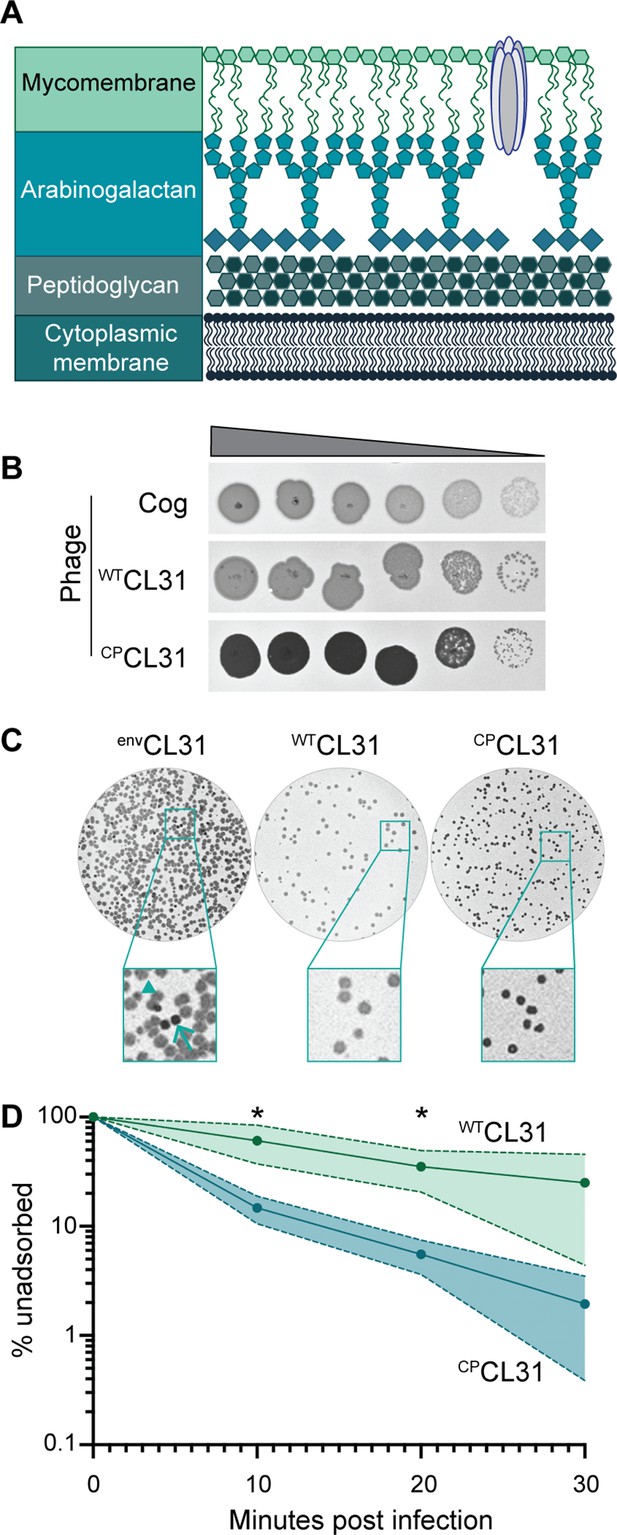
The Corynebacterium glutamicum (Cglu) envelope and adaptation of corynephages to the lab strain.
(A) Cartoon of the multi-layered cell envelope of Cglu and other members of the Corynebacteriales. The mycomembrane is composed of mycolic acids covalently attached to the arabinogalactan layer or conjugated with trehalose. (B) Phages adapted to the MB001 lab strain of Cglu. Ten-fold serial dilutions of lysates for the indicated phages were prepared and 3 µL of each dilution spotted on a lawn of MB001. Large zones of growth inhibition are observed at high phage concentrations and individual plaques are observed at the lowest concentrations. (C) The CL31 phage forms plaques with different morphologies. (Left) Phage grown on the environmental host strain (envCL31) were plated on MB001 and formed both turbid (arrowhead) and clear (arrow) plaques. Both plaque types were purified and replated on MB001, retaining their turbid (WTCL31) or clear phenotype (CPCL31). The CPCL31 phage were found to have a mutation in a gene encoding a tail protein. (D) Phage adsorption assay. The fraction of unadsorbed WTCL31 and CPCL31 phages was determined at the indicated timepoint after mixing with MB001. Shaded regions indicate standard deviation and solid lines indicate the mean from three independent experiments. Significance was determined by two-tailed t test. *p<0.05.
