Obesogenic diet induces circuit-specific memory deficits in mice
Figures
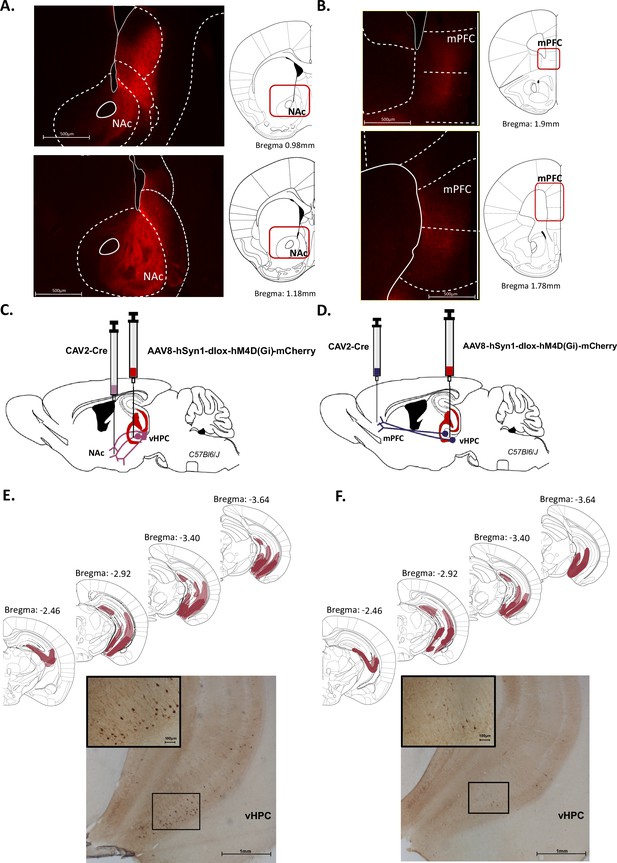
Characterization of ventral hippocampus (vHPC) projections to nucleus accumbens (NAc) and medial prefrontal cortex (mPFC).
Representative images illustrating expression of TdTomato in fibres in the NAc (A) and the mPFC (B) after AAV-CaMKII-Cre injection in the vHPC of Ai14(RCL-tdT)-D mice. Schematics adapted from Figures 21 and 23 (A) and Figures 14 and 16 (B) from Paxinos and Franklin, 2004, indicating the levels of fibres labelling. (C, D) Schema of intersectional chemogenetic approach. An AAV-hSyn1-dlox-hM4D(Gi)-mCherry vector was injected into the vHPC, while a retrograde CAV2-Cre vector was injected in the NAc (C) or the mPFC (D). (E, F) Expression of mCherry is depicted for each condition after amplification using immunohistochemistry. Schematics adapted from Figures 51, 55, 59 and 61 from Paxinos and Franklin, 2004, indicating the largest (light red) or the smallest (dark red) viral infection. Representative images illustrating mCherry expression after CAV2-Cre injection in (E) the NAc or (F) the mPFC. Scale bar is set to 100 µm, 500 µm, or 1 mm.

Labelling in the ventral hippocampus (vHPC).
Parasagittal (A) and frontal (B) schema illustrating AAV-CaMKII-Cre injection in the vHPC of Ai14(RCL-tdT)-D mice and representative image illustrating expression of TdTomato in vHPC neurons (C). Representative example of mCherry labelling in the vHPC after injections of AAV-hSyn1-dloxhM4D(Gi)-mCherry in the vHPC and retrograde CAV2-Cre vector in the nucleus accumbens (NAc, D) or the medial prefrontal cortex (mPFC, E).
