Bacillus thuringiensis toxins divert progenitor cells toward enteroendocrine fate by decreasing cell adhesion with intestinal stem cells in Drosophila
Figures

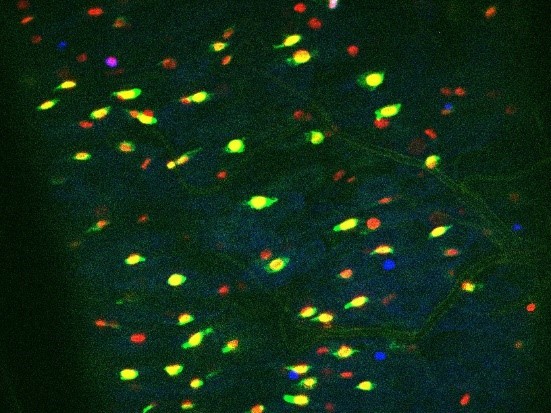
Crystals of Btk Cry protoxins induce EC death and stimulate proliferation of intestinal stem cells.
(A) EC apoptosis was monitored by expressing the Caspase 3 sensor (Casp:: GFP) using the myo1A-GAL4 EC driver (myo1A>Casp::GFP). With this transgenic combination, the GFP is detectable only when the Caspase 3 is activated in ECs. Left panel: ×40 magnification of a R4 subregion. Green stars mark GFP-positive dying ECs. Scale bar = 20 µm. Right panel: quantification of dead ECs 24 hr post ingestion (PI) in the posterior midgut (R4 region). (B) Quantification of mitoses using the anti-PH3 antibody in the whole midgut 24 hr PI. (C) ISC density in the R4 region of esg >GFP flies 24, 72, and 120 hr PI. (D) EC density in the R4 region of myo1A>GFP flies 24, 72, and 120 hr PI. Data is reported as mean ± SEM. ns = not significant; * (p≤0.05); ** (p≤0.01), *** (p≤0.001).
-
Figure 1—source data 1
Cell type counting.
- https://cdn.elifesciences.org/articles/80179/elife-80179-fig1-data1-v1.zip
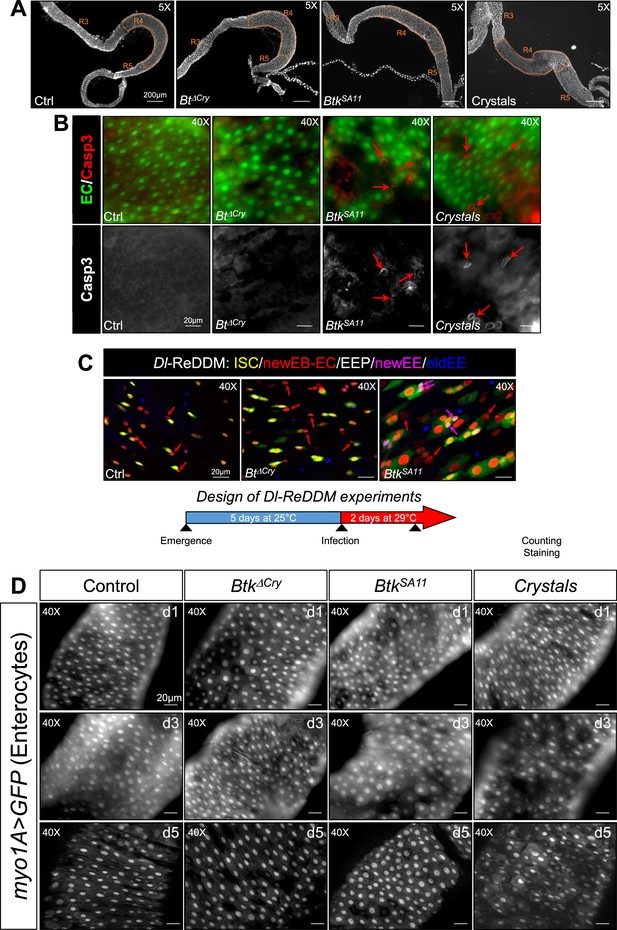
Crystals of Btk Cry protoxins disturb intestinal homeostasis.
(A) Posterior midgut views of wild type flies fed with water (Ctrl), Btk∆Cry or BtkSA11 spores or crystals. The R4 region we analyzed in all this study is detoured in orange. ×5 magnification. Scale bar = 200 µm. (B) Upper panels: myo1A>GFP midguts labeled with the anti-cleaved Caspase 3 antibody (Casp3, red). ECs are labelled by the GFP (green). Lower panels: single anti-cleaved Caspase 3 channel. Red arrows point cleaved Caspase-3-positive cells. Scale bare = 20 µm. (C) Monitoring of the ISC daughter cell commitment in posterior midguts of Dl-ReDDM flies. Drosophila were fed with water, Btk∆Cry spores or BtkSA11 spores. The experimental design is shown below the panels. Anti-Pros (blue) marks the EEPs and EEs. Red arrows point to EBs or ECs (expressing only the RFP). Pink arrows point the EEPs (expressing the GFP, the RFP and Pros) ×40 magnification. Scale bar = 20 µm (D) myo1A>GFP posterior midguts of Drosophila fed with water (control), Btk∆Cry spores, BtkSA11 spores or crystals at 1, 3, and 5 days PI. GFP marks the ECs. ×40 magnification. Scale bar = 20 µm.
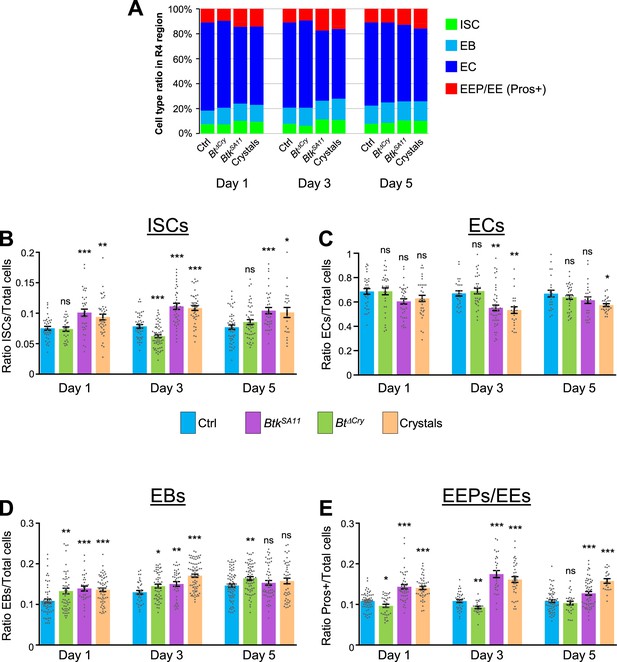
Cell ratio analysis in the R4 region.
(A) Graph representing the proportion of the different cell types in the R4 region of the midgut of flies fed with water (Ctrl), Btk∆Cry spores, BtkSA11 spores or crystals 1, 3, or 5 days PI. (B–E) ISC (B), ECs (C), EBs (D) and EEPs/EEs (E) ratio over the total number of cells in the R4 region 1, 3, or 5 days PI. Data is reported as mean ± SEM. ns (non-significant), * (p≤0.05), ** (p≤0.01), *** (p≤0.001).
-
Figure 1—figure supplement 2—source data 1
Cell ratio analysis in the R4 region.