-
Figure 1—source data 1
Single nucleotide polymorphisms (SNPs) detected in phages adapted to MB001.
This table contains the whole-genome sequencing data for the lab-adapted phages.
- https://cdn.elifesciences.org/articles/79981/elife-79981-fig1-data1-v2.docx
-
Figure 1—source data 2
Adsorption source data.
This spreadsheet contains the data used to create Figure 1C.
- https://cdn.elifesciences.org/articles/79981/elife-79981-fig1-data2-v2.xlsx

Adaptation of Cog phage to lab Corynebacterium glutamicum (Cglu) strain MB001 and genome sequencing.
(A) Cog forms plaques on its environmental corynebacterial host LP-6 (top left plate) but fails to form a plaque on the lab Cglu strain MB001 grown on brain-heart infusion (BHI) medium (middle plate on left). However, when using LB 0.4% glucose (LBG) top agar and an MB001 derivative lacking a restriction system (ΔRM), Cog plaques were observed. After a round of plaque purification using the same medium and host, plaques were isolated on wild-type MB001 grown in LBG. Lysates prepared from clear plaques isolated from this lawn were then able to plaque efficiently on MB001 grown in BHI medium. Phage preparations amplified from these plaques on a wild-type MB001 host were sequenced. (B) Cartoon of the Cog genome with the functional modules labeled.
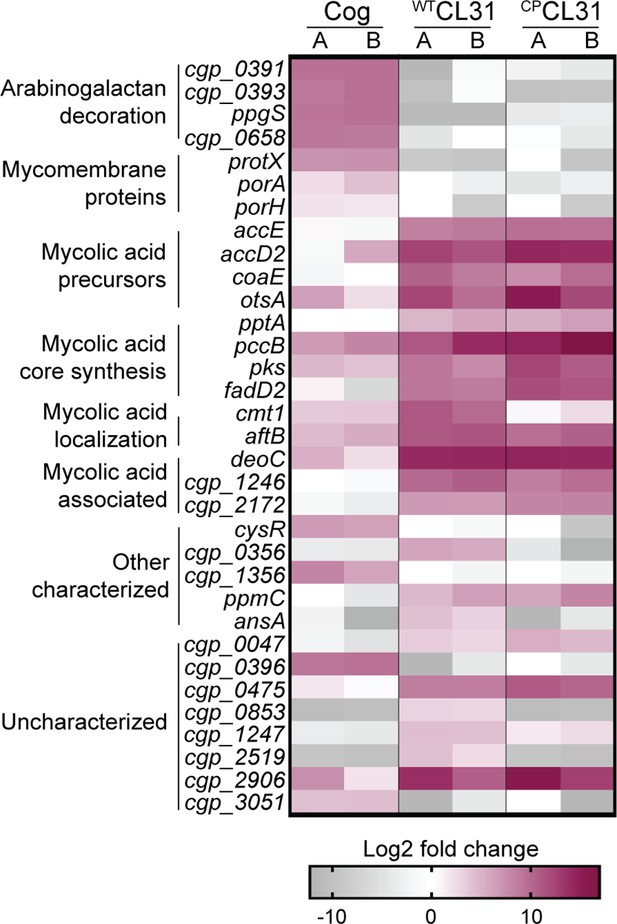
CL31 and Cog have distinct requirements for host envelope components.
The heatmap shows genes that displayed a log2 fold change in transposon insertion reads of >5 in the transposon sequencing (Tn-seq) analysis following challenge with the indicated phage. Each phage challenge was performed independently twice, and the log2 fold changes of each challenge are shown in the A and B columns, respectively. Genes were clustered based on predicted or published functions, which are indicated on the left. Depletion in transposon insertion reads is unlikely to be meaningful given that the experiment was a selection for phage resistance and that transposon mutants in genes that do not promote survival when inactivated are expected to be depleted by phage killing.
-
Figure 2—source data 1
Phage challenge transposon sequencing (Tn-Seq) metadata.