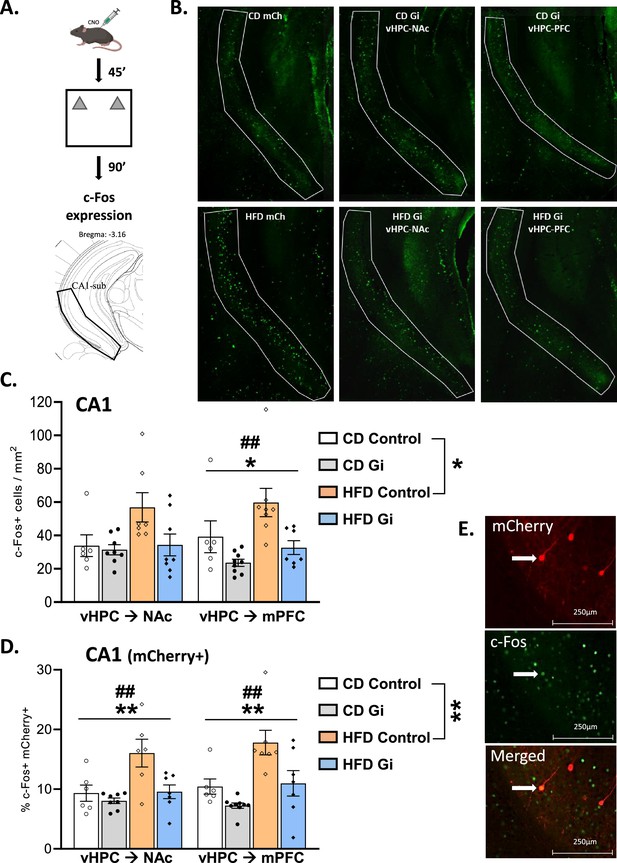
Effects of high-fat diet and chemogenetic silencing of ventral hippocampus (vHPC)–nucleus accumbens (NAc) or vHPC–medial prefrontal cortex (mPFC) pathway on c-Fos expression in ventral CA1/subiculum.
(A) Schema of exposure to a novel arena containing two identical and novel objects. (B) Representative images of c-Fos-positive (Fos+) cells in the ventral CA1/subiculum for each group. (C) Quantification of c-Fos+ cells in the ventral CA1/subiculum of the different groups. Data are shown as the number of c-Fos+ cells per mm2. The area corresponding to ventral CA1/subiculum area is delineated. Scale bar is set at 500 nm. (D) Representative example of mCherry and c-Fos labelling in the ventral CA1/subiculum. (E) Percentage of mCherry+ and c-Fos+ cells over total number of mCherry+ in the different groups. Scale bar is set to 250 nm and data are shown as a percentage of cells. Each point represents a single animal value. Diet effect: *p < 0.05, **p < 0.01; DREADD effect: ##p < 0.01 (two-way analysis of variance [ANOVA]).
-
Figure 2—source data 1
Effects of high-fat diet and chemogenetic silencing of ventral hippocampus (vHPC)–nucleus accumbens or vHPC–medial prefrontal cortex pathway on c-Fos expression in ventral CA1/subiculum.
- https://cdn.elifesciences.org/articles/80388/elife-80388-fig2-data1-v1.xlsx

Effect of high-fat diet and chemogenetic manipulation on c-Fos expression in hippocampus, nucleus accumbens (NAc) and medial prefrontal cortex (mPFC).
Quantification of c-Fos-positive cells in the ventral CA3 (A), the NAc (B) and the mPFC (C) of the different groups. Data are shown as the number of c-Fos-positive cells per mm². Diet effect: *p < 0.05, **p < 0.01, ***p < 0.001; DREADD effect: ##p < 0.01.
-
Figure 2—figure supplement 1—source data 1
Effect of high-fat diet and chemogenetic manipulation on c-Fos expression in hippocampus, nucleus accumbens and medial prefrontal cortex.