- https://cdn.elifesciences.org/articles/80179/elife-80179-fig1-figsupp2-data1-v1.zip
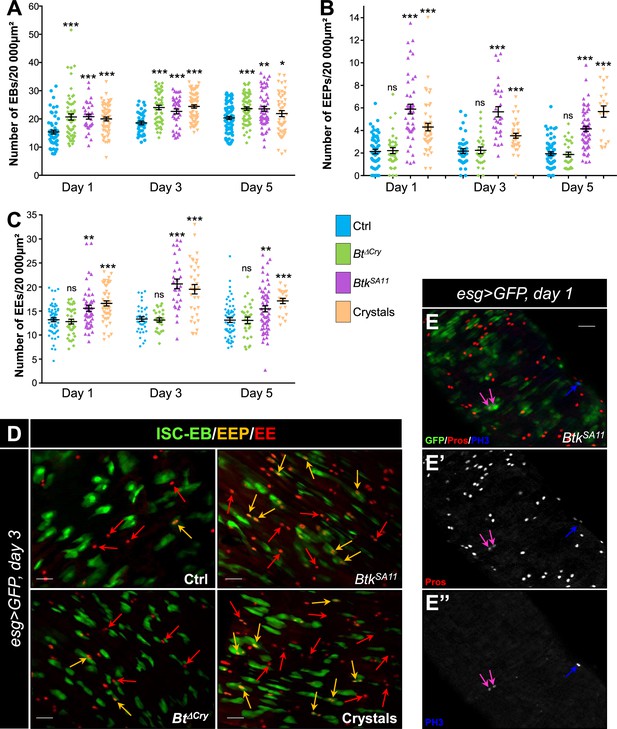
BtkSA11 spores induce an increase in EB, EEP and EE numbers.
(A–D) Flies were fed with water, BtkΔCry spores, BtkSA11 spores or Crystals. (E-E") Flies were fed with BtkSA11 spores. (A–C) Control (water-ctrl): blue; BtkΔCry spores: green; BtkSA11 spores: purple; Crystals: beige. (A) EB density in the R4 region of Su(H)>CD8::GFP flies 24, 48, and 72 h PI. (B and C) EEP (B) and EE (C) density in the R4 region of esg >GFP flies 24, 48, and 72 hr PI. (D-E") R4 region of esg >GFP flies labeled with anti-Pros (Red). GFP was expressed in ISCs, EBs and EEPs, and Pros was expressed in EEPs (yellow arrows in D) and EEs (red arrows in D). (E-E’’) PH3 staining (blue) marks mitosis. Pink arrows point to dividing EEPs and blue arrows point to dividing ISCs. ×40 magnification. Scale bar = 20 µm. Data is reported as mean ± SEM. ns = not significant; * (p≤0.05); ** (p≤0.01), *** (p≤0.001).
-
Figure 2—source data 1
Cell type counting.
- https://cdn.elifesciences.org/articles/80179/elife-80179-fig2-data1-v1.zip
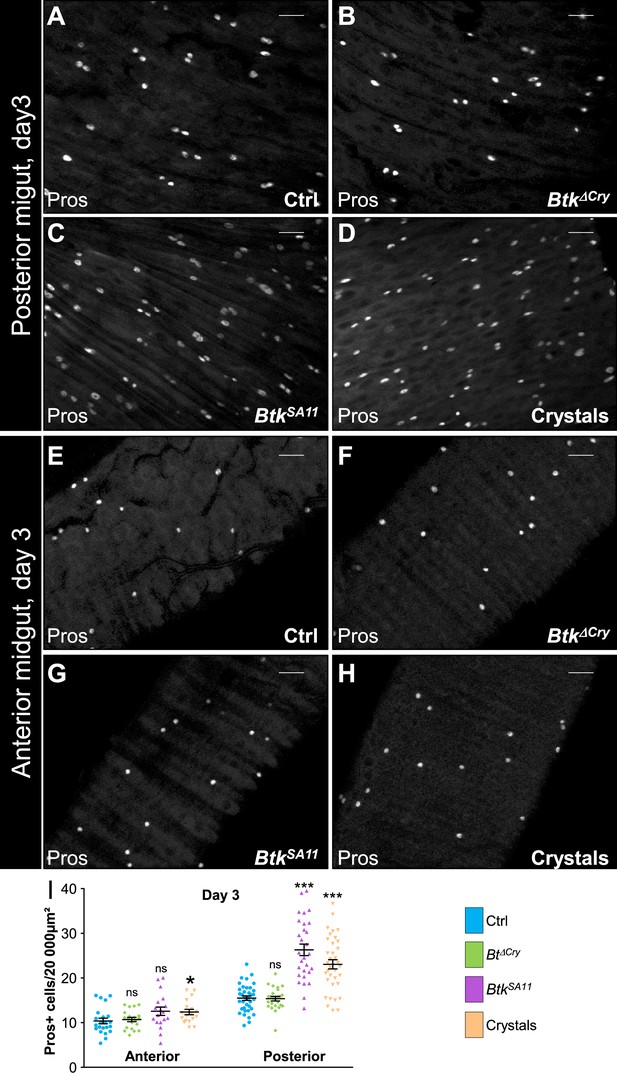
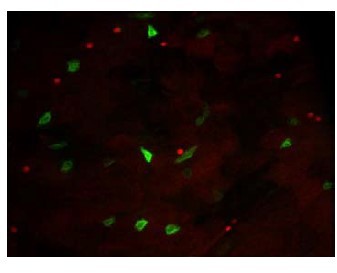
BtkSA11 crystals induce an increase in EEP and EE number in the posterior midgut.
(A–H) Anti-Pros was used to mark the EEs and EEPs. ×40 magnification. Scale bar = 20 µm. (A–D) R4 region of esg >GFP midguts 72 hr PI of water (A), Btk∆Cry spores (B), BtkSA11 spores (C) or crystals (D). (E–H) R2 of region of esg >GFP midguts 72 hr PI of water (E), Btk∆Cry spores (F), BtkSA11 spores (G) or crystals (H). (I) Pros + cell density in the anterior R2 and posterior R4 parts of the midgut 72 hr PI of water (blue), Btk∆Cry spores (green), BtkSA11 spores (purple) or Crystals (beige). Data is reported as mean ± SEM. ns = not significant; * (p≤0.05); *** (p≤0.001).
-
Figure 2—figure supplement 1—source data 1
Cell counting.
- https://cdn.elifesciences.org/articles/80179/elife-80179-fig2-figsupp1-data1-v1.zip

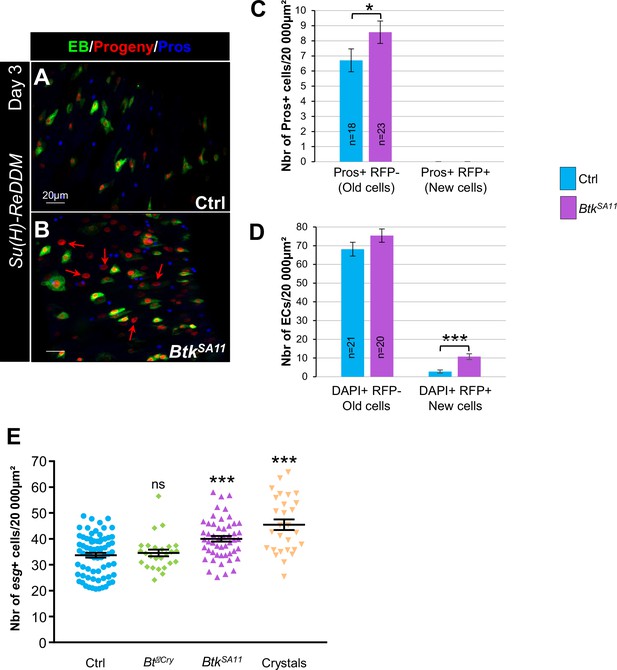
EE excess arises from newborn EEPs after ingestion of BtkSA11 crystals.