This table contains the metadata associated with the Tn-Seq runs.
- https://cdn.elifesciences.org/articles/79981/elife-79981-fig2-data1-v2.docx
-
Figure 2—source data 2
Transposon sequencing results.
This spreadsheet contains the data acquired from the transposon sequencing experiments.
- https://cdn.elifesciences.org/articles/79981/elife-79981-fig2-data2-v2.xlsx
-
Figure 2—source data 3
Transposon enrichment source data.
This spreadsheet contains the data used to create Figure 2.
- https://cdn.elifesciences.org/articles/79981/elife-79981-fig2-data3-v2.xlsx
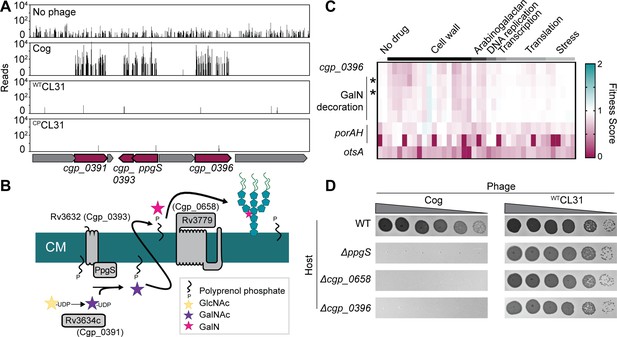
Cog requires genes implicated in arabinogalactan modification to infect Corynebacterium glutamicum (Cglu).
(A) Transposon sequencing (Tn-Seq) insertion profiles for a locus encoding genes with potential functions in the modification of the Cglu arabinogalactan with galactosamine (GalN). Genes enriched for insertions following Cog infection are colored in burgundy. (B) Schematic showing the arabinogalactan modification pathway characterized in Mycobacterium tuberculosis (Mtb). The names of the corresponding Cglu homologs are given in parentheses. (C) Heatmap showing the previously determined phenotypic profiles of the indicated genes following challenge of the transposon library with a variety of antibiotics and other stressors. The target categories of antibiotics used for the challenge are indicated above the heatmap. Note the similarity of the profile for cgp_0396 to that of the gene implicated in GalN modification of arabinogalactan, including the glycosyltransferases ppgS and cgp_0658, indicated with an asterisk. These genes are not clustered in the phenotypic profile near other genes like porAH, or otsA, which have different phenotypic fingerprints that are shown for comparison. Similarity determined by correlation, *r>0.9. Data from Sher et al., 2020. (D) Validation of the Tn-Seq results for the predicted arabinogalactan modification genes. Ten-fold serial dilutions of lysates for the indicated phages were prepared and 3 µL of each dilution were spotted on a lawn of the indicated host strain.
-
Figure 3—source data 1
Phenotypic profiling data.
This spreadsheet contains the data from Sher et al., 2020 used to make Figure 3C.
- https://cdn.elifesciences.org/articles/79981/elife-79981-fig3-data1-v2.xlsx
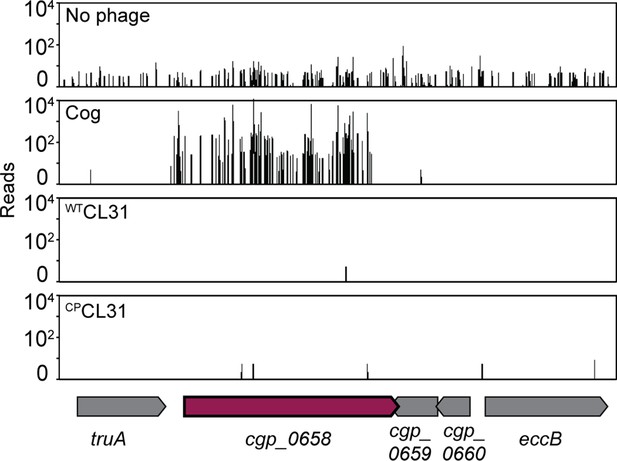
Transposon sequencing (Tn-Seq) insertion profiles for the cgp_0658 locus with and without Cog challenge.
The cgp_0658 gene encodes a putative glycosyltransferase involved in the galactosamine (GalN) modification of arabinogalactan.
