A moonlighting function of a chitin polysaccharide monooxygenase, CWR-1, in Neurospora crassa allorecognition
Figures
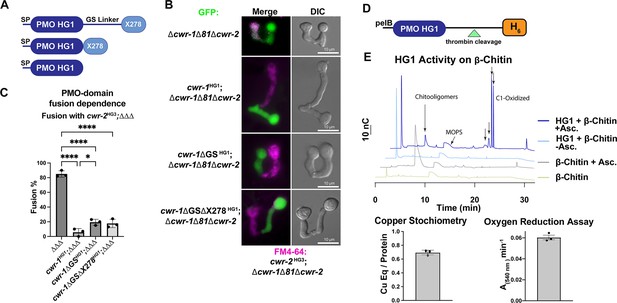
Characterization of the polysaccharide monooxygenase (PMO) domain from CWR-1 from a haplogroup 1 strain (FGSC2489) and functional dissection of CWR-1 domains in vivo.
(A) A schematic diagram depicting the series of truncated cwr-1 constructs studied. SP indicates signal peptide; GS linker indicates the glycine/serine-rich region that connects the PMO catalytic domain to the presumptive chitin-binding module, X278. (B) Cell fusion assays between Δcwr-1Δ81Δcwr-2 germlings alone or bearing either HG1 cwr-1 (FGSC2489) or truncated versions cwr-1ΔGS, cwr-1ΔGSΔX278 (all expressing cytoplasmic GFP) and subsequently paired with FM4-64-stained Δcwr-1Δ81Δcwr-2 germlings expressing an HG3 cwr-2 allele (from JW258). (see Figure 1—figure supplement 3 for fusion rates with all haplogroups [HGs] with HG1 and fusion controls). (C) Quantification of cell fusion frequencies shown in (B) of Δcwr-1Δ81Δcwr-2 (GFP) germlings (ΔΔΔ), or ΔΔΔ germlings bearing HG1 cwr-1HG1 or ΔΔΔ germlings bearing truncated versions cwr-1ΔG/S or cwr-1ΔGSΔX278 and paired with FM4-64-stained ΔΔΔ germlings expressing an HG3 cwr-2 allele. Experiments were performed in biological triplicate, assessing fusion of 100 germling pairs for each replicate. A one-way ANOVA followed by Tukey’s post-hoc test was used for statistical analysis, error bars represent SD, *p<0.05, ****p<0.0001. Individual p-values are reported in Figure 1—source data 4. (D) Schematic depiction of the E. coli expression constructs. pelB indicates the signal peptide, the PMO domain from an HG1 strain (FGSC2489), and a thrombin-cleavable hexahistidine tag showing cleavage site at the indicated triangle (see Figure 1—figure supplement 4 for protein gel and MS of purified protein). (E) Initial characterization of the PMO domain from HG1 strain (FGSC2489). This PMO exhibited C1-oxiding activity on chitin in the presence of ascorbate, reduced oxygen in the absence of chitin, and bound one copper atom per protein, all properties consistent with previously characterized AA11 PMOs. ICP analyses were done in technical triplicate, each datapoint in the oxygen reduction assay represents a biological replicate. All HPAEC-PAD assays were done in at least biological triplicate with a typical trace shown. Asc. means ascorbate. Black arrows denote peaks that elute in the region of the method corresponding to C1-oxidized oligosaccharides. The copper stoichiometry and oxygen reduction assay error bars are SEM where n=3. Source data for this figure can be found in Figure 1—source data 1, Figure 1—source data 2, and Figure 1—source data 3. See Figure 1—figure supplement 5 for oxidized standards and Figure 1—figure supplement 6 for MS/MS spectra on PMO products from α-chitin.
-
Figure 1—source data 1
HPAEC-PAD source data.
Blank subtracted HPAEC-PAD traces. The samples were prepared as described in ‘Materials and methods'. The time and nC response are listed in the table.
- https://cdn.elifesciences.org/articles/80459/elife-80459-fig1-data1-v2.xlsx
-
Figure 1—source data 2
ICP source data.
The amount of bound copper that was calculated for each polysaccharide monooxygenase (PMO) construct. The amount of copper was determined using ICP-MS and then divided by the protein concentration to generate a ratio. The protein concentration was determined using the absorbance at 280 nm with a NanoDrop UV-Vis with extinction coefficients predicted by Benchling (all Cys oxidized). The results from the technical triplicates are listed.
- https://cdn.elifesciences.org/articles/80459/elife-80459-fig1-data2-v2.xlsx
-
Figure 1—source data 3
Horseradish peroxidase (HRP)-oxygen reduction assay source data.
The slopes from monitoring polysaccharide monooxygenase (PMO)-catalyzed oxygen reduction using an HRP-coupled assay. A PMO sample (2 µM) was incubated at room temperature with 100 µM Amplex red, 1.3 µM HRP (Sigma), 2 mM ascorbate in 50 mM MOPS pH 7.0. The change in absorbance at 540 nm vs. time (min) is listed for each biological replicate in this table.
- https://cdn.elifesciences.org/articles/80459/elife-80459-fig1-data3-v2.xlsx
-
Figure 1—source data 4
p-Values.
Fusion test of the truncated versions of CWR-1 paired with cwr-2HG3; ΔΔΔ.
