RPG acts as a central determinant for infectosome formation and cellular polarization during intracellular rhizobial infections
Figures
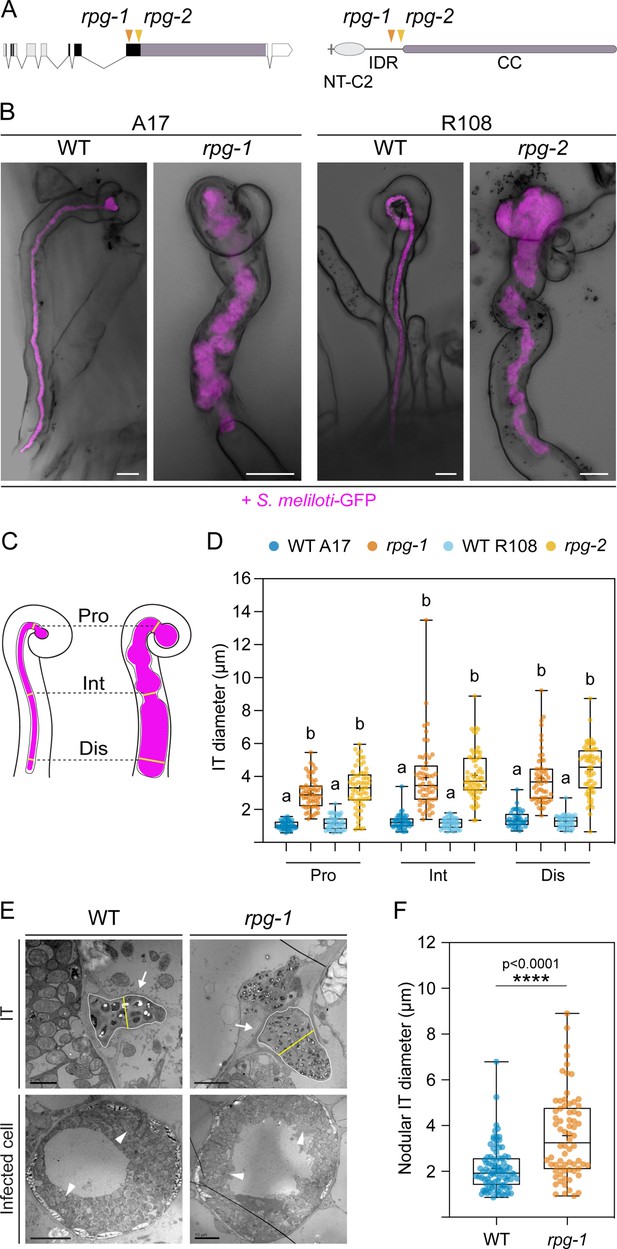
Rhizobium-directed polar growth (RPG) is required for the maintenance of infection thread morphology.
(A) Gene (left) and protein (right) structure of RPG showing the position of the mutations of two different mutant alleles (rpg-1, Arrighi et al., 2008, orange arrowhead; rpg-2, this study, yellow arrowhead). Both mutations map to the third exon of the RPG gene, corresponding to the disordered region of the protein located upstream of the coiled-coil domain. NT-C2=N-terminal C2 domain; IDR = intrinsically disordered region; CC = coiled coil domain. (B) Representative confocal images of aberrant ITs formed 35 dpi within root hairs of rpg-1 and rpg-2 roots compared to thin and elongated infection threads (ITs) of the corresponding wild-types (WT). Images are overlaid with intensity projections of fluorescence and bright field channels. S. meliloti-GFP is shown in magenta. Scale bars = 10 µm. (C) Schematic visualization of the method used to quantify morphological IT defects. The IT diameter was measured on the fluorescent channel at three different points along the IT length: in the proximity of the infection chamber (Pro), in the intermediate part of the IT (Int), and in the distal part of the IT (Dis). (D) IT diameters scored on roots of rpg mutants and corresponding WTs at the three measured points shown in (C). Letters indicate statistically significant differences according to Kruskal-Wallis multiple comparison analysis followed by Dunn’s post-hoc test. Data are from two independent experiments with 10 (WT A17); 11 (rpg-1); 10 (WT R108); 10 (rpg-2) plants analyzed. n=44 (WT A17), 53 (rpg-1), 44 (WT R108), 61 (rpg-2) ITs. (E) Representative transmission electron microscopy (TEM) sections obtained from WT and rpg-1 nodules showing IT structures (arrows) with a thicker appearance in rpg-1 compared to WT. The organization of infected cells and symbiosome (arrowheads) morphology are similar in the two genotypes. The yellow line in the upper panel indicates the IT diameter, positioned at the center of the IT area (white outline), perpendicular to its longest axis. Scale bars = 2 µm (left, upper panel), 5 µm (right, upper panel), 10 µm (lower panels). (F) Nodular IT diameters were measured on TEM sections from nodules formed on roots of rpg-1 and WT. Asterisks indicate statistical significance based on a Mann-Whitney test with p-values <0.05 (*), <0.01 (**), <0.001 (***), and <0.0001 (****). Data are from two independent experiments, with six (WT) and five (rpg-1) nodules analyzed, each harvested from a different plant. n=35 (WT) and 31 (rpg-1) infected cells; n=90 (WT) and 70 (rpg-1) nodular ITs. In each box plot, the top and bottom of each box represent the 75th and 25th percentiles, the middle horizontal bar indicates the median and the whiskers represent the range of minimum and maximum values. Crosses represent sample means.
-
Figure 1—source data 1
IT diameter and statistical analysis.
- https://cdn.elifesciences.org/articles/80741/elife-80741-fig1-data1-v2.zip
-
Figure 1—source data 2
Nodular IT diameter and statistical analysis.