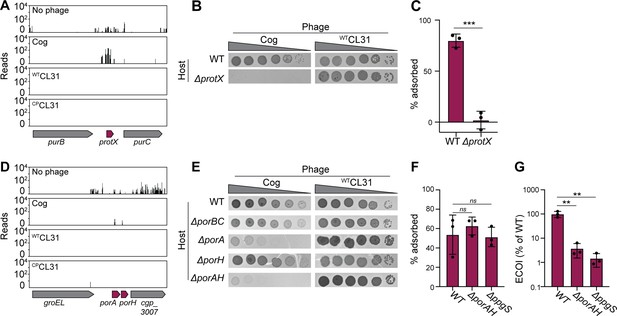
Mycomembrane proteins are required for Cog infection.
(A,D) Transposon sequencing (Tn-Seq) insertion profile for the protX locus and the porAH locus, respectively. (B,E) Validation of the Tn-Seq results. Ten-fold serial dilutions of lysates for the indicated phages were prepared and 3 µL of each dilution were spotted on a lawn of the indicated host strain. (C,F) Percent of Cog adsorbed after 30 min of incubation with the indicated Corynebacterium glutamicum (Cglu) strain. (G). Efficiency by which Cog formed centers of infection (ECOI) on a lawn of wild-type cells following 15 min of adsorption to the indicated host cells. Circles represent individual data points, bar height represents mean, and error bars indicate standard deviation of three independent experiments. Significance was determined by two-tailed t tests. **p<0.005, ***p<0.001, ns, not significant.
-
Figure 4—source data 1
Adsorption and COI data.
This spreadsheet contains the data used to make Figure 4C and D.
- https://cdn.elifesciences.org/articles/79981/elife-79981-fig4-data1-v2.xlsx
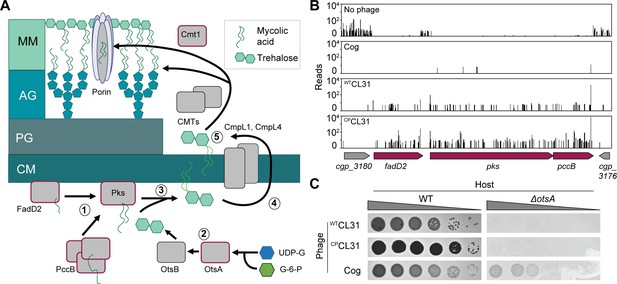
Mycolic acids are required for CL31 infection.
(A) Cartoon of the mycolic acid synthesis pathway. Proteins outlined in burgundy were enriched for transposon insertions following CL31 challenge. (1) Long-chain fatty acids are synthesized by FadD2 and the acyl-CoA carboxylase complex (ACC), including PccB, for condensation by Pks. Concurrently, (2) OtsAB synthesizes trehalose from UDP-glucose (UDP-G) and glucose-6-phosphate (G-6-P). (3) The mycolic acid is attached to trehalose to form trehalose monomycolate (TMM) that is acetylated and reduced before (4) being transported across the membrane by the transporters CmpL1 and CmpL4. Once in the periplasm, (5) TMM is then used as a substrate of the corynemycoloyltransferases (CMTs), including Cmt1, to transfer the mycolate moieties to the arabinogalactan or proteins. (B) Transposon sequencing (Tn-Seq) insertion profile for a locus encoding core mycolic acid synthesis genes. (C) Validation of the Tn-Seq results for ostA. Ten-fold serial dilutions of lysates for the indicated phages were prepared and 3 µL of each dilution were spotted on a lawn of the indicated host strain.

CL31 phage requires genes involved in mycolic acid to infect Corynebacterium glutamicum (Cglu).
(A–C) Transposon sequencing (Tn-Seq) insertion profiles with and without challenge with the indicated phage for the ostA genetic locus.