- https://cdn.elifesciences.org/articles/80459/elife-80459-fig1-data4-v2.xlsx
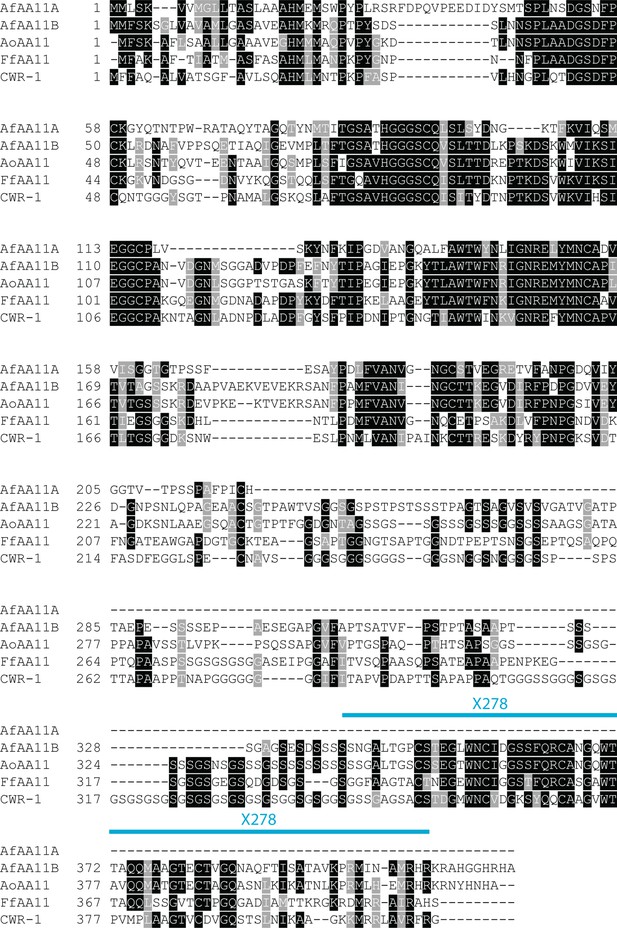
Alignment of characterized AA11s.
All of the AA11s besides AfAA11A have a very similar architecture to CWR-1, including the GS linker and X278 domain.
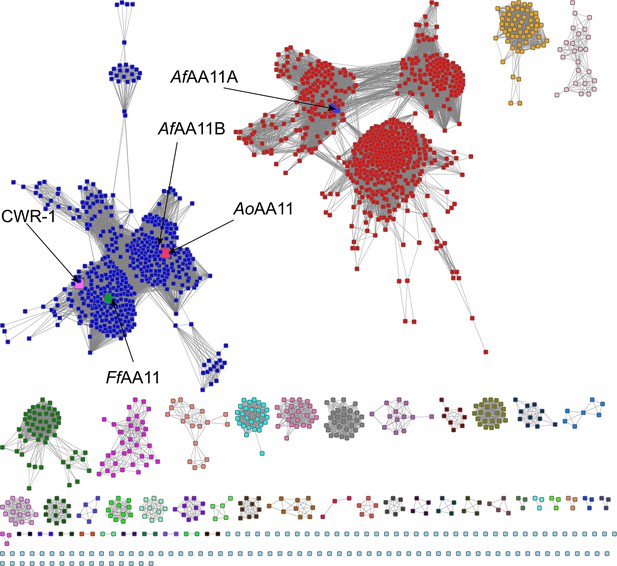
AA11 sequence similarity network (SSN) annotated with characterized proteins.
This SSN was generated using the EFI-EST BLAST feature with the CWR-1HG1 sequence as the search peptide. This search yielded approximately 1700 sequences (Supplementary file 1g), which were visualized using an alignment score of 63 in this network corresponding to ~40% sequence ID. The five labeled nodes are the characterized AA11s. The four found in the blue cluster contain the GS linker and X278 domain, while AfAA11A is a single-domain enzyme consisting of only the AA11 domain. The CWR-1 and FfAA11 polysaccharide monooxygenases (PMOs) cluster away from the others at a higher alignment score and may have a unique role within the blue cluster. The other clusters are proteins with similar sequences and presumably similar functions, but do not contain characterized proteins characterized. Some species contain genes that belong to several clusters, but most often the red and blue clusters. Raw data used for SSN construction are provided in Figure 1—figure supplement 2—source data 1.
-
Figure 1—figure supplement 2—source data 1
Sequence similarity network (SSN) data from the EFI-EST tool as described in ‘Materials and methods’ used to construct the figure.
- https://cdn.elifesciences.org/articles/80459/elife-80459-fig1-figsupp2-data1-v2.zip
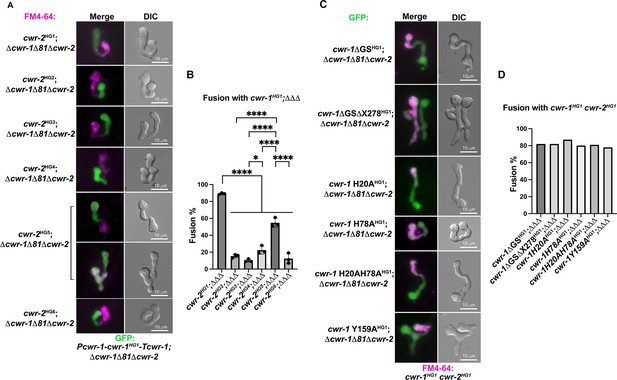
Germling fusion phenotype and percentages among engineered stains.
(A) Pairings between a cwr-1HG1 strain and strains carrying cwr-2 alleles from the six different haplogroups. A strain harboring a cwr-1HG1 allele and expressing cytoplasmic GFP (his-3::Pcwr-1-cwr-1HG1-Tcwr-1; Δcwr-1Δ81Δcwr-2::hph; csr-1-Pccg-1-gfp) was paired with strains carrying cwr-2 alleles from the different haplogroups (HG2-HG6) (his-3:: Ptef-1-cwr-2HG-X-V5-Tccg-1; Δcwr-1Δ81Δcwr-2) stained with FM4-64; fusion percentages were determined microscopically. (B) Quantification of cell fusion percentages depicted in (A). The experiments were performed in biological triplicate, assessing fusion outcome of 100 germling pairs for each replicate. For statistical analysis, a one-way ANOVA followed by Tukey’s post-hoc test was used, error bars represent SD, *p<0.05, ****p<0.0001. Individual p-values are reported in Figure 1—figure supplement 3—source data 1. (C) Strains carrying cwr-1HG1 alleles bearing a deletion of the glycine serine-rich (GS) domain (cwr-1ΔGS) or the GS linker and the X278 domains (cwr-1ΔGSΔX278), the catalytic domain mutants (cwr-1H20A, cwr-1H78A, cwr-1H20A;H78A, and cwr-1Y159A, all in a Δcwr-1Δ81Δcwr-2 background and expressing cytoplasmic GFP) were paired with isogenic cwr-1HG1cwr-2HG1 germlings stained with FM4-64. The genetic background for all the cwr-1 mutants is his-3::Pcwr-1-cwr-1MUTANT-Tcwr-1; Δcwr-1Δ81Δcwr-2::hph; csr-1-Pccg-1-gfp. (D) Quantification of cell fusion percentages between 100 germling pairs depicted in (C).