(A) Schema of the experimental design for the esg-ReDDM cell lineage used in this entire figure. (B-E") R4 region of esg-ReDDM flies. Midguts were stained for Pros (blue) and DAPI which marks nuclei (white in B, C, D and E). (B-E’’) show the different cell types which either existed before the ingestion (green and red) or arise after the ingestion (red only) of water (B-B", Ctrl), Btk∆Cry spores (C-C") or BtkSA11 spores (D-D") and Crystals (E-E’’). ISCs were GFP + RFP + DAPI +with small nuclei; EBs were GFP + RFP + DAPI +with bigger nuclei; EEPs were GFP + RFP + Pros +DAPI + ; new EEs were RFP+, Pros + DAPI + ; old EEs were Pros +DAPI + ; new ECs were RFP + DAPI + with polyploid big nuclei and old ECs were DAPI +with very big nuclei. 40 X magnification. Scale bar = 20 µm. (F) Counting old EEs (Pros + RFP-) and new EEs (Pros + RFP + ) in the conditions described in (B–E). (G) Counting old ECs (DAPI+) and new ECs (DAPI + RFP + ) in the conditions described in (B–E) n=number of 40 x images analyzed Data is reported as mean ± SEM. ns = not significant; * (p≤0.05); *** (p≤0.001).
-
Figure 3—source data 1
Cell type counting.
- https://cdn.elifesciences.org/articles/80179/elife-80179-fig3-data1-v1.zip
EBs do not give birth to EEs.
(A and B) R4 region of Su(H)- ReDDM flies. Flies were fed with either water (A, Ctrl) or BtkSA11 spores (B). Midguts were labelled for Pros (blue). ×40 magnification. Scale bar = 20 µm. (C) Counting of old EEs (Pros + RFP-) and new EEs (Pros + RFP + ) in the conditions described in (A and B). n=number of 40 X images analyed. (D) Counting of old (DAPI+, RFP-, large nucleus) and new ECs (DAPI+, RFP+, large nucleus) in the condition described in (A and B). n=number of 40 X images analyzed. (E) Density of the esg+ (GFP+) cells in the esg-ReDDM experiments shown in Figure 3A–E”.Data is reported as mean ± SEM. ns (non-significant), * (p≤0.05), *** (p≤0.001).
-
Figure 3—figure supplement 1—source data 1
Cell type counting.
- https://cdn.elifesciences.org/articles/80179/elife-80179-fig3-figsupp1-data1-v1.zip

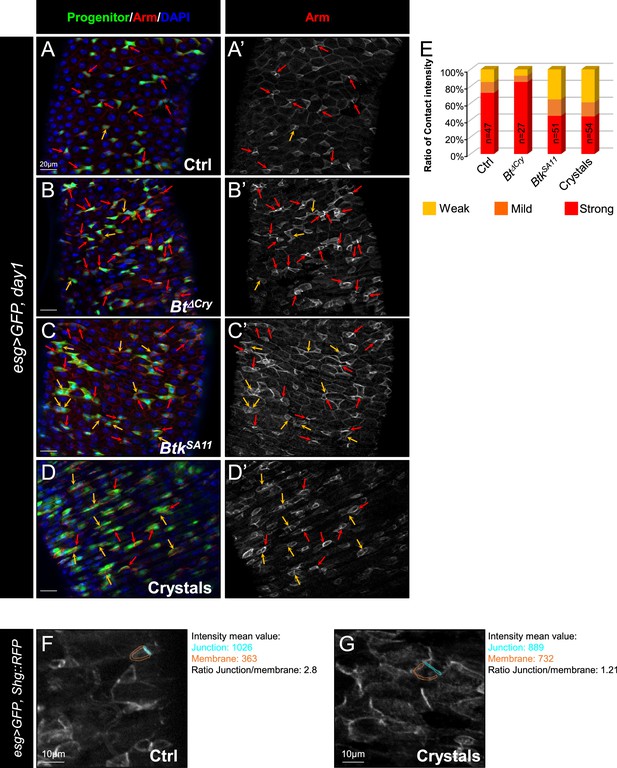
Btk crystals decrease ISC-Progenitor cell-cell adhesion.
(A–E) esg >UAS GFP, Tomato::shg Drosophila midgut R4 region 24 hr PI of water (A-A” Ctrl), Btk∆Cry spores (B-B”), BtkSA11 spores (C-C”) or crystals (D-D”). (A-D") Midguts are labelled for Pros (blue), DE-Cadherin (red) and ISCs and progenitors (green). Red arrows point to the high intensity of adherens junctions staining between ISC and progenitors. Yellow arrows point to the weak intensity of adherens junction staining. Note that the high intensity of adherens junction staining is associated with ISC/EB interaction while the weak intensity of adherens junction staining is associated with ISC/EEP interaction (blue stars mark EEPs). ×40 magnification. Scale bar = 10 µm. (E) Graph representing the percentage of the different categories of cell contact intensity between ISCs and progenitors. n=number of cell pairs analyzed. Weak = Contact Intensity/Membrane Intensity <1.4; Mild = 1.4 < Contact Intensity/Membrane Intensity <1.6; Strong = Contact Intensity/Membrane Intensity >1.6.
-
Figure 4—source data 1
Ratio of contact intensity.
- https://cdn.elifesciences.org/articles/80179/elife-80179-fig4-data1-v1.zip
Btk bioinsecticide decreases ISC-Progenitor cell-cell adhesion.
(A–E) esg-GAL4 UAS-GFP (esg >GFP) Drosophila midgut R4 region 24 hr PI of water (A-A’ Ctrl), Btk∆Cry spores (B-B’), BtkSA11 spores (C-C’) or crystals (D-D’). Midguts are labeled with anti-Armadillo (Arm) (red) which strongly marks the adherens junctions between ISC and progenitors (green). Dapi (blue) marks the nuclei. (A', B', C', and D') correspond to the single Arm channel. Red arrows point to the high intensity of adherens junctions staining between pairs of GFP + cells (ISCs and progenitors). Yellow arrows point to the weak intensity of adherens junction staining. ×40 magnification. Scale bar = 20 µm. (E) raph representing the percentage of the different categories of cell contact intensity between pairs of GFP + cells (ISCs and progenitors) in the experimental conditions shown in (A-D’). n=number of GFP + cell pairs analyzed. Weak = Contact Intensity/Membrane Intensity <1.4; Mild = 1.4 < Contact Intensity/Membrane Intensity <1.6; Strong = Contact Intensity/Membrane Intensity >1.6. (F and G) Example of adherens junction intensity measurements in esg >GFP, shg::RFP midgut R4 region of flies fed with water (Ctrl, F) of Crystals (G). The ratio correspond to the average intensity of the junction (turquoise)/average intensity of the rest of the plasma membrane (blue). ×40 magnification. Scale bar = 10 µm.