- https://cdn.elifesciences.org/articles/80388/elife-80388-fig2-figsupp1-data1-v1.xlsx
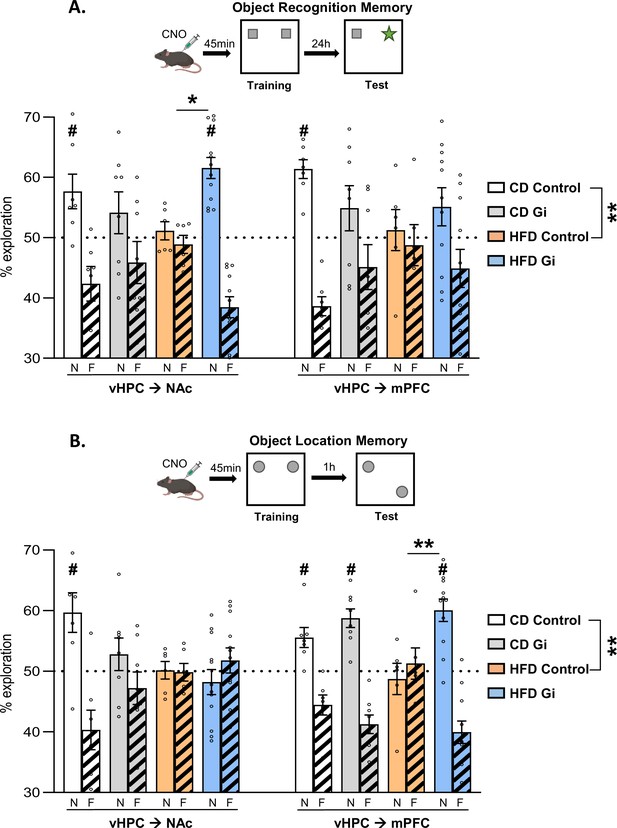
Impacts of high-fat diet and chemogenetic silencing of ventral hippocampus (vHPC)–nucleus accumbens (NAc) or vHPC–medial prefrontal cortex (mPFC) pathway on object-based memory.
(A) Schema of object recognition memory (ORM) task (top) and ORM performance expressed as percentage of exploration time of novel (empty bars) or familiar (striped bars) object over both objects (bottom). (B) Schema of object location memory (OLM) task (top) and OLM performance expressed as percentage of exploration time of novel (empty bars) or familiar (striped bars) location over both objects (bottom). Each point represents a single animal value. Diet effect: **p < 0.01 (two-way analysis of variance [ANOVA], diet × pathway). Difference between groups: *p < 0.05, **p < 0.01 (two-way ANOVA, diet × DREADD for each pathway, significant interaction followed by post hoc test). Different from 50%: #p < 0.05 (one-sample t-test).
-
Figure 3—source data 1
Impacts of high-fat diet and chemogenetic silencing of ventral hippocampus (vHPC)–nucleus accumbens or vHPC–medial prefrontal cortex pathway on object-based memory.
- https://cdn.elifesciences.org/articles/80388/elife-80388-fig3-data1-v1.xlsx
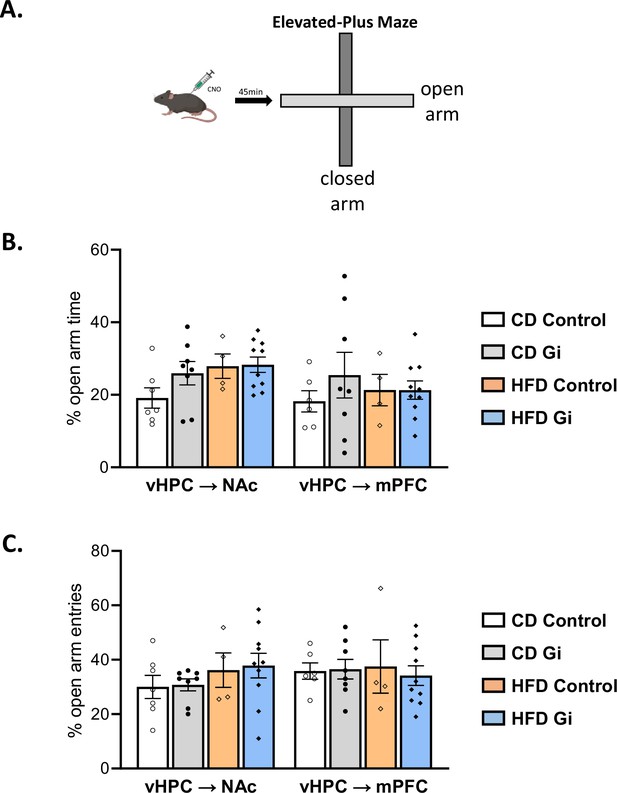
Effect of high-fat diet and chemogenetic manipulation on anxiety-like behaviours.