- https://cdn.elifesciences.org/articles/80741/elife-80741-fig1-data2-v2.zip
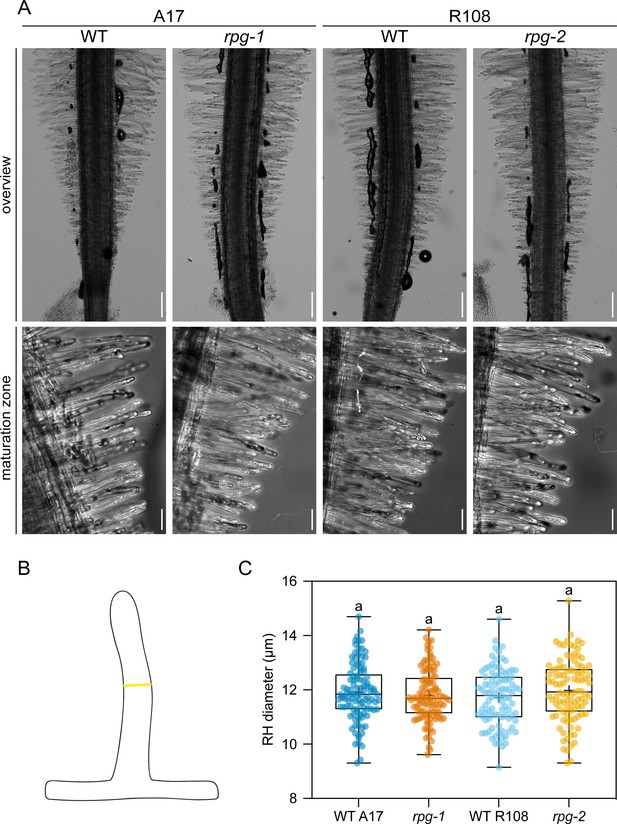
Rhizobium-directed polar growth (RPG) is not required to maintain root hair morphology in un-inoculated conditions.
(A) Representative bright field images of the root tip area (overview, single focal plane) and of the root hair maturation zone (maximum projection) of rpg mutant alleles and their corresponding wild-types (WT). Scale bars = 200 µm (upper panel), 50 µm (lower panel). The root hair diameter was scored on differentiated root hairs from the maturation zone, in the middle region along their length, as shown in the schematic representation in (B). (C) Quantification of root hair diameters on roots of rpg mutant alleles and corresponding wild-types. In the box plot, the top and bottom of each box represent the 75th and 25th percentiles, the middle horizontal bar indicates the median and the whiskers represent the range of minimum and maximum values. Crosses represent sample means. Letters indicate statistically significant differences according to Kruskal-Wallis multiple comparison analysis followed by Dunn’s post-hoc test. Data are from two independent experiments with 21 (WT A17), 21 (rpg-1), 19 (WT R108), 20 (rpg-2) plants analyzed and five root hair cells per plant being examined. n=105 (WT A17), 105 (rpg-1), 95 (WT R108), 100 (rpg-2) root hairs.
-
Figure 1—figure supplement 1—source data 1
RH diameter and statistical analysis.
- https://cdn.elifesciences.org/articles/80741/elife-80741-fig1-figsupp1-data1-v2.zip
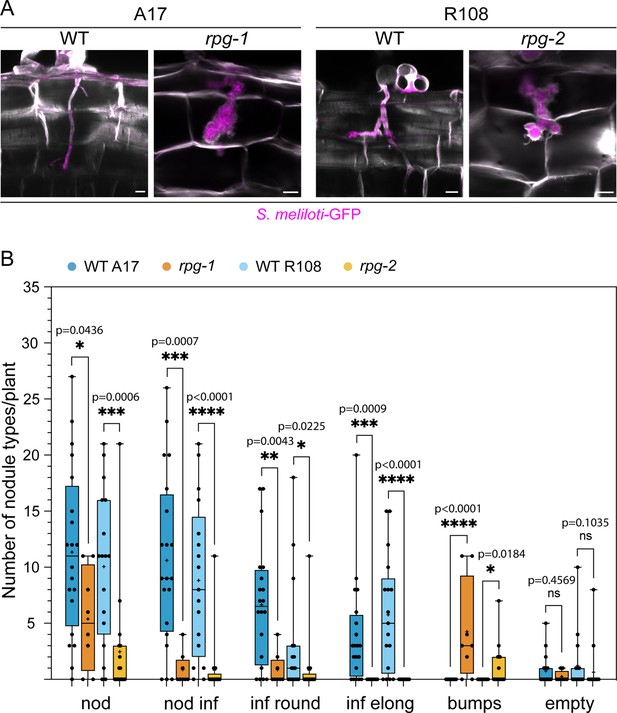
Cortical infection threads and nodule phenotype of rpg mutants.
(A) Confocal images of cortical infection threads (ITs) formed in roots of rpg mutants compared to their corresponding wild-types (WT) at 14 dpi with S. meliloti-GFP (magenta). Cell walls were stained with Calcofluor white (white). Images are merges of maximum intensity projections of the fluorescent channels. Samples are representative of two independent experiments, with three (WT A17, WT R108) and four (rpg-1, rpg-2) plants analysed. n=3 (WT A17), 7 (rpg-1), 6 (WT R108), 6 (rpg-2) cortical ITs. Scale bars = 10 µm. (B) Number and type of nodules formed on rpg mutant alleles compared to their corresponding wild-types at 21 dpi with S. meliloti-LacZ. Data are presented as box plots where the top and bottom of each box represent the 75th and 25th percentiles, the middle horizontal bar indicates the median and the whiskers represent the range of minimum and maximum values. Crosses represent sample means. Asterisks indicate statistical significance based on a Mann-Whitney test with p-values <0.05 (*), <0.01 (**), <0.001 (***), and <0.0001 (****). ns = not significant. The number of plants analyzed per genotype was 20 (WT A17), 8 (rpg-1), 17 (WT R108), 17 (rpg-2). Two independent experiments were performed. Nod = total nodule structures; nod inf = total infected nodules; inf round = infected round nodules; inf elong = infected elongated nodules; bumps = bump like nodules with the arrested infection on the top; empty = empty nodules.
-
Figure 1—figure supplement 2—source data 1
Nodule number and statistical analysis.