-
Figure 4—figure supplement 1—source data 1
Ratio of contact intensity.
- https://cdn.elifesciences.org/articles/80179/elife-80179-fig4-figsupp1-data1-v1.zip
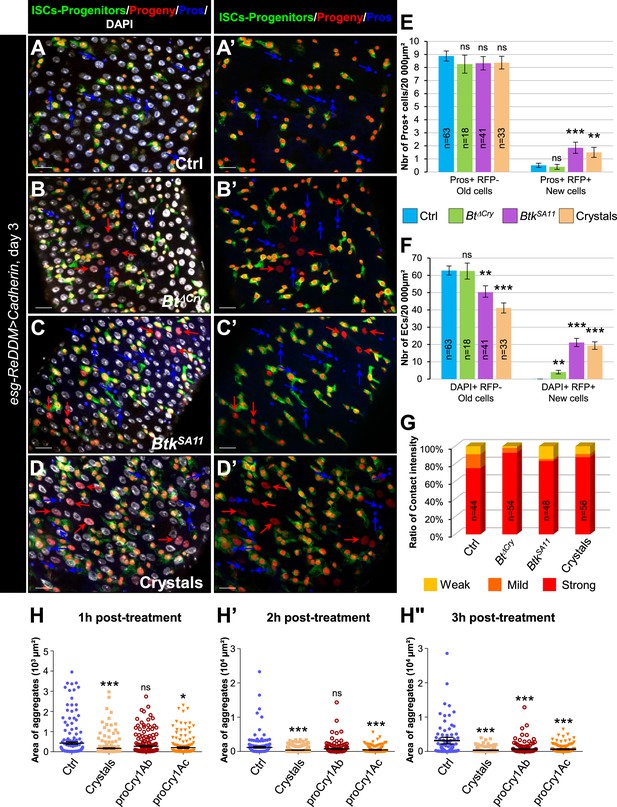
Increasing adherens junction strength rescues crystal-dependent cell fate diversion.
(A–G) esg-ReDDM >DE Cad Drosophila midgut R4 region. These flies specifically overexpress the DE-Cad in ISCs and progenitors. Flies were fed with water (A-A', and blue in E and F, Ctrl), Btk∆Cry spores (B, B’, and green in E and F), BtkSA11 spores (C, C’ and purple in E and F) or Crystal (D-D' and beige in E and F) and observed 72 h PI (see Figure 3A for the experimental design). In (A-D') blue arrows point to old EEs and red arrows newborn ECs. (E) Number of old EEs (Pros + RFP-) and new EEs (Pros + RFP + ) and (F) number of old ECs (DAPI +RFP-) and new ECs (DAPI + RFP + ) in the conditions described in (A–D). (A-D') 40 X magnification. Scale bar = 20 µm. (G) Graph representing the percentage of the different categories of cell contact intensity between ISCs and progenitors in the experimental conditions shown in Figure 5—figure supplement 1A–D’. Weak = Contact Intensity/Membrane Intensity <1.4; Mild = 1.4 < Contact Intensity/Membrane Intensity <1.6; Strong = Contact Intensity/Membrane Intensity >1.6. n=number of cell pairs analysed. (H-H") Cell aggregation assays on S2 cells expressing the DE-Cadherin::GFP. Cells placed under constant rotation were incubated with or without Bt crystals or purified Cry protoxins (Cry1Ab or Cry1Ac) for 1 hr (G), 2 hr (G') or 3 hr (G"). Each scatter plot represents the area (µm2) of all objects (aggregates or individual cells) obtained from three independent experiments. Representative images of cell aggregates formed in aggregation assays are shown in Figure 5—figure supplement 1F data. In (E and F), n=number of 40 X images analyzed. In (G), n=number of cell pairs analyzed. Data is reported as mean ± SEM. ns (non-significant), * (p≤0.05), ** (p≤0.01), *** (p≤0.001).
-
Figure 5—source data 1
Assessment of ISC division.
- https://cdn.elifesciences.org/articles/80179/elife-80179-fig5-data1-v1.zip
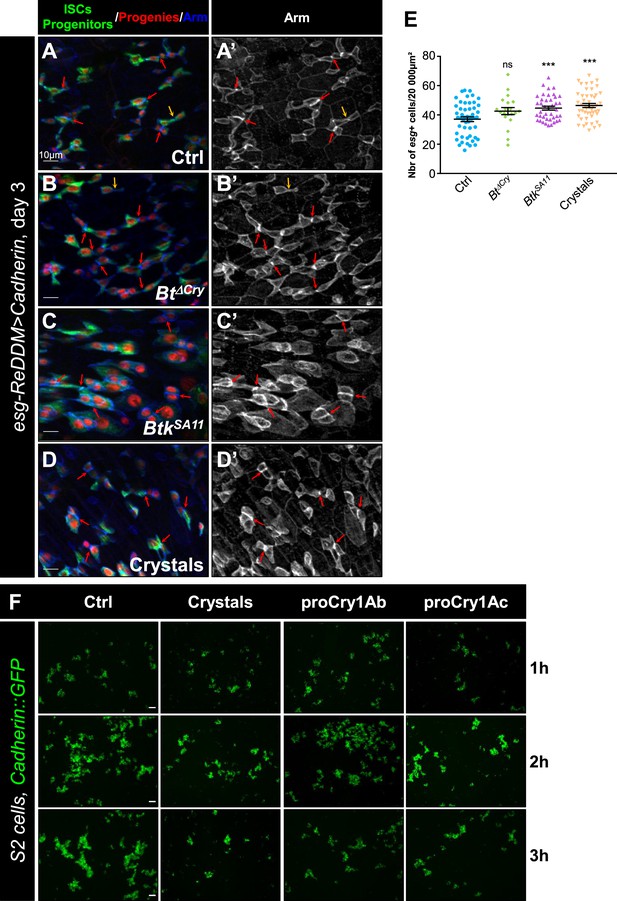
Cry1A protoxins reduced homophilic interactions of DE-cadherin.
(A–E) esg-ReDDM >DE Cad Drosophila midgut R4 region. Flies were fed with water (A and A', Ctrl), Btk∆Cry spores (B and B’), BtkSA11 spores (C and C’) or Crystals (D and D') and observed 72 hr PI (see Figure 3A for the experimental design). Midguts were labeled with anti-Arm (blue) which strongly marks the adherens junctions between GFP + pairs of cells (ISC and progenitors, green). (A’, B’, C’ and D’) correspond to the single Arm channel. Red arrows point to the high intensity of adherens junction staining. Yellow arrows point to the weak intensity of adherens junction staining. ×40 magnification. Scale bar = 10 µm. (E) Counting of esg+ (GFP + cells) corresponding to ISCs and progenitors. Data is reported as mean ± SEM. ns = not significant; *** (p≤0.001). (F) Representative images of cell aggregation assays obtained from three independent experiments. S2 cells transiently expressing the DE-Cadherin::GFP were placed under constant rotation and incubated for 1, 2, or 3 hr with or without Btk crystals or purified Cry1Ab or Cry1Ac protoxins. Scale bars = 50 µm.
-
Figure 5—figure supplement 1—source data 1
Assessment of ISC division.