-
Figure 1—figure supplement 3—source data 1
p-Values.
Comparison of the fusion test of the cwr-1HG1 paired with strains harboring cwr-2 alleles from six different haplogroups.
- https://cdn.elifesciences.org/articles/80459/elife-80459-fig1-figsupp3-data1-v2.xlsx

Purification and size of polysaccharide monooxygenase (PMO) domains of CWR-1 haplogroup (HG) proteins.
(A) An SDS-PAGE gel showing purity of CWR-1 PMO domains encoded by cwr-1 alleles from the six different haplogroups. The haplogroup is denoted above each lane. Original data provided in Figure 1—figure supplement 4—source data 1. (B). Deconvoluted protein mass spectrum of purified wild-type PMOs between 20 kDa and 25 kDa. The predicted mass of each PMO is shown on the right in teal. Each mass observed is 6 or 5 Da lower than predicted, consistent with three pairs of disulfide bonds present in these PMOs. The HGs of each PMO domain and the strain it was derived from are noted on the left. The HG for (A) and (B) correspond to it. Source data for the SDS-PAGE gel are provided in Figure 1—figure supplement 4—source data 1, and the source data for the whole-protein MS is found in Figure 1—figure supplement 4—source data 2.
-
Figure 1—figure supplement 4—source data 1
Purified polysaccharide monooxygenase (PMO) domain from the six haplogroups (HGs).
An SDS-PAGE stain-free gel of the purified CWR-1 PMO domains from each haplogroup. The order of the lanes from left to right is: ladder (Precision Plus Protein Standards unstained Bio-Rad) HG 6, HG4, HG3, HG5, HG2, HG1.
- https://cdn.elifesciences.org/articles/80459/elife-80459-fig1-figsupp4-data1-v2.zip
-
Figure 1—figure supplement 4—source data 2
Whole-protein MS data.
Deconvoluted mass spectra of the purified polysaccharide monooxygenase (PMO) domains from each haplogroup and selected mutants. The whole-protein mass spectrometry is described in ‘Materials and methods'.
- https://cdn.elifesciences.org/articles/80459/elife-80459-fig1-figsupp4-data2-v2.xlsx
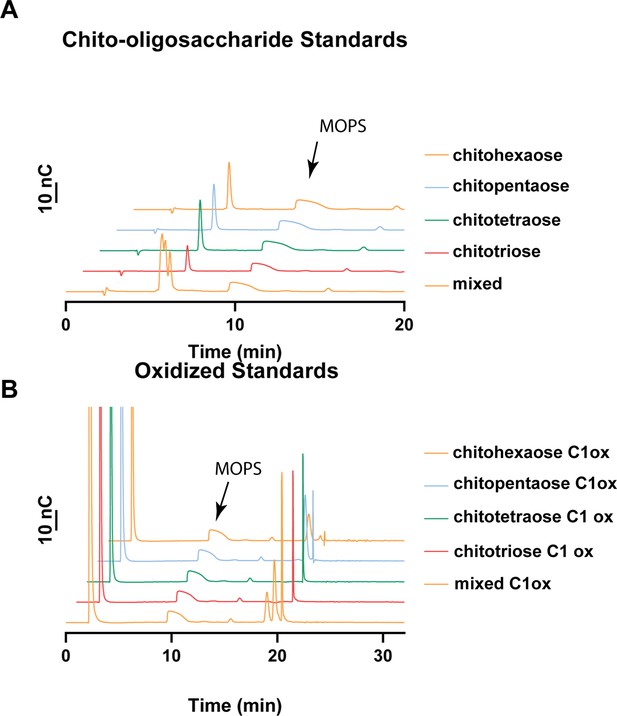
Oligosaccharide standard chromatograms and elution times.
(A) Chitooligosaccharide elution times. Chitooligosaccharides elute early on in the HPAEC-PAD trace. The chitotetraose and chitotriose elute at very similar times. An arrow denotes where the buffer, MOPS, elutes as a tailing peak. (B) C1-oxidized chitooligosaccharides (ChitO-treated chitooligosaccharides) elute ~18–22 min. The chitotriose and chitotetraose elute at very similar retention times. There is degradation in the form of smaller oxidized oligosaccharides present in the chitopentaose and chitohexaose samples. An arrow denotes where the buffer elutes. Source data are provided in Figure 1—figure supplement 5—source data 1 and Figure 1—figure supplement 5—source data 2.
-
Figure 1—figure supplement 5—source data 1
HPAEC-PAD source data.
Blank subtracted HPAEC-PAD traces. The samples were prepared as described in ‘Materials and methods'. The time and nC response are listed in the table.
- https://cdn.elifesciences.org/articles/80459/elife-80459-fig1-figsupp5-data1-v2.xlsx
-
Figure 1—figure supplement 5—source data 2
HPAEC-PAD source data.
Blank subtracted HPAEC-PAD traces. The samples were prepared as described in ‘Materials and methods'. The time and nC response are listed in the table.
- https://cdn.elifesciences.org/articles/80459/elife-80459-fig1-figsupp5-data2-v2.xlsx
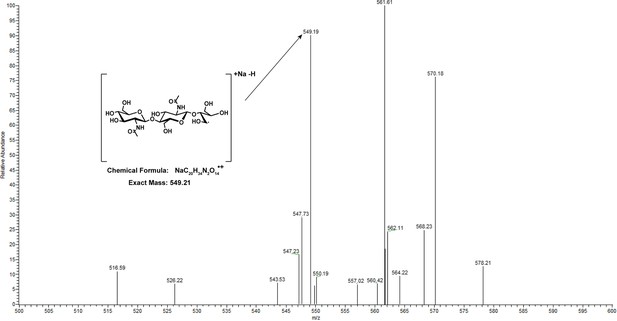
MS/MS fragment is consistent with C1 oxidation.
A MS/MS spectrum from the product of PMOHG1-treated α-chitin. This protein was used as a representative sample for the others as the products do not appear different by HPAEC-PAD between the polysaccharide monooxygenases (PMOs) from the six different haplogroups. The inset shows an expanded region of the fragmented oxidized chitotriose ion with an exact mass of 666.2327. This precursor ion was chosen as it was the most abundant oxidized oligosaccharide ion in the MS run. There is some ambiguity between C1 and C4 fragments; however, one peak can be assigned as a fragment consistent with C1 and not C4 oxidation. Source data for this experiment can be found in Figure 1—figure supplement 6—source data 1.