- https://cdn.elifesciences.org/articles/80741/elife-80741-fig1-figsupp2-data1-v2.zip
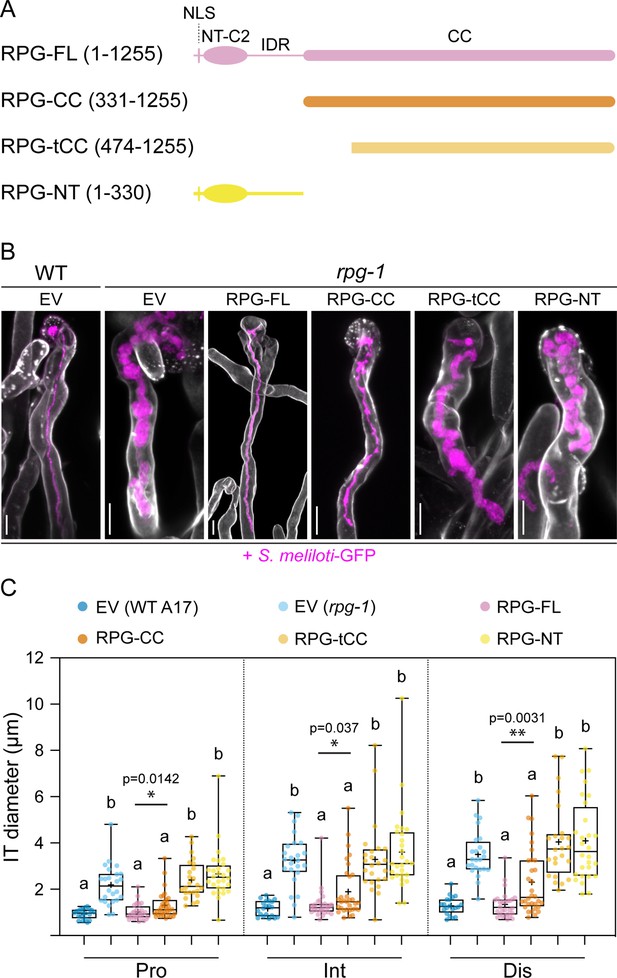
The coiled-coil domain of Rhizobium-directed polar growth (RPG) is necessary to restore WT-like infection thread morphology in rpg-1 root hairs.
(A) Schematic representation of the RPG full-length protein and deletion derivatives used for complementation assays. Numbers indicate amino acids included in each fragment. NLS = nuclear localization signal; NT-C2=N-terminal C2 domain; IDR = intrinsically disordered region; CC = coiled coil domain. (B) Representative confocal images of infection threads (ITs) developed within root hairs of wild-type (WT) and rpg-1 transgenic roots expressing the different constructs 21 dpi with S. meliloti-GFP (magenta). Cell walls were stained with Calcofluor white (white). Images are merges of maximum-intensity projections of fluorescent channels. EV = empty vector. Scale bars = 10 µm. (C) IT diameters scored at the three different points schematically represented in Figure 1C. In the box plot, the top and bottom of each box represents the 75th and 25th percentiles, the middle horizontal bars indicate the median and the whiskers represent the range of minimum and maximum values. Crosses represent sample means. Letters indicate statistically significant differences according to Kruskal-Wallis multiple comparison analysis followed by a Dunn’s post-hoc test. A Mann-Whitney test was performed to compare RPG-FL and RPG-CC with p-values <0.05 (*), <0.01 (**), and <0.001 (***). Data are from one of two independent experiments showing similar tendencies, with five (EV in WT), six (EV in rpg-1), seven (RPG-FL,), eight (RPG-CC), six (RPG-tCC), seven (RPG-NT) composite plants analyzed (one root per plant) and four ITs per roots. n=20 (EV in WT), 24 (EV in rpg-1), 28 (RPG-FL), 32 (RPG-CC), 24 (RPG-tCC), 28 (RPG-NT) ITs.
-
Figure 2—source data 1
IT diameter and statistical analysis.
- https://cdn.elifesciences.org/articles/80741/elife-80741-fig2-data1-v2.zip

Sequence conservation among Rhizobium-directed polar growth (RPG) homologs.
Amino acid conservation between the NT-C2 domains and N-terminal parts of the coiled-coil region of RPG and homologous proteins from rice (Os) and Arabidopsis (At). Sequence alignment was performed using MUSCLE and visualized with Jalview to calculate conservation scores (Procter et al., 2021). Amino acids are colored according to Clustal X Colour Scheme. The sequence of M. truncatula RPG was used as a reference for the numbering of residues. NLS = nuclear localization signal; NT-C2=N-terminal C2 domain; IDR = intrinsically disordered region; CC (partial) = coiled coil region, partial.
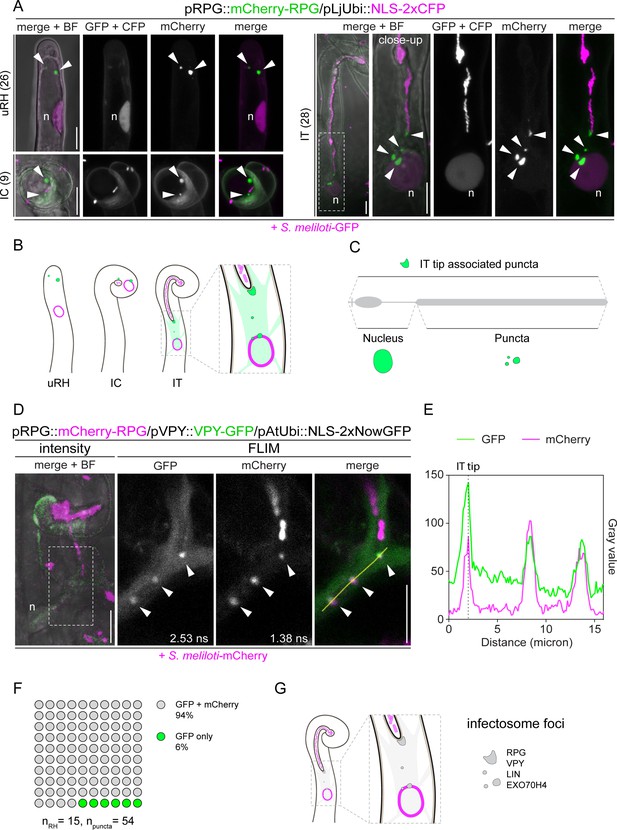
Rhizobium-directed polar growth (RPG) is a key component of the VPY-LIN-EXO70H4 infectosome complex.