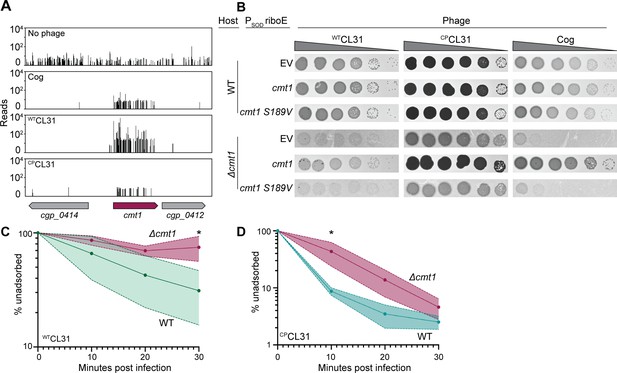
CL31 likely requires a post-translationally modified protein for efficient infection.
(A) Transposon sequencing (Tn-Seq) insertion profiles of cmt1 following phage challenge. (B) Ten-fold serial dilutions of lysates for the indicated phages were prepared and 3 µL of each dilution were spotted on a lawn of the indicated host strain. The strains contain an empty vector (EV) or the indicated gene expressed from the PSOD promoter with translation controlled by the riboE riboswitch. Protein production was induced with 1 mM theophylline. (C–D) Adsorption assay. The fraction of unadsorbed WTCL31 (C) or CPCL31 (D) phages was determined at the indicated timepoints following incubation with wild-type (WT) and Δcmt1 cells of Cglu. Solid line indicates the mean and the shaded regions indicate the standard deviation from three independent experiments. Significance was determined by two-tailed t test. *p<0.05.
-
Figure 6—source data 1
Adsorption data.
This spreadsheet contains the data used to make Figure 6C and D.
- https://cdn.elifesciences.org/articles/79981/elife-79981-fig6-data1-v2.xlsx
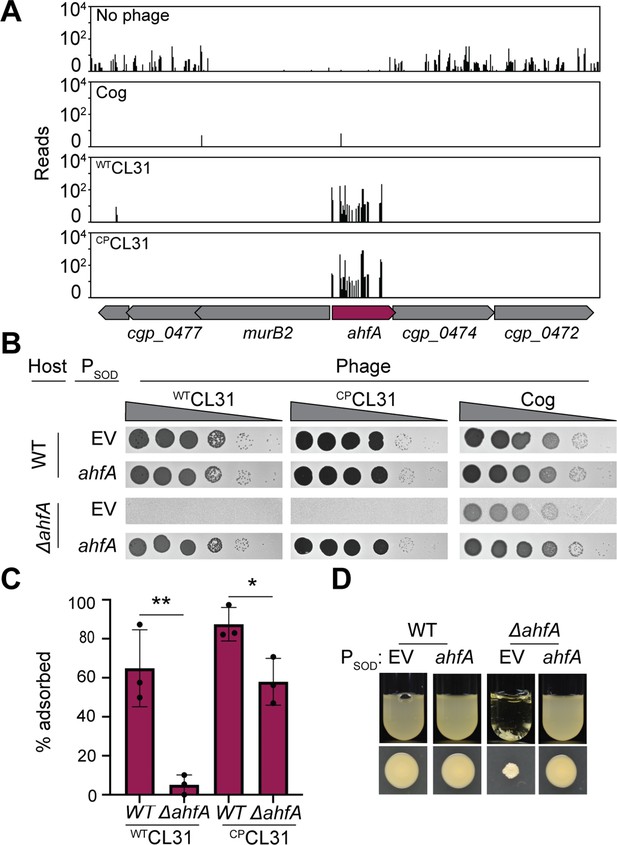
An uncharacterized gene, actinobacterial helix-grip fold A (ahfA) (cgp_0475), is required for CL31 adsorption.
(A) Transposon sequencing (Tn-Seq) insertion profile for the ahfA (cgp_0475) locus. (B) Ten-fold serial dilutions of lysates for the indicated phages were prepared and 3 µL of each dilution were spotted on a lawn of the indicated host strain. (C) The fraction of the indicated CL31 variant adsorbed was determined following a 30 min incubation with the indicated host strain. Dots indicate individual data points, bar height indicates mean, and error bars indicate standard deviation of three independent experiments. Significance was determined by two-tailed t test. *p<0.05, **p<0.005. (D) Representative images of overnight cultures and single colonies of the indicated strains after 3 days incubation at 30°C. Note the clumping and small colony phenotype of cells lacking ahfA.The strains in panels B and D contained an empty vector (EV) or the indicated gene constitutively expressed from the PSOD promoter.