-
Figure 1—figure supplement 6—source data 1
Tandem MS source data.
Raw MS data from tandem MS experiment as described in ‘Materials and methods'.
- https://cdn.elifesciences.org/articles/80459/elife-80459-fig1-figsupp6-data1-v2.zip
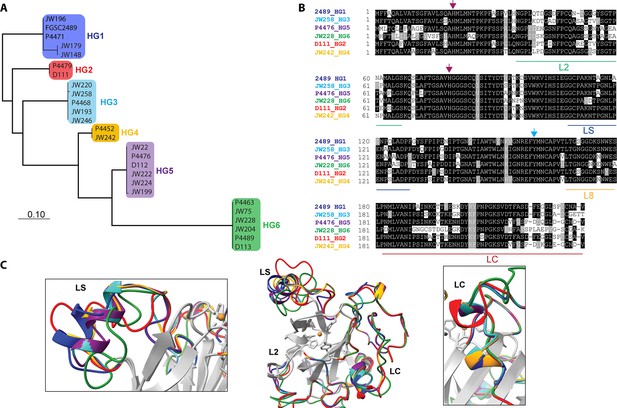
Comparison of CWR-1 polysaccharide monooxygenase (PMO) domains.
In all panels, blue indicates cwr-1 predicted proteins from HG1 isolates, red indicates HG2, light blue indicates HG3, orange indicates HG4, purple indicates HG5, and green indicates HG6 isolates. (A) A phylogenetic tree was constructed using the predicted CWR-1 PMO domain from 26 wild-type isolates. The phylogenetic tree was made using PhyML Phylogenetic Maximum Likelihood and edited in MEGA11. (B). Alignment of the six CWR-1 PMO domain protein sequences from representative isolates from HG1-6. The red arrows show the two histidine residues of the histidine brace that are involved in copper coordination. The blue arrow shows the position of the tyrosine involved in the secondary coordination sphere. L2, LS, and LC correspond to loops that exhibited sequence variation between the CWR-1 PMO domains among the six different haplogroups. (C) A SwissProt homology model of the PMO domain from the six haplogroups. There are apparent differences in the outer loops, whereas the core of the protein is not predicted to have significant differences between haplogroups. The four most affected loops are the loops that correspond to AA9 LS, L2, LC, and L8 loops (middle panel). A portion of the LS loop (left panel) has the most striking differences between haplogroups, and a portion of the LC loop (right panel) contains the second-best region for allelic differences.
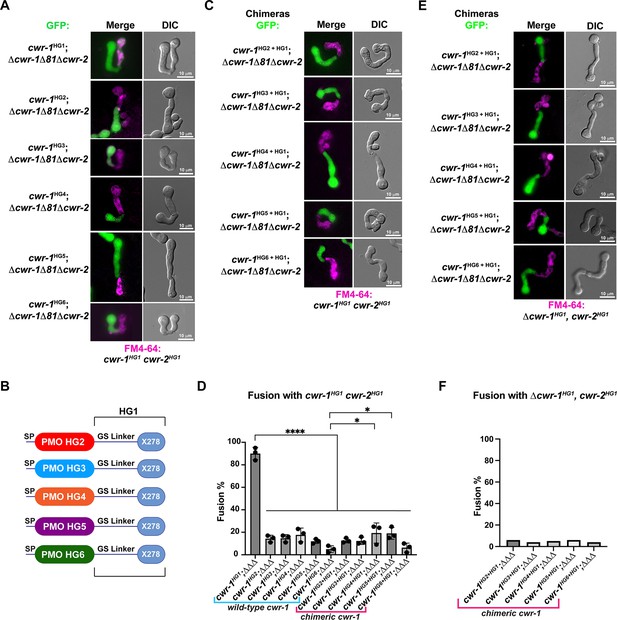
The polysaccharide monooxygenase (PMO) domain in CWR-1 functions to confer the allorecognition fusion checkpoint.
(A) Micrographs of the dominant fusion events between germlings expressing cwr-1 alleles from each haplogroup in a Δcwr-1Δ81Δcwr-2 GFP background when paired with cwr-1HG1 cwr-2HG1 germlings (FGSC2489) stained with FM4-64. (B) CWR-1 chimeras with the PMO domain from the different cwr haplogroups (cwr-1HG2 from D111, cwr-1HG3 from JW258, cwr-1HG4 from JW242, cwr-1HG5 from P4476, and cwr-1HG6 from JW228), with the glycine serine linker and X278 domains from cwr-1HG1 (from FGSC2489) are schematically shown. (C) Micrographs of the dominant fusion events between germlings expressing the chimeric CWR-1 proteins paired with cwr-1HG1 cwr-2HG1 germlings stained with FM4-64. (D) Quantification of cell fusion frequencies depicted in (A) and (C). The experiments were performed in biological triplicate, assessing fusion of 100 germling pairs for each replicate. A one-way ANOVA followed by Tukey’s post-hoc test was used for statistical analysis, error bars represent SD, *p<0.05, ****p<0.0001. Individual p-values are reported in Figure 3—source data 1. (E) Micrographs of the dominant fusion event between GFP germlings expressing the chimeric CWR-1 proteins paired with a Δcwr-1 cwr-2HG1 strain stained with FM4-64. (F) Quantification of cell fusion frequencies between 100 germlings depicted in (E). See Figure 3—figure supplement 1 for control experiments.
-
Figure 3—source data 1
p-Values. Figure 3D.
Fusion test of the different CWR-1 full-length and CWR-1 chimeras paired with cwr-1HG1cwr-2HG1.