Schema of elevated plus-maze task (A). Results are presented as percentage of open arm time (calculated as open arm time over total time spent in both open and closed arms × 100; B) and percentage of open arm entries (calculated as open arm entries with respect to the total number of entries in both open and closed arms × 100; C).
-
Figure 3—figure supplement 1—source data 1
Effect of high-fat diet and chemogenetic manipulation on anxiety-like behaviours.
- https://cdn.elifesciences.org/articles/80388/elife-80388-fig3-figsupp1-data1-v1.xlsx
Tables
Body weight and fat mass measurements.
Parameter/group | CD mCherry vHPC–NAc (n = 7) | CD mCherry vHPC–mPFC (n = 7) | CDGi vHPC–NAc(n = 8) | CDGi vHPC– mPFC(n = 9) | HFD mCherry vHPC–NAc(n = 8) | HFD mCherry vHPC–mPFC(n = 7) | HFDGi vHPC–NAc(n = 22) | HFDGi vHPC– mPFC(n = 22) |
|---|---|---|---|---|---|---|---|---|
| Body weight (g) | 29.3 ± 0.6 | 30.1 ± 0.9 | 30.5 ± 0.6 | 30.3 ± 0.7 | 30.4 ± 0.9 | 32.6 ± 0.8 | 31.7 ± 0.6 | 33.3 ± 0.7 |
| % of fat content | 7.7 ± 1.2 | 7.1 ± 1.6 | 9.8 ± 1.5 | 8.7 ± 1.8 | 16.8 ± 3.5 | 20.0 ± 4.9 | 20.6 ± 2.1 | 23.3 ± 2.4 |
-
Table 1—source data 1
Body weight and fat mass measurements.
- https://cdn.elifesciences.org/articles/80388/elife-80388-table1-data1-v1.xlsx
Object training before cellular imaging.
| Parameter/group | CD mCherry vHPC–NAcCNO(n = 6) | CD mCherry vHPC– mPFCCNO(n = 6) | CDGi vHPC–NAcCNO (n = 8) | CDGi vHPC–mPFCCNO (n = 9) | HFDGi vHPC–NAc/mPFCsaline(n = 7) | HFD mCherry vHPC–NAc/mPFCCNO(n = 8) | HFDGi vHPC– NAcCNO (n = 7) | HFDGi vHPC–mPFCCNO (n = 7) |
|---|---|---|---|---|---|---|---|---|
| Time (s) object 1 | 23.5 ± 2.9 | 20.9 ± 1.2 | 18.4 ± 2.0 | 20.2 ± 1.7 | 22.9 ± 2.3 | 23.1 ± 1.2 | 20.8 ± 2.4 | 23.1 ± 2.4 |
| Time (s) object 2 | 22.3 ± 1.8 | 21.9 ± 1.2 | 19.7 ± 2.3 | 22.1 ± 2.1 | 20.5 ± 1.0 | 22.2 ± 1.9 | 20.0 ± 2.3 | 22.1 ± 1.3 |
| Total time (s) 1 + 2 | 45.8 ± 3.0 | 42.8 ± 1.6 | 38.1 ± 2.5 | 42.3 ± 2.6 | 43.4 ± 3.1 | 45.4 ± 3.0 | 40.8 ± 4.1 | 45.3 ± 3.3 |
| mCherry+ cells (total) | 1061 | 817 | 1328 | 1153 | 1222 | 1452 | 1088 | 839 |
| mCherry+ cells/mouse | 177 ± 53 | 136 ± 32 | 166 ± 29 | 128 ± 26 | 153 ± 17 | 182 ± 37 | 155 ± 19 | 120 ± 30 |
-
Table 2—source data 1
Object training before cellular imaging.