(A) Live-cell confocal images showing mCherry-RPG localization in root hairs of WT transgenic roots at 4–7 dpi with S. meliloti-GFP. mCherry-RPG accumulates in cytoplasmic punctate structures (arrowheads) which are found close to the apex of uninfected root hairs (uRH), in the vicinity of rhizobia entrapped within the infection chamber of curled root hairs (IC) and associated to the perinuclear cytoplasm and the tip of infection threads (IT). The dashed white line box indicates the region shown in the close-up, which is a projection of Z-stacks acquired at higher magnification. A nuclear-localized tandem CFP was used as a transformation marker. Images are maximum intensity projections. Individual channels are false colored in gray. When channels are merged, the GFP/CFP channel is shown in magenta and the mCherry channel is shown in green. The bright field (BF) is overlaid with merged fluorescent channels. The number of cells analyzed per each infection stage is reported at the left side of each panel. At least seven composite plants from three independent experiments were analyzed. (B–C) Schematic representations of the subcellular patterning of RPG (green irregular round shapes and diffuse green coloring) in root hairs upon inoculation with rhizobia (light magenta) (B) and contribution of RPG protein (gray) domains to its localization pattern (C). The nucleus is represented as an oval outlined in magenta (B) or colored in green (C). (D) Co-localization of mCherry-RPG with VPY-GFP in cytoplasmic and IT tip-associated puncta (arrowheads) in root hairs of wild-type (WT) transgenic roots imaged at 4–7 dpi with S. meliloti-mCherry. The intensity-based image is a merge of maximum intensity projections of GFP (green) and mCherry (magenta) channels overlaid with the BF channel. The dashed white line box indicates the region shown in Fluorescence Lifetime Imaging Microscopy (FLIM) images. FLIM images are maximum intensity projections of two focal planes (20 repetitions, threshold = 10 photons). Lifetime values (ns) obtained from the exponential reconvolution of decay profiles of GFP and mCherry channels are reported. Individual channels are shown in gray. When components are merged, GFP is shown in green and mCherry in magenta. A nuclear-localized tandem GFP was used as a transformation marker (not visible in these images - see Figure 3—figure supplement 5). (E) Intensity profile of GFP and mCherry signal along the yellow transect is shown in (D). The intensity peaks at the infection thread tip (IT tip) are marked by a dashed black line. (F) Quantification of the co-localization in infected root hairs (n=15) displaying the discrete signal from VPY-GFP and/or mCherry-RPG in punctate structures (n=54). Six composite plants from two independent replicas were analyzed. (G) Schematic representation of the co-localization of RPG with the infectosome components VAPYRIN (VPY) and LUMPY INFECTION (LIN) in distinct subcellular foci (gray irregular round shapes). The nucleus is represented as a magenta-outlined oval. Rhizobia within infection threads are colored in light magenta. n=nucleus. Scale bars = 10 µm.
-
Figure 3—source data 1
FLIM fit values, intensity profile values, and percentage of co-localization.
- https://cdn.elifesciences.org/articles/80741/elife-80741-fig3-data1-v2.zip
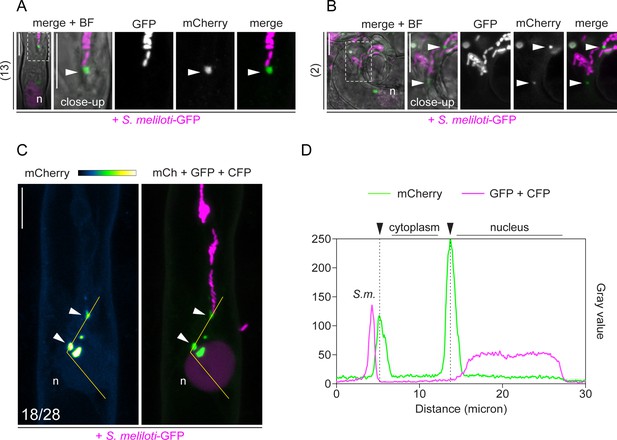
Rhizobium-directed polar growth (RPG) sub-cellular localization pattern in root hairs.
(A–C) Live-cell confocal imaging of infected root hairs of wild-type (WT) transgenic roots co-expressing mCherry-RPG and a nuclear-localized tandem CFP as transformation marker. Images were acquired at 4–7 dpi with S. meliloti-GFP. (A) Patch labeled by mCherry-RPG (arrowhead) adjacent to the infection thread (IT) tip ahead of S. meliloti-GFP colonizing the tube. (B) mCherry-RPG punctate structures localize at the tip of ITs harboring multiple branches. Numbers on the left side of (A) and (B) indicate the amount of cells analyzed, from eight and two composite plants, respectively, in at least two independent replicas. (C) Alongside with a prominent signal located to puncta (arrowheads), a weak signal of mCherry-RPG is observed in the nucleus (n) and cytoplasm of infected root hairs (enlarged panels of Figure 3A -IT close-up). Numbers indicate frequencies of observation. (D) Intensity profile along the yellow transect in (C). Black arrowheads indicate intensity peaks on cytoplasmic puncta. The dashed white line boxes in (A) and (B) indicate the region shown in the respective close-ups. Images are maximum intensity projections. Individual channels are shown in gray; in merged images, the GFP/CFP channel is shown in magenta and the mCherry channel is shown in green. The mCherry channel in (C) is shown in Green Fire Blue when isolated, with yellow indicating the maximum intensity and blue a low level of fluorescence. n=nucleus. S.m.=S. meliloti. Scale bars = 10 µm.
-
Figure 3—figure supplement 1—source data 1
Intensity profile values.