- https://cdn.elifesciences.org/articles/80459/elife-80459-fig3-data1-v2.xlsx
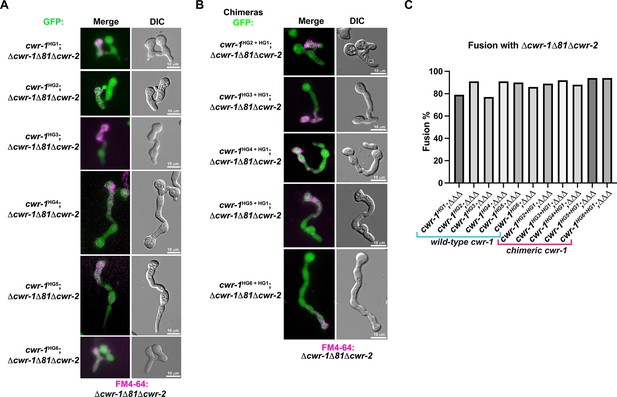
Germling fusion phenotype and fusion percentages in strains bearing cwr-1 from the different haplogroups and cwr-1 chimeras.
(A) Representative micrographs of fusion phenotypes of cells expressing cwr-1 alleles (from HG1-6) and paired the permissive Δcwr-1Δ81Δcwr-2 mutant stained with FM4-64. (B) Representative micrographs of cwr-1 chimeras containing the polysaccharide monooxygenase (PMO) domain from cwr-1 alleles from HG1-6 in Δcwr-1Δ81Δcwr-2 GFP background and paired with a Δcwr-1Δ81Δcwr-2 mutant stained with FM4-64. (C) Quantification of cell fusion percentages between 100 germling pairs depicted in (A) and (B).
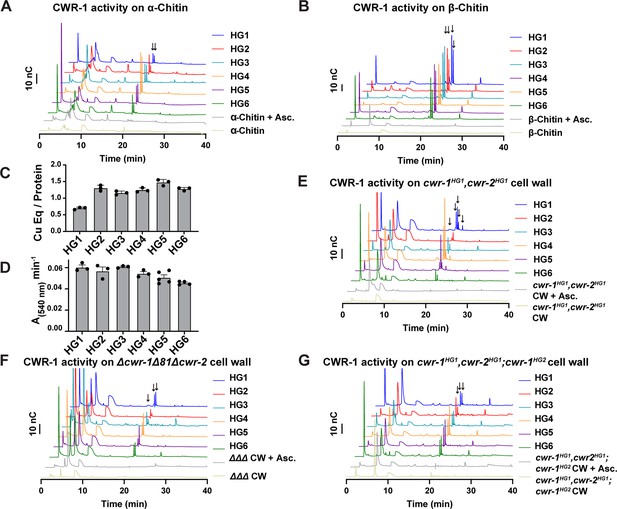
Catalytic activity of CWR-1 haplogroups.
(A, B) A comparison of the reaction products from CWR-1 from each of the six haplogroups on α-chitin and β-chitin. There were minor differences, but each haplogroup generates the same C1-oxidized fragments. (C, D) Comparison of bound copper and oxygen reduction of CWR-1 from the six different haplogroups. All of the CWR-1 proteins bound ~1 copper atom per polysaccharide monooxygenase (PMO) and reduced oxygen at similar rates. ICP experiments were done in technical triplicate, and oxygen reduction assays were done with each point representing a biological replicate. All error bars are SEM. Each experiment was performed with an n=3 except for the HG5 and HG6 oxygen reduction assays that have n=5 and n=4, respectively. (E–G) A comparison of reaction products of CWR-1 from each haplogroup on purified cell wall from the wild-type HG1 strain (FGSC2489), the Δcwr-1Δ81Δcwr-2 mutant strain, and a strain expressing a CWR-1HG2 PMO protein in an HG1 strain (cwr-1HG1 cwr-2HG1; cwr-1HG2) (Gonçalves et al., 2019). There were minor differences between the alleles and substrates, but all contain the same C1-oxidized chitin fragments. Asc. means ascorbate only. Black arrows denote peaks that elute in the region where C1-oxidized products elute. All HPAEC-PAD assays were done in at least biological triplicate. Source data for this figure can be found in Figure 4—source data 1, Figure 4—source data 2, Figure 4—source data 3, Figure 4—source data 4, Figure 4—source data 5, Figure 4—source data 6, and Figure 4—source data 7.
-
Figure 4—source data 1
HPAEC-PAD source data.
Blank subtracted HPAEC-PAD traces. The samples were prepared as described in ‘Materials and methods.’ The time and nC response are listed in the table.
- https://cdn.elifesciences.org/articles/80459/elife-80459-fig4-data1-v2.xlsx
-
Figure 4—source data 2
HPAEC-PAD source data.
Blank subtracted HPAEC-PAD traces. The samples were prepared as described in ‘Materials and methods.’ The time and nC response are listed in the table.
- https://cdn.elifesciences.org/articles/80459/elife-80459-fig4-data2-v2.xlsx
-
Figure 4—source data 3
ICP source data.
The amount of bound copper that was calculated for each construct. The amount of copper was determined using ICP-MS and then divided by the protein concentration to generate a ratio. The protein concentration was determined using the absorbance at 280 nm with a NanoDrop UV-Vis with extinction coefficients predicted by Benchling (all Cys oxidized). The results from the technical triplicates are listed.
- https://cdn.elifesciences.org/articles/80459/elife-80459-fig4-data3-v2.xlsx
-
Figure 4—source data 4
Horseradish peroxidase (HRP)-oxygen reduction assay source data.
The slopes from monitoring polysaccharide monooxygenase (PMO)-catalyzed oxygen reduction using an HRP-coupled assay. A PMO sample (2 µM) was incubated at room temperature with 100 µM Amplex red, 1.3 µM HRP (Sigma), 2 mM ascorbate in 50 mM MOPS pH 7.0. The change in absorbance at 540 nm vs. time (min) is listed for each biological replicate in this table.
- https://cdn.elifesciences.org/articles/80459/elife-80459-fig4-data4-v2.xlsx
-
Figure 4—source data 5
HPAEC-PAD source data.
Blank subtracted HPAEC-PAD traces. The samples were prepared as described in ‘Materials and methods.’ The time and nC response are listed in the table.
- https://cdn.elifesciences.org/articles/80459/elife-80459-fig4-data5-v2.xlsx
-
Figure 4—source data 6
HPAEC-PAD source data.
Blank subtracted HPAEC-PAD traces. The samples were prepared as described in ‘Materials and methods.’ The time and nC response are listed in the table.
- https://cdn.elifesciences.org/articles/80459/elife-80459-fig4-data6-v2.xlsx
-
Figure 4—source data 7
HPAEC-PAD source data.