- https://cdn.elifesciences.org/articles/80179/elife-80179-fig5-figsupp1-data1-v1.zip
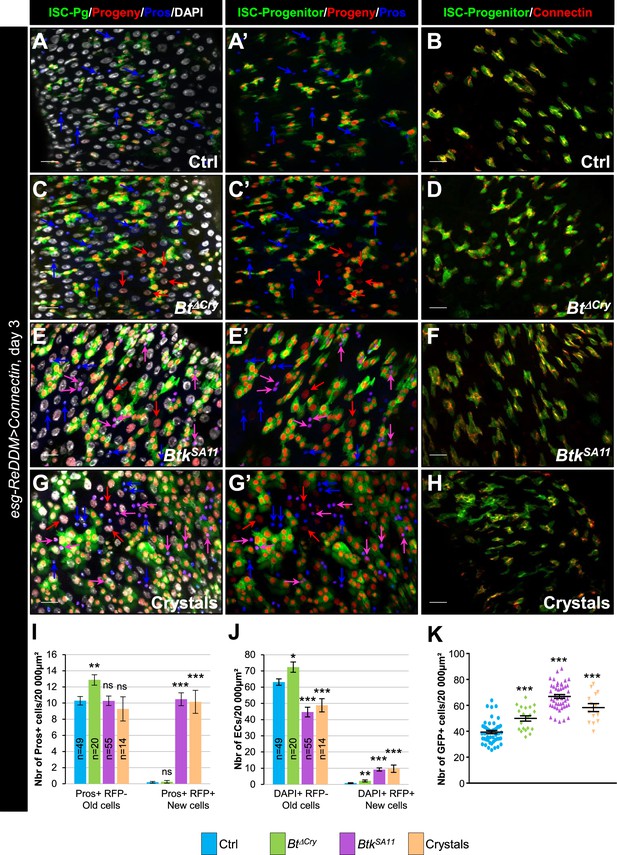
Connectin overexpression does not rescue cell adhesion disturbance induced by Btk crystals of toxins.
(A–K) esg-ReDDM >connectin. These flies specifically overexpress Connectin in ISCs and progenitors. Flies were fed with water (A-A’, B and blue in I, Ctrl), Btk∆Cry spores (C, C’, D and green in I), BtkSA11 spores (E, E’, F and purple in I) or Crystals (G-G’, H and pink in I) and observed 72 hr PI. Midguts were stained for Pros (blue in A, A', C, C', E, E', G and G') and for Connectin (red in B, D, F, and H). DAPI marks the nuclei (white in A; C, E and G). Blue arrows point to old EEs, pink arrows point to new EEs and red arrows newborn ECs. ×40 magnification. Scale bar = 20 µm. (I) Number of old EEPs-EEs (Pros + RFP-) and new EEPs-EEs (Pros + RFP + ) in the conditions described in (A–H). n=number of 40 x images analyzed. (J) Number of old ECs (DAPI+, RFP-, large nucleus) and new ECs (DAPI + RFP + , large nucleus) in the conditions described in (A–H). n=number of 40 x images analyzed. (K) Counting of esg+ cells (GFP+, ISCs and progenitors). Data is reported as mean ± SEM. ns (non-significant), * (p≤0.05), ** (p≤0.01), *** (p≤0.001).
-
Figure 5—figure supplement 2—source data 1
Counting of cell types.
- https://cdn.elifesciences.org/articles/80179/elife-80179-fig5-figsupp2-data1-v1.zip
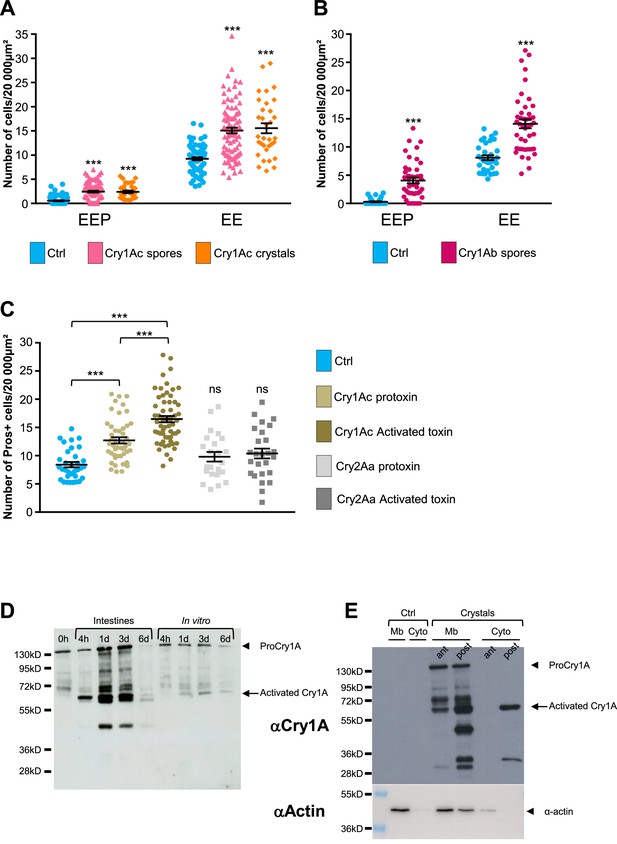
Cry1A toxins mimic Btk crystal effects.
(A–C) esg >GFP flies fed with water (blue, Ctrl), BtkCry1Ac spores (fuchsia in A), Cry1Ac crystals (orange in A), BtkCry1Ab spores (rose in B), Cry1Ac protoxins (light khaki in C), Cry1Ac activated toxins (khaki in C), Cry2Aa protoxins (light grey in C) and Cry2Aa activated toxins (grey in C). ns (non-significant). Data is reported as mean ± SEM. *** (p≤0.001). (A and B) Density of EEPs or EEs in the R4 region 72 hr PI. (C) Density of Pros + cells in the R4 region 72 hr PI. (D and E) Western Blot from dissected intestines using a polyclonal Anti-Cry1A antibody detecting both the protoxins and the activated forms of Cry1A family of toxins.(D) (left lane) 0 h corresponds to BtkSA11 spores extemporaneously resuspended in water. (Right part of the blot) BtkSA11 spores incubated ex vivo (control) in water at 25 °C for the period of the experiment. We mainly detect the protoxin forms of Cry1A at 130 kDa (arrowhead). (Left part of the blot). Proteins extract from midguts of flies fed by the same BtkSA11 preparation (T 0 h) at 4 hr and 1, 3, and 6 days PI. The 130 kDa protoxins are still visible. The 67 kDa activated form appears as early as 4 hr (arrow). 6 days PI no more toxins are detected in the midgut. (E) Flies fed 2 days with water (Ctrl, left part) or with purified crystals (right part). Protoxins (130 kDa) are present in the insoluble fraction (Mb) in both the anterior (ant) and the posterior (post) midgut. The 67 kDa activated forms are present in the insoluble fraction of both the anterior and posterior midgut and in the soluble fraction (Cyto) of the posterior midgut. Actin was used as western blot loading control, especially for the insoluble fraction.
-
Figure 6—source data 1
Counting of cell types.