- https://cdn.elifesciences.org/articles/80741/elife-80741-fig3-figsupp1-data1-v2.zip
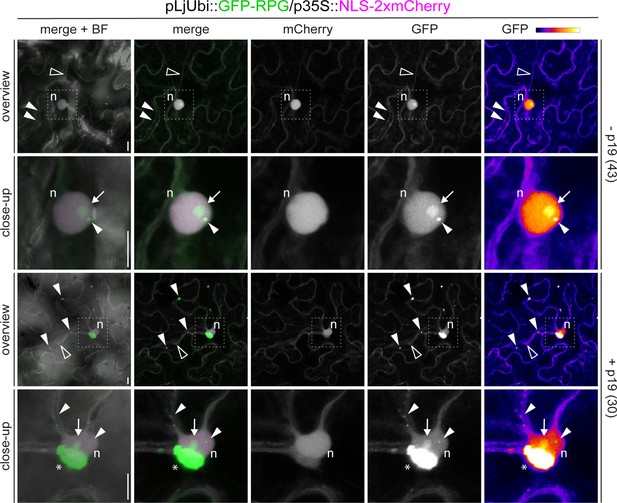
Subcellular localization of Rhizobium-directed polar growth (RPG) in N. benthamiana epidermal cells.
Live-cell confocal images of N. benthamiana epidermal cells ectopically expressing GFP-RPG. In the absence of the silencing suppressor p19 (- p19, upper panels), GFP-RPG signal is detected in the cytoplasm (empty arrowheads), in the nucleus (n), in the nucleolus (arrow) and in discrete puncta (white arrowhead) located in the perinuclear region or along cytoplasmic strands. Co-infiltration with p19 (+p19, lower panels) enhances the accumulation of GFP-RPG in punctate structures and leads to the formation of large perinuclear accumulations (asterisks). The dashed white line boxes in the overview images indicate the region shown in the respective close-ups. A nuclear-localized tandem mCherry was used as a transformation marker. Images are maximum intensity projections. Individual channels are false colored in gray and the GFP channel is additionally shown in Fire, with white showing the maximum intensity and purple with a low level of fluorescence. When channels are merged, the GFP channel is shown in green and the mCherry channel is shown in magenta. The bright field (BF) is overlaid with merged fluorescent channels. Images are representative of three independent agroinfiltrations. The number of cells analyzed per each condition is reported on the right side of each panel. Scale bars = 10 µm.
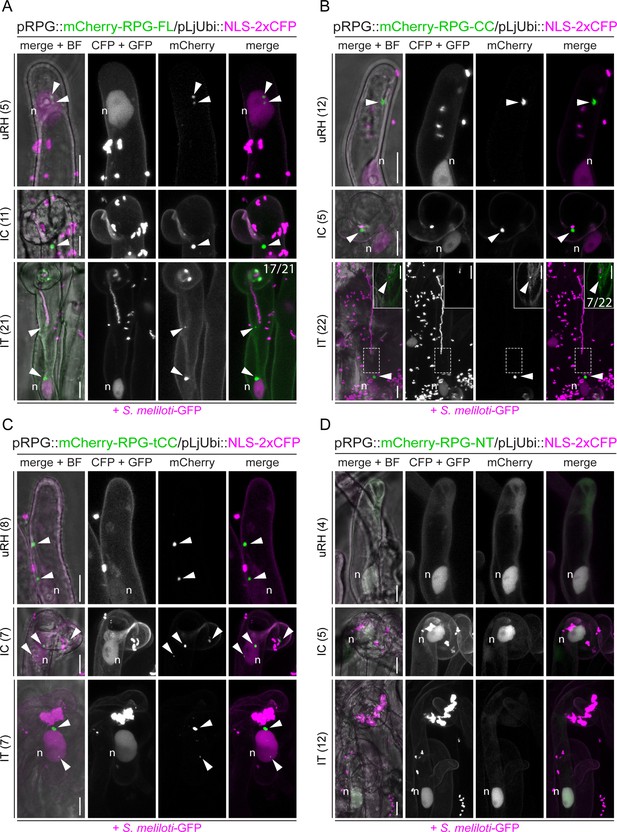
The coiled-coil region is necessary and sufficient to drive Rhizobium-directed polar growth (RPG) localization to puncta.
Live-cell confocal images of root hairs from rpg-1 transgenic roots expressing mCherry-RPG and its deletion derivatives imaged at 4–7 dpi with S. meliloti-GFP. For each construct, uninfected root hairs (uRH), curled root hairs hosting infection chambers (IC), and root hairs harboring infection threads (IT) were imaged. mCherry-RPG-FL (A), mCherry-RPG-CC (B), and mCherry-RPG-tCC (C) preferentially accumulate in bright punctate structures (arrowheads) in root hairs at different stages, while mCherry-RPG-NT (D) appears diffuse in the cytoplasm and accumulates in the nucleus. Images are maximum intensity projections. Individual channels are shown in gray; in merged images, the GFP/CFP channel is shown in magenta and the mCherry channel is shown in green. A nuclear-localized tandem CFP was used as a transformation marker. The dashed white line boxes in (B) indicate the region shown in the corresponding insets. Images in the insets are FLIM-based scans (20 repetitions) of single focal planes (CFP +GFP channel = 2.43 ns; mCherry channel = 1.38 ns; threshold = 10 photons). In (A) and (B) numbers on the rightmost images of the lower panels (IT) indicate frequencies of IT tip labeling. The number of cells analyzed per each construct at each infection stage is reported on the left side of each panel. A minimum of three composite plants from at least two independent experiments were analyzed. n=nucleus. Scale bars = 10 µm.