Blank subtracted HPAEC-PAD traces. The samples were prepared as described in ‘Materials and methods.’ The time and nC response are listed in the table.
- https://cdn.elifesciences.org/articles/80459/elife-80459-fig4-data7-v2.xlsx
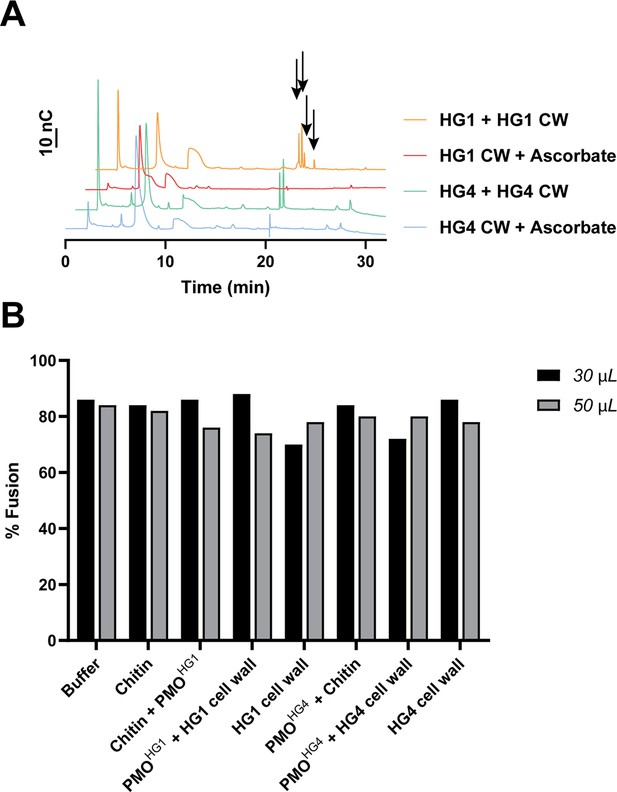
HPAEC-PAD traces of reaction before fusion experiment and cell fusion percentages.
(A) Purified CWR-1 polysaccharide monooxygenase (PMO) (from HG1 or HG4) was mixed with conidial cell wall sample, with or without ascorbate. All solutions used reagents at the same concentration as other activity assays (1 mM ascorbate, 50 mM MOPS pH 7.0, 10 µL PMO, 20 mg/mL substrate) and purified PMOHG1 or PMOHG4. The traces with ascorbate present show that chitin degradation has occurred in both cases with new peaks that appear between 18 and 20 min in the sample with PMO present consistent with PMO degradation of chitin from the cell wall. Black arrows denote peaks that elute in the 18–20 min window where C1-oxidized products elute. (B) Crude reaction mixtures from the enzymatic activity of the HG1 or HG4 purified PMO acting on either chitin or purified conidial cell walls from HG1 or HG4 strains was obtained; the reaction was allowed to proceed for 1 hr, insoluble material was removed through centrifugation at 10,000 × g for 10 min. The resulting supernatant was used in a germling fusion assay between Δcwr-1 (stained with FM4-64) and Δcwr-1 germlings expressing cytoplasmic GFP to determine whether addition of reaction products affected fusion percentages. Quantification of cell fusion frequencies between 50 germling pairs and represented in percentages. 30 and 50 µL denote the amount of completed reaction mixture added. Source data for the HPAEC-PAD traces are provided in Figure 4—figure supplement 1—source data 1.
-
Figure 4—figure supplement 1—source data 1
HPAEC-PAD source data.
Blank subtracted HPAEC-PAD traces. The samples were prepared as described in ‘Materials and methods.’ The time and nC response are listed in the table.
- https://cdn.elifesciences.org/articles/80459/elife-80459-fig4-figsupp1-data1-v2.xlsx
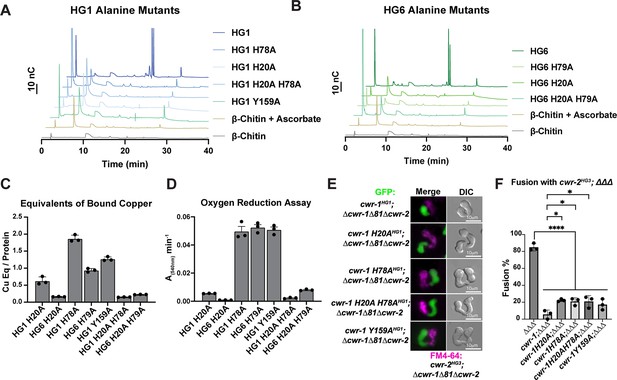
Characterization of CWR-1 polysaccharide monooxygenase (PMO) active-site variants.
(A, B) Comparison of the ability of active-site variants of the PMOHG1 and PMOHG6 domains and their ability to degrade β-chitin. Mutation of one or both of the histidine brace residues abolished chitin-oxidizing activity. All HPAEC-PAD assays were done in at least biological triplicate. (C) Comparison of bound copper of active-site variants. Mutating the second histidine alone did not abolish copper-binding; however, mutation of the first histidine dramatically reduced the amount of copper bound, and mutation of both histidine residues almost eliminated copper-binding entirely. ICP experiments were performed in technical triplicate. Error bars represent SEM. (D) Comparison of oxygen-reduction activity of active site mutants. All constructs that contained the first histidine to alanine variant had significantly reduced oxygen reduction activity. Each data point represents a biological replicate. Error bars represent SEM. (E) Fusion test of the different CWR-1HG1 histidine variants and tyrosine variant paired with a strain expressing cwr-2HG3 in the Δcwr-1Δ81Δcwr-2 mutant background. (F) Quantification of cell fusion percentages depicted in (E). The experiments were performed in biological triplicate, counting 100 germling pairs for each replicate. A one-way ANOVA followed by Tukey’s post-hoc test was used for statistical analysis, error bars represent SD, *p<0.05, ****p<0.0001. Individual p-values are reported in Figure 5—source data 5. The fusion percentages with ΔΔΔ and cwr-1HG1; ΔΔΔ are the same as in Figure 1C. Source data for this figure can be found in Figure 5—source data 1, Figure 5—source data 2, Figure 5—source data 3, and Figure 5—source data 4.
-
Figure 5—source data 1
HPAEC-PAD source data.