- https://cdn.elifesciences.org/articles/80179/elife-80179-fig6-data1-v1.zip
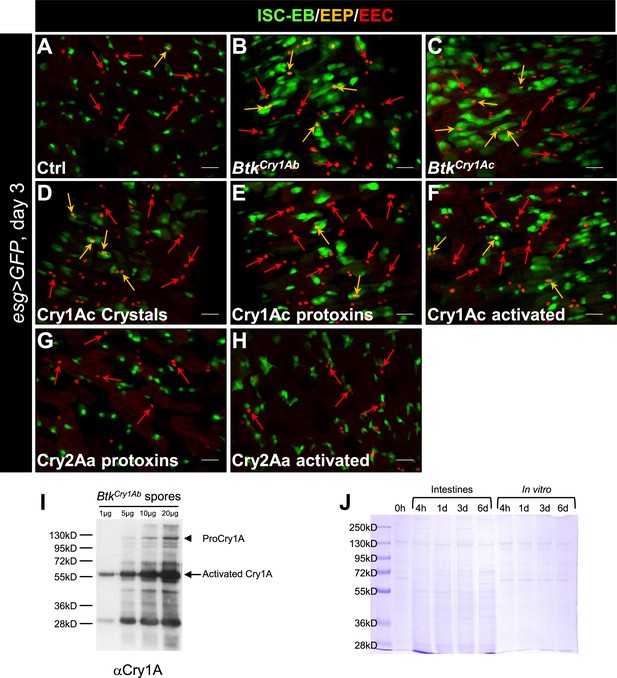
Cry1A toxins mimic Btk crystal effects.
(A–H) esg >GFP Drosophila midgut R4 region 72 hr PI of water (A, Ctrl), BtkCry1Ab spores, (B), BtkCry1Ac spores (C), Cry1Ac crystals (D), Cry1Ac protoxins (E), Cry1Ac activated toxins (F), Cry2Aa protoxins (G) and Cry2Aa activated toxins (H). GFP labels ISCs and progenitors, Pros labels EEPs and EEs (red). Orange arrows point to EEPs and red arrows points to the EEs. ×40 magnification. Scale bar = 20 µm. (I) Western blot on BtkCry1Ab spores using the Anti-Cry1A polyclonal antibody. Spores of BtkCry1Ab were resuspended in water to obtain a concentration of 1 µg/µL. 1, 5, 10, and 20 µg of proteins were loaded on a 10% gel of acrylamide. The protoxins (130 kDa, arrowhead) are present but in very low quantities. The activated 67 kDa forms (arrow) are more abundant.
-
Figure 6—figure supplement 1—source data 1
The uncropped images of Western blots.
- https://cdn.elifesciences.org/articles/80179/elife-80179-fig6-figsupp1-data1-v1.zip
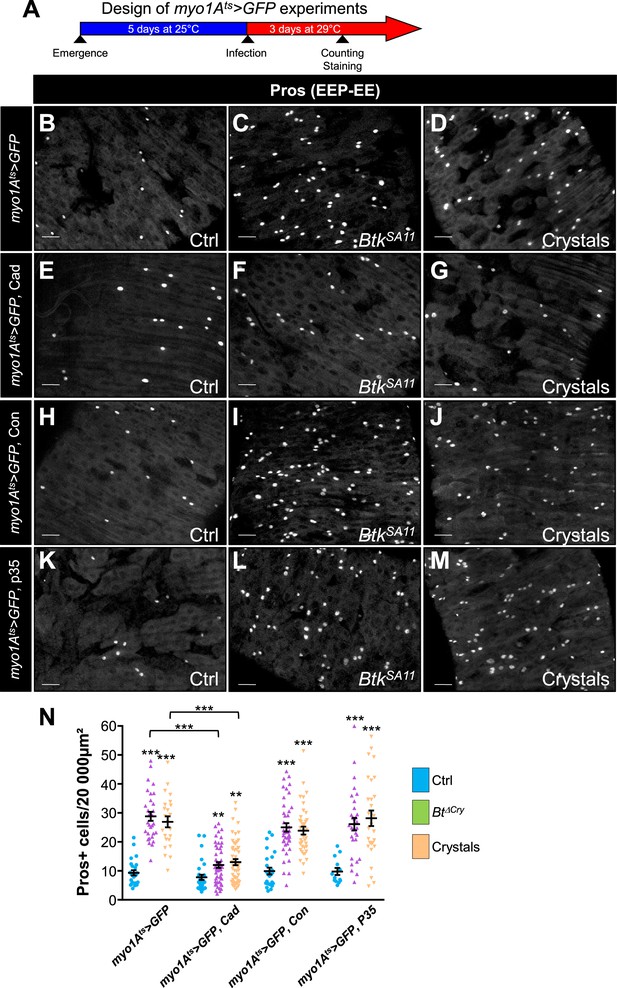
Cry1A toxins likely cross the intestinal barrier through EC transcytosis.
(A) Schema of the experimental design for the myo1A-GAL4 UAS-GFP tub-GAL80ts (myo1Ats >GFP) overexpression in ECs used in this entire figure. (B–N) R4 region of midguts of flies fed with water (Ctrl, B, E, H, K and blue in N), BtkSA11 spores (C, F, I, L and green in N) and crystals (D, G, J,M and beige in N) and labelled for Pros. ×40 magnification. Scale bar = 20 µm. (B–D) myo1Ats >GFP midguts. (E–G) myo1Ats >GFP midguts overexpressing DE-Cad. (H–J) myo1Ats >GFP midguts overexpressing Connectin (Con). (K–L) myo1Ats >GFP midguts overexpressing the anti-apoptotic p35 factor. (N) Counting of EEPs/EEs (Pros + cells) in the different conditions described in (B–M). Data is reported as mean ± SEM. ** (p≤0.01), *** (p≤0.001).
-
Figure 7—source data 1
Counting of Prospero positive cells.
- https://cdn.elifesciences.org/articles/80179/elife-80179-fig7-data1-v1.zip
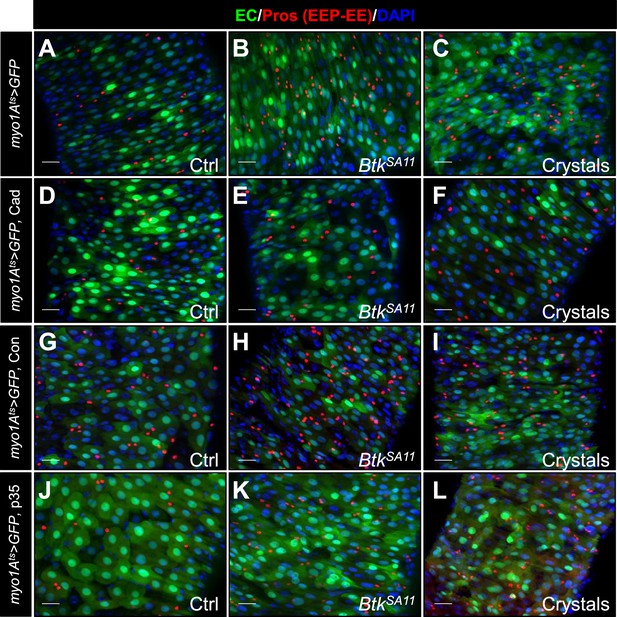
Cry1A toxins likely cross the intestinal barrier through EC transcytosis.