-
Figure 7—source data 1
Single nucleotide polymorphisms (SNPs) in Corynebacterium glutamicum (Cglu) following challenge with WTCL31.
This table contains the whole-genome sequencing results from MB001 survivors following WTCL31 challenged.
- https://cdn.elifesciences.org/articles/79981/elife-79981-fig7-data1-v2.docx
-
Figure 7—source data 2
Adsorption data.
This spreadsheet contains the data used to make Figure 7D.
- https://cdn.elifesciences.org/articles/79981/elife-79981-fig7-data2-v2.xlsx
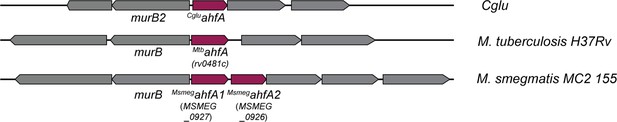
Actinobacterial helix-grip fold A (AhfA) is required for efficient CL31 infection and has homologs in other mycolic acid synthesizing bacteria.
(A) Diagram of the ahfA locus in Corynebacterium glutamicum (Cglu), Mycobacterium tuberculosis (Mtb), and Mycobacterium smegmatis (Msmeg). Notably, there are two ahfA alleles encoded in this locus in Msmeg. Gene numbers of the relevant Mtb and Msmeg alleles are listed in parentheses.
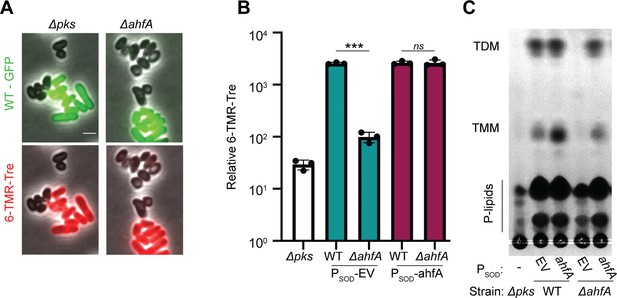
Actinobacterial helix-grip fold A (AhfA) is required for mycolic acid synthesis.
(A) Wild-type (WT) cells expressing gfp or cells of the indicated strains lacking a fluorescent protein marker were grown to an OD600 between 0.15 and 0.4 depending on strain fitness, stained with 6-TAMRA-Trehalose (6-TMR-Tre) for 30 min, washed, mixed together, and then applied to an agarose pad for visualization by fluorescence microscopy. Scale bar represents 2 µm. (B) Quantification of the extent of mycolic acid staining with 6-TMR-Tre as measured by a fluorescence plate reader. Dots indicate individual data points, bar height indicates the mean, and error bars indicate the standard deviation of three independent experiments. Significance was determined by two-tailed t test. ***p<0.001, ns, not significant. (C) TLC analysis of lipids extracted with chloroform/methanol from the indicated Corynebacterium glutamicum (Cglu) strains. The plate was developed in chloroform:methanol:water (30:8:1), dipped in primuline, and spots visualized with UV. Key species are labeled: TMM, trehalose monomycolate; TDM, trehalose dimycolate; P-lipids, phospholipids. The strains in panels B and C contained an empty vector (EV) or the indicated gene constitutively expressed from the PSOD promoter.
-
Figure 8—source data 1
Uncropped microscopy images.
Wild-type (WT) cells expressing gfp or unlabeled Δpks (left) or ΔahfA (right) cells lacking a fluorescent protein marker were grown to an OD600 between 0.15 and 0.4 depending on strain fitness, stained with 6-TMR-Tre for 30 minutes, washed, mixed together, and then applied to an agarose pad for visualization by fluorescence microscopy. Scale bar represents 10 µm and white box indicates area cropped for enlarged figure.
- https://cdn.elifesciences.org/articles/79981/elife-79981-fig8-data1-v2.pdf
-
Figure 8—source data 2
6-TAMRA-Trehalose (6-TMR-Tre) data.
This spreadsheet contains the data used to make Figure 8B.
- https://cdn.elifesciences.org/articles/79981/elife-79981-fig8-data2-v2.xlsx
-
Figure 8—source data 3
TLC analysis of lipids extracted with chloroform/methanol from the indicated Corynebacterium glutamicum (Cglu) strains.