Blank subtracted HPAEC-PAD traces. The samples were prepared as described in ‘Materials and methods.’ The time and nC response are listed in the table.
- https://cdn.elifesciences.org/articles/80459/elife-80459-fig5-data1-v2.xlsx
-
Figure 5—source data 2
HPAEC-PAD source data.
Blank subtracted HPAEC-PAD traces. The samples were prepared as described in ‘Materials and methods.’ The time and nC response are listed in the table.
- https://cdn.elifesciences.org/articles/80459/elife-80459-fig5-data2-v2.xlsx
-
Figure 5—source data 3
ICP source data.
The amount of bound copper that was calculated for each construct. The amount of copper was determined using ICP-MS and then divided by the protein concentration to generate a ratio. The protein concentration was determined using the absorbance at 280 nm with a NanoDrop UV-Vis with extinction coefficients predicted by Benchling (all Cys oxidized). The results from the technical triplicates are listed.
- https://cdn.elifesciences.org/articles/80459/elife-80459-fig5-data3-v2.xlsx
-
Figure 5—source data 4
Horseradish peroxidase (HRP)-oxygen reduction assay source data.
The slopes from monitoring polysaccharide monooxygenase (PMO)-catalyzed oxygen reduction using an HRP-coupled assay. A PMO sample (2 µM) was incubated at room temperature with 100 µM Amplex red, 1.3 µM HRP (Sigma), 2 mM ascorbate in 50 mM MOPS pH 7.0. The change in absorbance at 540 nm vs. time (min) is listed for each biological replicate in this table.
- https://cdn.elifesciences.org/articles/80459/elife-80459-fig5-data4-v2.xlsx
-
Figure 5—source data 5
p-Values.
Fusion test of the different cwr-1HG1 histidine variants and tyrosine variant paired with a strain expressing cwr-2HG3; ΔΔΔ.
- https://cdn.elifesciences.org/articles/80459/elife-80459-fig5-data5-v2.xlsx
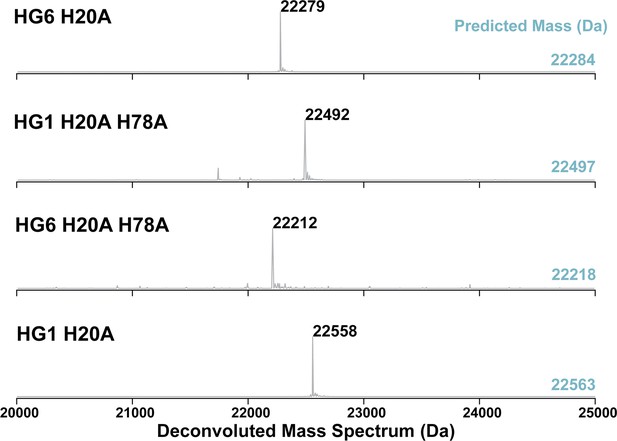
Deconvoluted mass spectrum of polysaccharide monooxygenase (PMO) histidine variants.
These PMOHG1 variants have a histidine to alanine substitution of the first histidine (His20; the first residue after the signal peptide), a second variant containing a His→Ala substitution at H78 and double His→Ala substitutions (H20A; H78A). All proteins were processed correctly in the pelB system. The predicted mass of each PMOHG1 variant is shown on the right in teal. Each mass observed is 6 or 5 Da lower, consistent with three pairs of disulfide bonds present in these PMOs. Source data are provided in Figure 5—figure supplement 1—source data 1.
-
Figure 5—figure supplement 1—source data 1
Whole-protein MS data.
Deconvoluted mass spectra of the purified polysaccharide monooxygenase (PMO) domains from each haplogroup and selected mutants. The whole-protein mass spectrometry is described in ‘Materials and methods’.
- https://cdn.elifesciences.org/articles/80459/elife-80459-fig5-figsupp1-data1-v2.xlsx
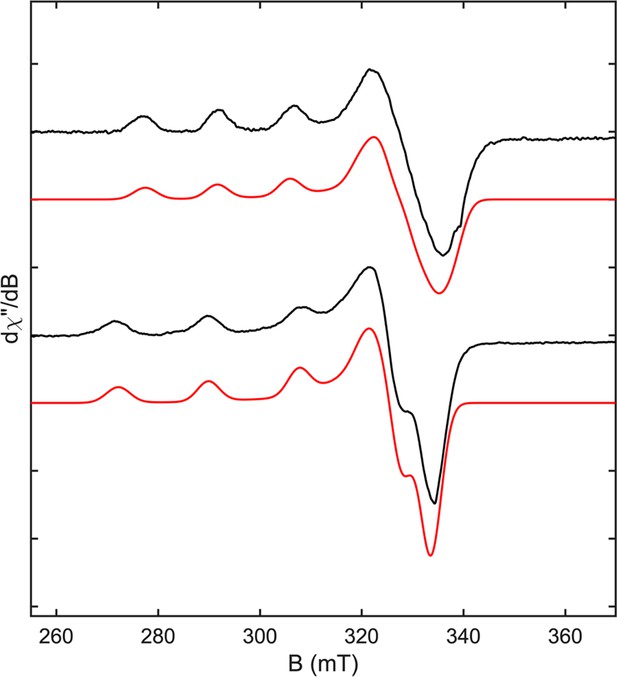
X-band EPR and simulated spectra of PMOHG1 and PMOHG1 H78A variant.
The top black trace is the X-band continuous-wave (CW) spectrum from Cu(II) from PMOHG1. The top red trace is the simulated fit for WT PMOHG1 with the following parameters: g1 = 2.245, g2 = 2.065, g3 = 2.005, A1 = 436 MHz, gStrain1 = 0.03, gStrain1 = 0.03, gStrain1 = 0.03. The bottom black trace is the X-band CW spectrum from the Cu(II) bound to the PMOHG1 H78A variant. The lower red trace was simulated with the following parameters: g1 = 2.245, g2 = 2.08, g3 = 2.05, A1 = 546 MHz, gStrain1 = 0.03, gStrain1 = 0.035, gStrain1 = 0.02. All spectra were acquired at 40 K. Source data for the raw traces are provided in Figure 5—figure supplement 2—source data 1.
-
Figure 5—figure supplement 2—source data 1
EPR source data.