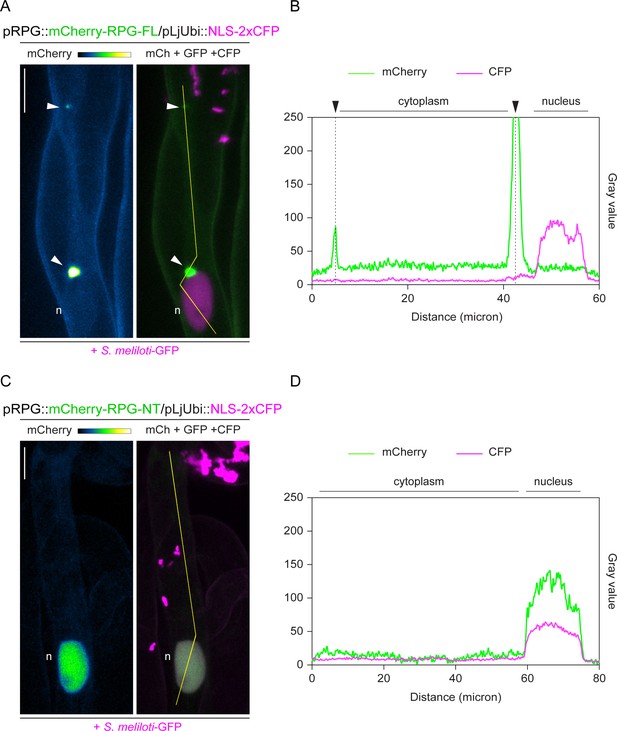
The coiled-coil domain determines the preferential retention of Rhizobium-directed polar growth (RPG) to cytoplasmic puncta.
(A–D) Close-ups of live cell confocal images are presented in Figure 3—figure supplement 3A, D and corresponding intensity profiles along the yellow transects drawn in the respective images. mCherry-RPG-FL preferentially accumulates to bright cytoplasmic puncta (arrowheads) and its weak and diffuse nucleo-cytoplasmic signal markedly differs from the consistent nuclear accumulation exhibited by mCherry-RPG-NT. A nuclear-localized tandem CFP was used as a transformation marker. Images are maximum intensity projections. The mCherry channel is shown in Green Fire Blue when isolated, with yellow indicating the maximum intensity and blue a low level of fluorescence, and in green when merged with the GFP/CFP channel (magenta). n=nucleus. Scale bars = 10 µm.
-
Figure 3—figure supplement 4—source data 1
Intensity profile values.
- https://cdn.elifesciences.org/articles/80741/elife-80741-fig3-figsupp4-data1-v2.zip
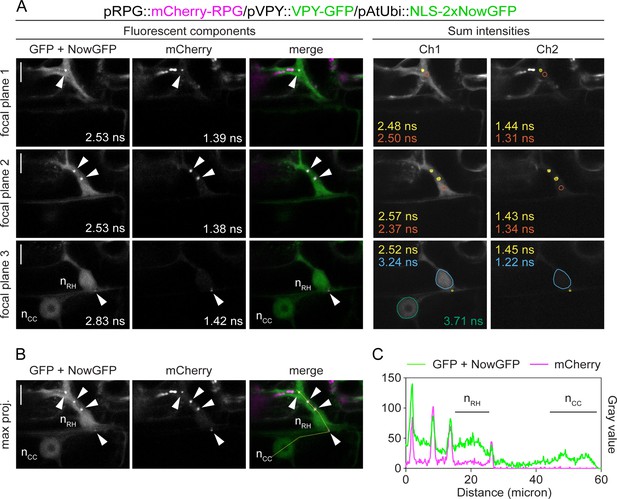
Rhizobium-directed polar growth (RPG) co-localizes with VAPYRIN (VPY) during root hair infection.
(A) Fluorescence Lifetime Imaging Microscopy (FLIM) images of single focal planes used for the close-up projection shown in Figure 3D (focal plane 1 + focal plane 2) and one additional focal plane (3) displaying the nuclei of the infected root hair cell (nRH) and underlying cortical cell (nCC) (20 repetitions, threshold = 10 photons). Lifetime-based separation of channels in their major fluorescent components (left panel) via global fitting of the intensity decay profile maintains the signals of VPY-GFP (~2.5–2.7 ns) and of the nuclear-localized tandem NowGFP (~4.0 ns) in a single component (see focal plane 3). This is most likely due to the high photobleaching rate of the NowGFP compared to the GFP (George Abraham et al., 2015), preventing the collection of enough photons originating from the nuclear marker to enable its discrimination as an isolated component. Determination of the spatial localization of the two different markers is achieved by analyzing the decay profile of the discrete region of interest (colored outlines, right panel) selected on the corresponding fluorescent channel (Ch1). The lifetime value (ns) of the signal from puncta (yellow) and from the cytoplasm (orange) is consistent with the predicted lifetime of the GFP, indicative of VPY accumulation. The signal in the nucleus of the cortical cell (green) exhibits a lifetime consistent with the predicted lifetime of the NowGFP, targeted to the nucleus via fusion with the NLS. Co-localization of VPY-GFP and NLS-2xNowGFP in the nucleus of the infected root hair cell (sky blue) is inferred from the shorter lifetime calculated in this region, resulting from the spatial overlap of the two fluorophores. For each region of interest, the lifetime values calculated in Ch2 are indicated, supporting the presence of mCherry-RPG signal (~1.4–1.5 ns) in cytoplasmic puncta, in the cytoplasm and in the nucleus of the root hair cell. Lifetime values obtained from the exponential reconvolution of decay profiles of GFP/NowGFP and mCherry channels (left panel) are reported. Images in the right panel are FLIM scans where the fluorescence intensities of each channel are summed without any global reconvolution of the decay profiles being applied. (B) Maximum projection of the three focal planes shown in (A). The intensity profile along the yellow transect is shown in (C). Arrowheads indicate cytoplasmic puncta. Scale bars = 10 µm.
-
Figure 3—figure supplement 5—source data 1
FLIM fit values and intensity profile values.