- https://cdn.elifesciences.org/articles/80388/elife-80388-table2-data1-v1.xlsx
Object recognition memory task.
| Parameter/group | CD mCherry vHPC–NAc(n = 7) | CD mCherry vHPC–mPFC(n = 7) | CDGi vHPC–NAc (n = 8) | CDGi vHPC–mPFC(n = 8) | HFD mCherry vHPC–NAc(n = 6) | HFD mCherry vHPC–mPFC(n = 6) | HFDGi vHPC–NAc(n = 12) | HFDGi vHPC–mPFC(n = 11) |
|---|---|---|---|---|---|---|---|---|
| ORM % exploration | 51.2 ± 2.9 | 50.9 ± 2.2 | 48.6 ± 4.4 | 50.3 ± 2.1 | 53.9 ± 2.0 | 45.9 ± 4.8 | 49.0 ± 2.7 | 50.9 ± 2.2 |
| Time (s) object to be changed | 10.2 ± 0.6 | 10.2 ± 0.4 | 9.7 ± 0.9 | 10.1 ±0.4 | 10.8 ± 0.4 | 9.2 ± 1.0 | 9.8 ± 0.5 | 10.2 ± 0.5 |
| Time (s) object not to be changed | 9.8 ± 0.6 | 9.8 ± 0.4 | 10.3 ± 0.9 | 9.9 ± 0.4 | 9.2 ± 0.4 | 10.8 ± 1.0 | 10.2 ± 0.5 | 9.8 ± 0.5 |
| Time (s) to reach criterium (training) | 400.1 ± 40.8 | 345.7 ± 42.7 | 285.6 ± 25.5 | 292.6 ± 31.9 | 395.7 ± 65.8 | 362 ± 79.8 | 298.5 ± 32.4 | 301.4 ± 34.7 |
| Time (s) to reach criterium (test) | 401.0 ± 44.6 | 292.0 ± 42.8 | 305.0 ± 26.1 | 296.6 ± 55.6 | 374.7 ± 79.6 | 343.7 ± 62.7 | 229.1 ± 20.6 | 285.7 ± 26.1 |
-
Table 3—source data 1
Object recognition memory task.
- https://cdn.elifesciences.org/articles/80388/elife-80388-table3-data1-v1.xlsx
Object location memory task.
| Parameter/group | CD mCherry vHPC–NAc(n = 7) | CD mCherry vHPC–mPFC(n = 7) | CDGi vHPC–NAc (n = 8) | CDGi vHPC–mPFC (n = 8) | HFD mCherry vHPC–NAc(n = 6) | HFD mCherry vHPC–mPFC(n = 6) | HFDGi vHPC–NAc (n = 12 ) | HFDGi vHPC–mPFC (n = 11) |
|---|---|---|---|---|---|---|---|---|
| OLM % exploration | 45.1 ± 1.1 | 48.1 ± 2.1 | 47.0 ± 1.6 | 50.6 ± 2.6 | 49.1 ± 1.0 | 47.05 ± 2.2 | 50.6 ± 1.5 | 47.7 ± 1.9 |
| Time (s) object to be displaced | 9.0 ± 0.2 | 9.6 ± 0.4 | 9.4 ± 0.3 | 10.1 ± 0.5 | 9.9 ± 0.1 | 9.5 ± 0.5 | 10.1 ± 0.3 | 9.5 ± 0.4 |
| Time (s) object not to be displaced | 11.0 ± 0.2 | 10.4 ± 0.4 | 10.6 ± 0.3 | 9.9 ± 0.5 | 10.1 ± 0.1 | 10.5 ± 0.5 | 9.9 ± 0.3 | 10.5 ± 0.4 |
| Time (s) to reach criterium (training) | 390.6 ± 59.9 | 279.3 ± 43.0 | 349.6 ± 40.1 | 316.0 ± 57.4 | 351.7 ± 28.0 | 327.8 ± 42.6 | 340.8 ± 37.3 | 358.4 ± 24.7 |
| Time (s) to reach criterium (test) | 407.9 ± 55.2 | 327.3 ± 30.6 | 343.8 ± 44.7 | 341.1 ± 45.7 | 292.7 ± 52.3 | 330.0 ± 44.9 | 337.9 ± 27.8 | 339.6 ± 36.9 |
-
Table 4—source data 1
Object location memory task.
- https://cdn.elifesciences.org/articles/80388/elife-80388-table4-data1-v1.xlsx