The plate was developed in chloroform:methanol:water (30:8:1), dipped in primuline, and spots visualized with UV. Key species are labeled: TMM, trehalose monomycolate; TDM, trehalose dimycolate; P-lipids, phospholipids. The strains in panels B and C contained an empty vector (EV) or the indicated gene constitutively expressed from the PSOD promoter.
- https://cdn.elifesciences.org/articles/79981/elife-79981-fig8-data3-v2.pdf
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Echerichia coli) | F– hsdR17 deoR recA1 endA1 phoA supE44 thi-1 gyrA96 relA1 Δ(lacZYA-argF)U169 Φ80dlacZΔM15 ****add pir | Gibco BRL | DH5α | |
| Strain, strain background (Cglu, MB001) | MB001 (ATCC 13032 ΔCGP1 (cg1507-gp1524) ΔCGP2 (cg1746-1752) ΔCGP3 (cg1890-cg2071)) | Baumgart et al., 2013 | H60 | |
| Strain, strain background (Cglu) | Environmental Cglu isolate ATCC 15990 | Félix d'Hérelle Reference Center for Bacterial Viruses | ATCC 15990 | |
| Strain, strain background (Cglu) | Environmental Cglu isolate LP-6 | Félix d'Hérelle Reference Center for Bacterial Viruses | LP-6 | |
| Strain, strain background (Cglu, MB001) | ΔRM (Δcgp_0844) | This paper | H722 | Bernhardt Lab |
| Strain, strain background (Cglu, MB001) | ΔppgS (Δcgp_0394) | This paper | H2258 | Bernhardt Lab |
| Strain, strain background (Cglu, MB001) | Δcgp_0658 | This paper | H2259 | Bernhardt Lab |
| Strain, strain background (Cglu, MB001) | Δcgp_0396 | This paper | H2260 | Bernhardt Lab |
| Strain, strain background (Cglu, MB001) | ΔprotX (Δcgp_2875) | This paper | H1111 | Bernhardt Lab |
| Strain, strain background (Cglu, MB001) | ΔporBC (Δcgp_1108c) | This paper | H1239 | Bernhardt Lab |
| Strain, strain background (Cglu, MB001) | ΔporAH (Δcgp_3008–3009) | This paper | H1152 | Bernhardt Lab |
| Strain, strain background (Cglu, MB001) | ΔporH (Δcgp_3009) | This paper | H1241 | Bernhardt Lab |
| Strain, strain background (Cglu, MB001) | ΔporA (Δcgp_3008) | This paper | H1247 | Bernhardt Lab |
| Strain, strain background (Cglu, MB001) | ΔotsA (Δcgp_2907) | This paper | H1666 | Bernhardt Lab |
| Strain, strain background (Cglu, MB001) | Δpks (Δcgp_3178) | This paper | H2261 | Bernhardt Lab |
| Strain, strain background (Cglu, MB001) | Δcmt1 (Δcgp_0413) | This paper | H1664 | Bernhardt Lab |
| Strain, strain background (Cglu, MB001) | ΔahfA (Δcgp_0475) | This paper | H2262 | Bernhardt Lab |
| Strain, strain background (Cglu, MB001) | WT PSOD-ahfA | This paper | H2272 | pK-PIM derivative encoding PSOD-ahfA isothermally assembled and directly transformed into Cglu. |
| Strain, strain background (Cglu, MB001) | ΔahfA PSOD-ahfA | This paper | H2273 | pK-PIM derivative encoding PSOD-ahfA isothermally assembled and directly transformed into Cglu. |
| Strain, strain background (Bacteriophage) | envCL31 (phage isolated on Cglu ATCC 15990) | Félix d'Hérelle Reference Center for Bacterial Viruses Trautwetter et al., 1987 | HER#229 | |
| Strain, strain background (Bacteriophage) | envCog (phage isolated on Cglu LP-6) | Félix d'Hérelle Reference Center for Bacterial Viruses (Sonnen et al., 1990) | HER#360 | |
| Strain, strain background (Bacteriophage) | WTCL31 (T19334G) | This paper | ACMΦ2 | Bernhardt Lab |
| Strain, strain background (Bacteriophage) | CPCL31 (T19334G, T24296A) | This paper | ACMΦ9 | Bernhardt Lab |