- https://cdn.elifesciences.org/articles/80741/elife-80741-fig3-figsupp5-data1-v2.zip
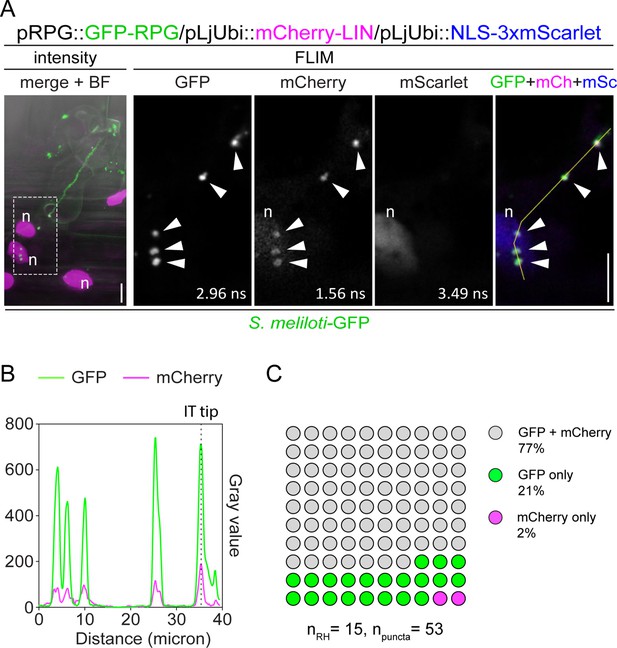
Rhizobium-directed polar growth (RPG) co-localizes with LUMPY INFECTION (LIN) during root hair infection.
(A) Live-cell confocal images of infected root hairs from transgenic wild-type (WT) roots showing co-localization of GFP-RPG with mCherry-LIN in punctate structures (arrowheads) located at the infection thread (IT) tip, in the perinuclear space and in the cytoplasm between these two poles. The intensity-based image is a merge of maximum intensity projections of GFP (green) and mCherry/mScarlet (magenta) channels overlaid with the bright field (BF) channel. The dashed white line box indicates the region shown in Fluorescence Lifetime Imaging Microscopy (FLIM) images. FLIM images are single focal planes (20 repetitions, threshold = 10 photons) with individual channels shown in gray. Lifetime values (ns) obtained from the exponential reconvolution of decay profiles of GFP and mCherry/mScarlet channels are reported. When components are merged, GFP is shown in green, mCherry in magenta, and mScarlet in blue. A nuclear-localized triple mScarlet was used as a transformation marker. n=nucleus. Scale bars = 10 µm. (B) Intensity profile along the yellow transect is shown in (A). The intensity peaks at the infection thread tip (IT tip) are marked by a dashed black line. (C) Quantification of the co-localization in infected root hairs (n=15) displaying the discrete signal from GFP-RPG and/or mCherry-LIN in punctate structures (n=53). Eight composite plants from two independent replicas were analyzed.
-
Figure 3—figure supplement 6—source data 1
FLIM fit values, intensity profile values, and percentage of co-localization.
- https://cdn.elifesciences.org/articles/80741/elife-80741-fig3-figsupp6-data1-v2.zip
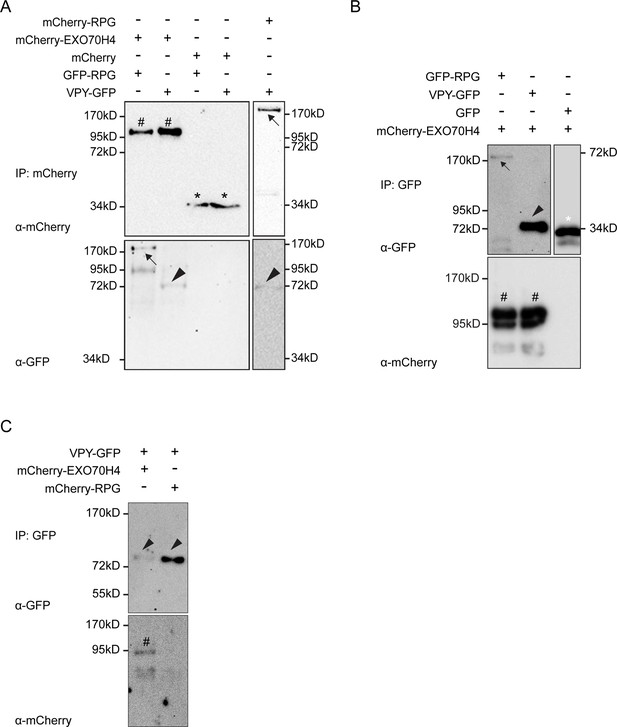
Rhizobium-directed polar growth (RPG) resides in a complex with the infectosome component EXO70H4.
(A–C) Co-immunoprecipitation analysis using anti-RFP (A) or anti-GFP (B and C) nanotraps showing the interaction between GFP-RPG and mCherry-EXO70H4 (A and B). Co-purification of mCherry-RPG and VPY-GFP was obtained when using mCherry-RPG as bait (A), while using VPY-GFP as bait (C) resulted in co-precipitation of EXO70H4 but not of mCherry-RPG (C). Total proteins were extracted from protoplasts obtained from N. benthamiana leaves co-expressing RPG, VPY, and EXO70H4 in different pairwise combinations. Black arrows indicate mCherry-RPG upper right panel in (A) and GFP-RPG (lower left panel in A) and upper left panel in (B), hashtags indicate mCherry-EXO70H4, arrowheads indicate VAPYRIN (VPY), black asterisks indicate mCherry, the white asterisk indicates GFP. Each experiment has been repeated at least two times independently with similar results.
-
Figure 3—figure supplement 7—source data 1
Raw images of Western Blot analysis.