Measured raw data in xy format from the EPR experiment as described in ‘Materials and methods’.
- https://cdn.elifesciences.org/articles/80459/elife-80459-fig5-figsupp2-data1-v2.xlsx
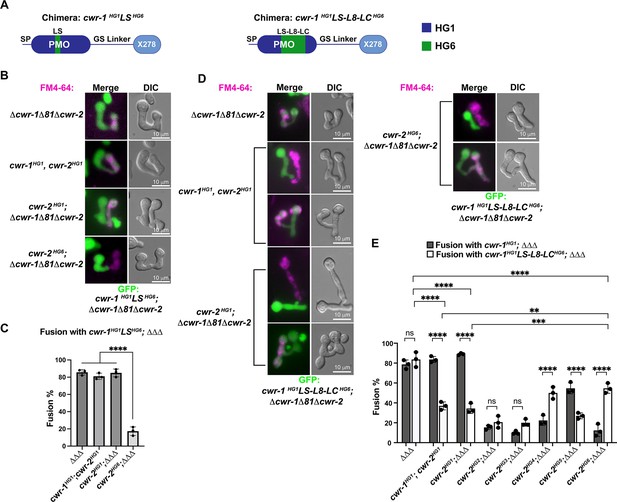
CWR-1 chimeras to define the polysaccharide monooxygenase (PMO) haplogroup specificity region.
(A) A schematic depicting where the loops from PMOHG1 domain that were replaced with loops from PMOHG6 (green). SP means signal peptide; GS linker means the glycine/serine-rich region that connects the catalytic domain to the X278, all of which were derived from CWR-1HG1. Navy is the region of the protein that is from PMOHG1, green is the region of the protein from PMOHG6. (B) Cells expressing the chimera cwr-1HG1LSHG6 (V86-T130) in a Δcwr-1Δ81Δcwr-2 GFP strain were paired with indicated FM4-64 stained germlings. (C) Cell fusion percentages between germlings depicted in (B). The experiments were performed in biological triplicate, counting 100 germling pairs for each replicate. For statistical analysis, a one-way ANOVA followed by Tukey’s post-hoc test was used, error bars represent SD, ****p<0.0001. Individual p-values are reported in Figure 6—source data 1. (D) Cells expressing the chimera cwr-1HG1LS-L8-LCHG6 (V86-D202) in a Δcwr-1Δ81Δcwr-2 GFP strain were paired with indicated FM4-64-stained germlings, with examples of blocked and fusing cells. (E) Cell fusion percentages between the chimeric strain (cwr-1HG1LS-L8-LCHG6) and strains harboring the cwr-2 alleles from all six haplogroups. Cell fusion percentages between cwr-1HG1 cells with strains carrying cwr-2 alleles from different haplogroups were shown in Figure 1—figure supplement 3A and B. The experiments were performed in biological triplicate, counting 100 germling pairs for each replicate. For statistical analysis, a two-way ANOVA followed by Tukey’s post-hoc test was used, error bars represent SD, **p<0.01, ***p<0.001, ****p<0.0001, ns, not significant. Individual p-values are reported in Figure 6—source data 2.
-
Figure 6—source data 1
p-Values.
Fusion test of the cwr-1HG1LSHG6 chimera paired with a strain expressing cwr-1HG1cwr-2HG1, cwr-2HG1; ΔΔΔ or cwr-2HG6; ΔΔΔ.
- https://cdn.elifesciences.org/articles/80459/elife-80459-fig6-data1-v2.xlsx
-
Figure 6—source data 2
p-Values.
Comparison of the fusion test of the cwr-1HG1 or cwr-1HG1LS-L8-LCHG6 chimera paired with strains harboring cwr-2 alleles from six different haplogroups.
- https://cdn.elifesciences.org/articles/80459/elife-80459-fig6-data2-v2.xlsx
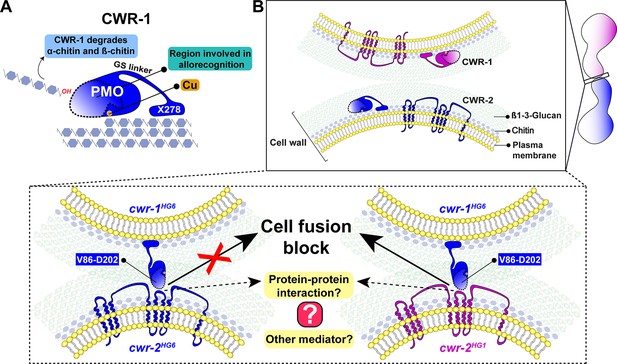
A schematic model that summarizes the role of CWR-1 and CWR-2 in allorecognition at the cell fusion checkpoint.
(A) The polysaccharide monooxygenase (PMO) domain, a glycine and serine-rich region, and a putative chitin-binding module, X278, of CWR-1 are shown. The PMO domain degrades chitin, and the region important for allorecognition is highlighted. (B) The top panel shows a schematic showing the approach of the two tips of germlings before cell–cell contact. Once the germlings approach each other, the bottom two panels show possible outcomes. In the left panel, both germlings express a CWR-1 and CWR-2 protein belonging to the same haplogroup. This pair of germlings will not trigger the cell fusion block signal and will undergo cell fusion. In the right panel, the germlings express incompatible CWR-1 and CWR-2 proteins from different haplogroups, thereby eliciting a cell-fusion-blocking signal. The LC-LS-L8 (V86-D202) region of the protein is represented by the area inside the dotted line. It remains to be answered how CWR-1 and CWR-2 interact either directly through a protein–protein interaction or a mediator and which regions of CWR-2 are involved.
Additional files
-
MDAR checklist
- https://cdn.elifesciences.org/articles/80459/elife-80459-mdarchecklist1-v2.docx
-
Supplementary file 1
Description of plasmids, strains and materials used in this study.
(a) Plasmids to transform N. crassa. (b) Plasmids to transform E. coli. (c) Primers used in this study. (d) N. crassa strains used in this work. (e) Intra-haplogroup sequence ID. (f) Substrates used to test CWR-1 activity. (g) Sequence similarity network (SSN) data set.
- https://cdn.elifesciences.org/articles/80459/elife-80459-supp1-v2.xlsx