(A–L) R4 region of flies midguts fed with water (Ctrl, A, D, G and J), BtkSA11 spores (B, E, H and K) and crystals (C, F, I and L) and labelled for Pros (red). GFP marks ISCs and progenitors (green). DAPI marks the nuclei (blue). These images correspond to the overlays of the Figure 7B–M. (A–C) myo1Ats > GFP midguts. (D–F) myo1Ats > GFP midguts overexpressing DE-Cad transgene. (G–I) myo1Ats > GFP midguts overexpressing Connectin (Con). (J–L) myo1Ats > GFP midguts overexpressing the anti-apoptotic p35 factor. ×40 magnification. Scale bar = 20 µm.
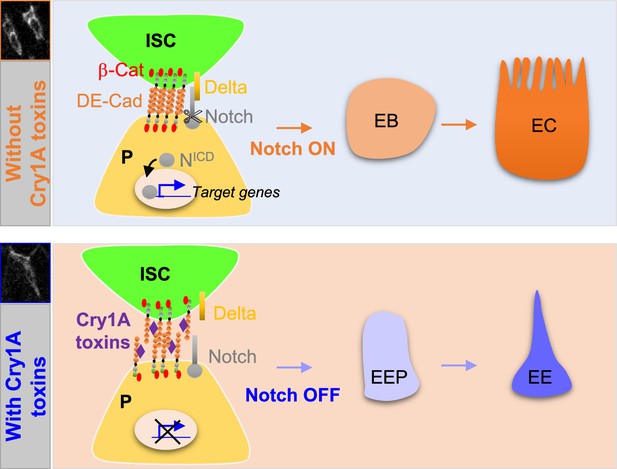
Cry1A toxins interfere with progenitor fate behavior.
Notch ON: in Drosophila, 90% of ISC daughter cells commit to the EB/EC fate owing to the strong activation of the Notch signaling pathway in the EBs. The adherens junction DE-Cadherin (DE-Cad)-dependent are required to permit the interaction between the Delta ligand in ISC and the Notch receptor in EB. Notch OFF: Ingestion of Cry1A toxins impedes the DE-Cad homophilic interaction between the ISCs and their progenitor daughter cells, reducing the activation of Notch signaling in progenitors. Consequently, progenitors adopt an EEP/EE fate.
Tables
Cell junction intensity ratio measurement between pairs of progenitors.
Cadherin::RFP labeling intensity were measured first at the cell junction between pairs of progenitors and second around the rest of the cell membrane (see Figure 4—figure supplement 1F–G). Ratio correspond to the Junction intensity/the rest of the membrane. Prospero positive progenitors were both GFP+/Pros+ (see Figure 4). Yellow highlight labels Pros + progenitors with a weak intensity ratio. Orange highlight labels Pros + progenitors with a medium intensity ratio. Red highlight labels Pros + progenitors with a strong intensity ratio.
| H2OIntensity ratio | Pros + | BtkΔCry Intensity ratio | Pros + | BtkSA11Intensity ratio | Pros + | Crystals Intensity ratio | Pros + |
|---|---|---|---|---|---|---|---|
| 2,29 | 1,46 | 1,49 | Yes | 1,27 | Yes | ||
| 1,37 | 2,27 | 1,38 | Yes | 2,03 | |||
| 1,43 | 2,25 | 1,91 | 0,78 | Yes | |||
| 1,75 | 1,92 | 1,52 | 1,18 | Yes | |||
| 1,87 | 1,51 | 1,89 | 1,89 | ||||
| 2,37 | 2,19 | 1,88 | 2,56 | ||||
| 2,54 | 1,96 | 1,19 | Yes | 2,20 | |||
| 1,77 | 1,91 | 2,27 | 3,12 | ||||
| 1,45 | 1,35 | Yes | 1,63 | 4,05 | |||
| 1,49 | 1,94 | 1,39 | 1,91 | ||||
| 1,83 | 2,46 | 1,54 | Yes | 1,70 | |||
| 1,76 | 2,93 | 1,87 | 1,76 | ||||
| 2,20 | 2,29 | 2,55 | 2,17 | ||||
| 1,25 | yes | 3,60 | 1,63 | 1,55 | |||
| 2,48 | 3,03 | 1,84 | 2,06 | ||||
| 1,12 | yes | 2,12 | 1,31 | Yes | 1,77 | ||
| 1,75 | 2,30 | 0,79 | Yes | 1,62 | |||
| 1,67 | 1,36 | Yes | 1,55 | ||||
| 1,43 | 2,91 | 1,98 | |||||
| 2,41 | 1,60 | 1,89 | |||||
| 2,64 | 2,82 | 1,65 | |||||
| 2,49 | 1,55 | 1,78 | |||||
| 1,60 | Yes | 3,00 | 1,21 | Yes | |||
| 2,95 | 1,88 | 1,86 | Yes | ||||
| 2,67 | 2,32 | 1,07 | Yes | ||||
| 2,31 | 3,69 | 2,24 | |||||
| 2,24 | 2,93 | 2,20 | |||||
| 2,45 | 1,87 | 1,08 | Yes | ||||
| 1,49 | 2,55 | 1,46 | |||||
| 2,06 | 1,70 | 1,72 | |||||
| 3,67 | 2,80 | 2,29 | |||||
| 2,57 | 3,74 | 1,74 | |||||
| 2,33 | 2,27 | 0,92 | Yes | ||||
| 1,50 | 1,92 | ||||||
| 2,10 | 1,79 | ||||||
| 1,47 | 2,39 | ||||||
| 1,76 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| strain, strain background (B. thuringiensis) | Btk SA-11 | Isolated from commercial product | Delfin | |
| strain, strain background (B. thuringiensis) | Btk∆Cry | BGSC | #4D22 | |
| strain, strain background (B. thuringiensis) | Btk producing Cry1Ac | BGSC | #4D4 | |
| strain, strain background (B. thuringiensis) | Btk producing Cry1Ab | This study | Materials and Methods: Generation of BtkCry1Ab | |
| strain, strain background (Escherichia coli) | producing Cry1Ab | BGSC | #ECE54 | |
| strain, strain background (Escherichia coli) | producing Cry1Ac | BGSC | #ECE53 | |
| strain, strain background (Escherichia coli) | producing Cry2Aa | BGSC | #ECE126 | |
| genetic reagent (D. melanogaster) | WT canton S | https://bdsc.indiana.edu/ | #64349 | |
| genetic reagent (D. melanogaster) | w; Sco/CyO; tub-GAL80ts/TM6b | https://bdsc.indiana.edu/ M. Vidal | #7018 | |
| genetic reagent (D. melanogaster) | w; tub-GAL80ts; TM2/TM6b | https://bdsc.indiana.edu/ | #7019 | |
| genetic reagent (D. melanogaster) | w;; Dl-GAL4/TM6b | S. Hou and X. Zeng; Zeng et al., 2010 | ||
| genetic reagent (D. melanogaster) | w; tub-GAL80ts; Dl-GAL4 UAS-GFP/TM6b | This study | Can be obtained from Gallet's lab | |
| genetic reagent (D. melanogaster) | w; esg-GAL4NP5130 | https://bdsc.indiana.edu/ N. Tapon | #67054 | |
| genetic reagent (D. melanogaster) | w; esg-GAL4NP5130 UAS-GFP | N. Tapon; Shaw et al., 2010 | ||
| genetic reagent (D. melanogaster) | w; esg-GAL4NP5130 UAS-GFP; tubGAL80ts | Y. Apidianakis; Apidianakis et al., 2009 | ||
| genetic reagent (D. melanogaster) | w; Su(H)GBE-GAL4, UAS-CD8::GFP | M. Vidal; Zeng et al., 2010 | ||
| genetic reagent (D. melanogaster) | w; Su(H)GBE-GAL4/SM6; tub-GAL80ts UAS-GFP/TM6b | This study | Can be obtained from Gallet's lab | |