| Strain, strain background (Bacteriophage) | Cog (A19202T) | This paper | ACMΦ8 | Bernhardt Lab |
| Recombinant DNA reagent (plasmid) | PSOD riboE EV, inducible (KanR, pK-PIM derivative encoding PSOD-riboE1 empty vector) | This paper | pACM185 | Bernhardt Lab, |
| Recombinant DNA reagent (plasmid) | PSOD riboE cmt1, inducible (KanR, pK-PIM derivative encoding PSOD-riboE1-cmt1) | This paper | pACM64 | Bernhartdt Lab, |
| Recombinant DNA reagent (plasmid) | KanR, pK-PIM derivative encoding PSOD-riboE1-cmt1 S189V (PSOD riboE cmt1 S189V, inducible) | This paper | pACM66 | Bernhardt Lab |
| Recombinant DNA reagent (plasmid) | PSOD EV, constitutive (KanR, pK-PIM derivative encoding PSOD-empty vector) | This paper | pEMH7 | Bernhardt Lab |
| Recombinant DNA reagent (plasmid) | PTAC-gfp, inducible (KanR, PTAC-eGFP, pGA1 mini replicon) | Ravasi et al., 2012 | pTGR5 | |
| Recombinant DNA reagent (plasmid) | KanR, pCRD206 derivative containing an insert covering upstream and downstream of cgp_0844 | This paper | pJWS51 | Bernhardt Lab |
| Recombinant DNA reagent (plasmid) | KanR, pCRD206 derivative containing an insert covering upstream and downstream of cgp_0394 (ppgS) | This paper | pACM285 | Bernhardt Lab |
| Recombinant DNA reagent (plasmid) | KanR, pCRD206 derivative containing an insert covering upstream and downstream of cgp_0658 | This paper | pACM286 | Bernhardt Lab |
| Recombinant DNA reagent (plasmid) | KanR, pCRD206 derivative containing an insert covering upstream and downstream of cgp_0396 | This paper | pACM284 | Bernhardt Lab |
| Recombinant DNA reagent (plasmid) | KanR, pCRD206 derivative containing an insert covering upstream and downstream of cgp_2875 (protX) | This paper | pEMH1 | Bernhardt Lab |
| Recombinant DNA reagent (plasmid) | KanR, pCRD206 derivative containing an insert covering upstream and downstream of cgp_1108-cgp_1109 (porBC) | This paper | pEMH52 | Bernhardt Lab |
| Recombinant DNA reagent (plasmid) | KanR, pCRD206 derivative containing an insert covering upstream and downstream of cgp_3008–3009 (porAH) | This paper | pEMH10 | Bernhardt Lab |
| Recombinant DNA reagent (plasmid) | KanR, pCRD206 derivative containing an insert covering upstream and downstream of cgp_3009 (porH) | This paper | pEMH9 | Bernhardt Lab |
| Recombinant DNA reagent (plasmid) | KanR, pCRD206 derivative containing an insert covering upstream and downstream of cgp_3008 (porA) | This paper | pEMH16 | Bernhardt Lab |
| Recombinant DNA reagent (plasmid) | KanR, pCRD206 derivative containing an insert covering upstream and downstream of cgp_2907 (otsA) | This paper | pACM30 | Bernhardt Lab |
| Recombinant DNA reagent (plasmid) | KanR, pCRD206 derivative containing an insert covering upstream and downstream of cgp_3178 (pks) | This paper | pACM175 | Bernhardt Lab |
| Recombinant DNA reagent (plasmid) | KanR, pCRD206 derivative containing an insert covering upstream and downstream of cgp_0413 (cmt1) | This paper | pACM20 | Bernhardt Lab |
| Recombinant DNA reagent (plasmid) | KanR, pCRD206 derivative containing an insert covering upstream and downstream of cgp_0475 (ahfA) | This paper | pACM230 | Bernhardt Lab |
| Chemical compound. drug | 6-TAMRA-Trehalose (6-TMR-Tre) | Tocris | 6802 |