| genetic reagent (D. melanogaster) | w; myo1A-GAL4 | N. Tapon; Shaw et al., 2010 | ||
| genetic reagent (D. melanogaster) | w; myo1A-GAL4 UAS-GFP/CyO | Y. Apidianakis; Apidianakis et al., 2009 | ||
| genetic reagent (D. melanogaster) | w; UAS-GFP/TM3 Sb | https://bdsc.indiana.edu/ | #5430 | |
| genetic reagent (D. melanogaster) | w; UAS-GFP::CD8; UAS-H2B::RFP/TM2 | T. Reiff and M. Dominguez; Antonello et al., 2015 | ||
| genetic reagent (D. melanogaster) | w; UAS-CD8::GFP; UAS-H2B::RFP, tub-GAL80ts/TM2 | T. Reiff and M. Dominguez; Antonello et al., 2015 | ||
| genetic reagent (D. melanogaster) | w; esg-GAL4, UAS-CD8::GFP/CyO; UAS-H2B::RFP, tub-GAL80ts/TM6b | T. Reiff and M. Dominguez; Antonello et al., 2015 | ||
| genetic reagent (D. melanogaster) | w; UAS-CD8::GFP; Dl-GAL4, UAS-H2B::RFP/TM6b | This study | Can be obtained from Gallet's lab | |
| genetic reagent (D. melanogaster) | w;; UAS-GC3AiG7S (UAS-Casp::GFP) | M. Suzanne; Schott et al., 2017 | ||
| genetic reagent (D. melanogaster) | w; UAS-shg-R (DE-Cadherin) | https://bdsc.indiana.edu/ | #58494 | |
| genetic reagent (D. melanogaster) | w; UAS-connectin | JP Boquete and B. Lemaitre; Zhai et al., 2017 | ||
| genetic reagent (D. melanogaster) | y w, shg::Tomato | https://bdsc.indiana.edu/ | #58789. | |
| genetic reagent (D. melanogaster) | w;UAS-p35 | Tony Ip; Amcheslavsky et al., 2009 | ||
| genetic reagent (D. melanogaster) | Dl-ReDDM (w/w; UAS-CD8::GFP/UAS-CD8::GFP; Dl-GAL4, UAS-H2B::RFP/UAS-H2B::RFP, tub-GAL80ts) | This study | Can be obtained from Gallet's lab | |
| genetic reagent (D. melanogaster) | esg-ReDDM (w/w+; esg-GAL4, UAS-CD8::GFP/+; UAS-H2B::RFP, tub-GAL80ts/+) | This study | Can be obtained from Gallet's lab | |
| genetic reagent (D. melanogaster) | Su(H)-ReDDM (w/w; Su(H)-GAL4/UAS-GFP::CD8; tub-GAL80ts UAS-GFP/UAS-H2B:RFP) | This study | Can be obtained from Gallet's lab | |
| cell line (D. melanogaster) | Drosophila melanogaster Schneider 2 (S2) cells | S2-DGRC Stock 181 | RRID:CVCL_Z992 | |
| antibody | Mouse monoclonal anti-Armadillo (ß-catenin) antibody | DSHB | Cat# N27A1 RRID:AB_528089 | 1/50 |
| antibody | Mouse monoclonal anti-Connectin antibody | DSHB | Cat# Connectin C1.427, RRID:AB_1066083 | 1/200 |
| antibody | Mouse monoclonal anti-Prospero antibody | DSHB | Cat# MR1A RRID:AB_528440 | 1/200 |
| antibody | Rabbit polyclonal anti-Cleaved Caspase-3 (Asp175) antibody | Cell Signalling | Cat# 9661 RRID:AB_2341188 | 1/600 |
| antibody | Rabbit polyclonal anti-phospho-Histone H3 (Ser10) antibody | Millipore | Cat# 06–570 RRID:AB_31017 | 1/1000 |
| antibody | Rabbit polyclonal anti-Cry1A antibody | Babin et al., 2020 | WB: 1/7500 i | |
| antibody | Mouse monoclonal anti-actin antibody (ACTN05, C4) antibody | Invitrogen | Thermo Fisher Scientific Cat# MA5-11866, RRID:AB_10985365 | WB: 1/2000 |
| antibody | Goat anti mouse IgG (H+L) secondary antibody, AlexaFluor-647 | Invitrogen | Molecular Probes Cat# A-21235, RRID:AB_2535804 | 1/500 |
| antibody | Goat polyclonal anti mouse IgG (H+L) secondary antibody, AlexaFluor-546 | Invitrogen | Molecular Probes Cat# A-11003, RRID:AB_141370 | 1/500 |
| antibody | Goat polyclonal anti-rabbit IgG (H+L) secondary antibody, AlexaFluor-647 | Invitrogen | Thermo Fisher Scientific Cat# A32733, RRID:AB_2633282 | 1/500 |
| antibody | Goat polyclonal anti-rabbit IgG (H+L) secondary antibody, AlexaFluor-546 | Invitrogen | Thermo Fisher Scientific Cat# A-11010, RRID:AB_253407 | 1/500 |
| recombinant DNA reagent | pUAST-DECadherintagged with GFP (DEFL) | Oda and Tsukita, 1999 | Materials and Methods: Cell aggregation assay | |
| recombinant DNA reagent | pWA-Gal4 | Gift from L. Ruel | ||
| Software, algorithm | Image J | http://imagej.nih.gov | RRID:SCR_003070 | |
| Software, algorithm | Fiji | http://fiji.sc | RRID:SCR_002285 | |
| Software, algorithm | ZEN 2 (blue edition) | Zeiss | ||
| Software, algorithm | Photoshop CS2 | Adobe | ||
| Software, algorithm | GraphPad Software | GraphPad Prism | RRID:SCR_002798 | GraphPad Prism 7.0 |
| Chemical compound, drug | PBS 10 x | Euromedex | ET330 | |
| Chemical compound, drug | Formaldehyde 16% | Thermo Fisher Scientific | Cat# 28908 | |
| Chemical compound, drug | Fluoroshield-DAPI | Sigma | Cat# F6057 | |
| Chemical compound, drug | Tween 20 | VWR | Cat# 28829.296 | |
| Chemical compound, drug | Acrylamide/Bis-acrylamide | Sigma | Cat# A3699 | |
| Commercial assay, kit | Invitrogen MycoFluor Mycoplasma Detection Kit | Thermo Fisher Scientific | Cat# 10063202 | |
| Other | Amersham Hyperfilm | GE Healthcare | Cat# 28906837 | Commercial product |
| Other | Bovin Serum Albumin | Sigma | Cat# A9647 | Commercial product |
| Other | Schneider’s insect medium | Sigma-Aldrich | Cat# S0146 | Commercial product |
| Other | TransIT–2020 | Mirus Bio | Cat# MIR5400 | Commercial product |
| other | Zeiss Axioplan Z1 with Apotome 2 microscope | Zeiss | Microscope | |
| other | Zeiss Confocal LSM 810 | Zeiss | Microscope |