- https://cdn.elifesciences.org/articles/80741/elife-80741-fig3-figsupp7-data1-v2.zip
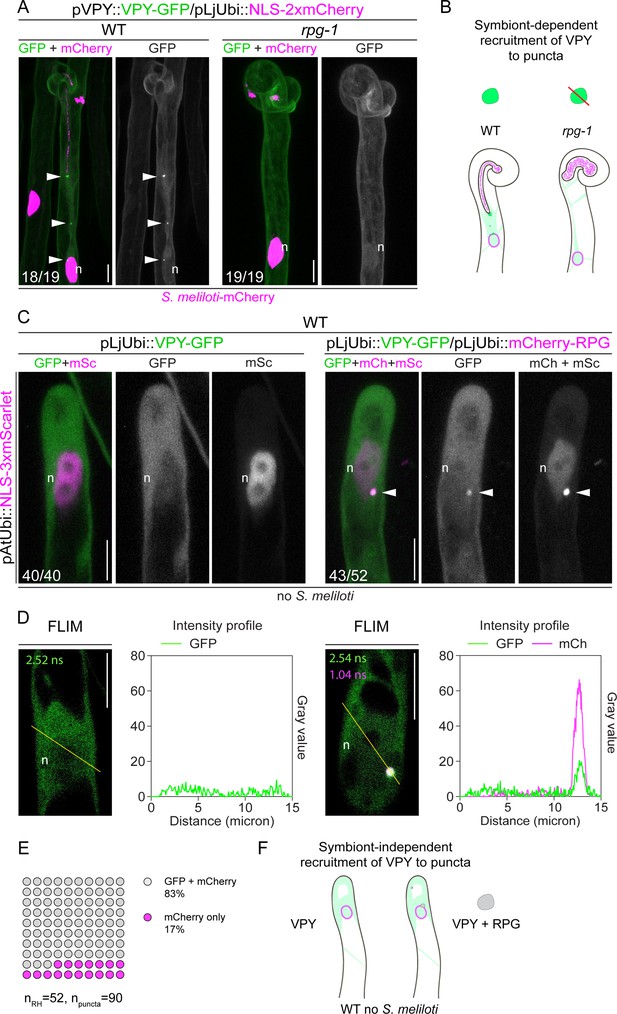
Rhizobium-directed polar growth (RPG) is necessary and sufficient for VAPYRIN (VPY) recruitment to infectosome foci.
(A) Localization pattern of VPY-GFP in root hairs of WT and rpg-1 transgenic roots imaged at 4–7 dpi with S. meliloti-mCherry. VPY-positive puncta (arrowheads) are systematically present in wild-type (WT) but not in rpg-1 infected root hairs. Numbers indicate frequencies of observations, made on a total number of eight (WT) and 10 (rpg-1) composite plants. Two independent replicas were performed. A nuclear-localized tandem mCherry was used as a transformation marker. Images are maximum intensity projections. The GFP channel is shown in gray when isolated, in green when merged with mCherry (magenta). (B) Schematic representation illustrating the dependency of VPY recruitment to infectosome foci by RPG. Upon infection with S. meliloti (light magenta), infectosome foci (green irregular round shape) visible in root hairs of WT transgenic roots are instead undetectable (red diagonal line) in root hairs of rpg-1, where a diffuse signal from VPY (light green coloring) labels the cytoplasm and nucleus (magenta outlined oval). (C) Subcellular patterning of ectopically expressed VPY-GFP either alone (left panel) or in combination with mCherry-RPG (right panel) in root hairs of WT transgenic roots in the absence of S. meliloti. VPY-GFP and mCherry-RPG co-localize in cytoplasmic puncta (arrowheads) which are detectable upon co-expression of the two transgenes, while coalescence of VPY-GFP in such puncta is not observed upon expression of the VPY transgene alone. Images are maximum intensity projections. Individual channels are shown in gray; in merged images, the GFP channel is shown in green and the mCherry/mScarlet channel is shown in magenta. A nuclear-localized triple mScarlet driven by the Arabidopsis Ubiquitin promoter was used as a transformation marker. Images are representative of 40 (VPY-GFP) and 52 (VPY-GFP/mCherry-RPG) root hairs from a minimum of 16 composite plants from two independent replicas. Numbers indicate frequencies of observations. (D) Fluorescence Lifetime Imaging Microscopy (FLIM) images (single focal planes, 20 repetitions, one photon threshold) of root hairs shown in (C) and intensity profiles along the yellow transects drawn on the respective images. Lifetime values (ns) obtained from the exponential reconvolution of decay profiles of each channel are reported. (E) Quantification of the co-localization in root hairs (n=52) displaying discrete signal from VPY-GFP and/or mCherry-RPG in punctate structures (n=90). 18 composite plants from two independent replicas were analyzed. (F) Schematic illustration showing the symbiont-independent recruitment of VPY to cytoplasmic puncta co-labeled by RPG (gray irregular round shape) upon ectopic co-expression of the two transgenes in root hairs of un-inoculated WT roots. The nucleus is represented as a magenta-outlined oval. The diffuse cytoplasmic signal from VPY is illustrated with light green coloring. n=nucleus. Scale bars = 10 µm.
-
Figure 4—source data 1
FLIM fit values, intensity profile values, and percentage of co-localization.
- https://cdn.elifesciences.org/articles/80741/elife-80741-fig4-data1-v2.zip
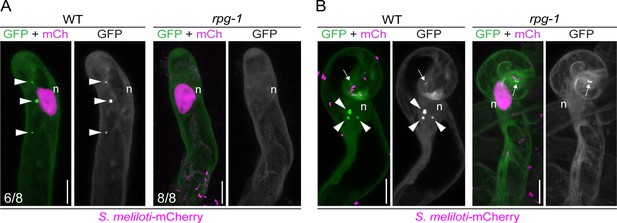
VAPYRIN (VPY) recruitment to puncta is dependent on Rhizobium-directed polar growth (RPG).
Live-cell confocal images of root hairs from transgenic wild-type (WT) and rpg-1 roots expressing VPY-GFP. (A) VPY-positive cytoplasmic puncta (arrowheads) visible in WT uninfected root hairs are not maintained in rpg-1. (B) Accumulation of VPY-GFP adjacent to the site of rhizobia infection (arrows) occurs in a few root hairs of rpg-1 transgenic roots but it is not accompanied by bright punctate structures (arrowheads) associated with the nucleus (n) as seen in root hairs of WT transgenic roots. The GFP channel is shown in gray when isolated, in green when merged with mCherry (magenta). A nuclear-localized tandem mCherry was used as a transformation marker. Numbers indicate frequencies of observations, made on a total number of six (WT) and seven (rpg-1) composite plants. Two independent replicas were performed. n=nucleus. Scale bars = 10 µm.
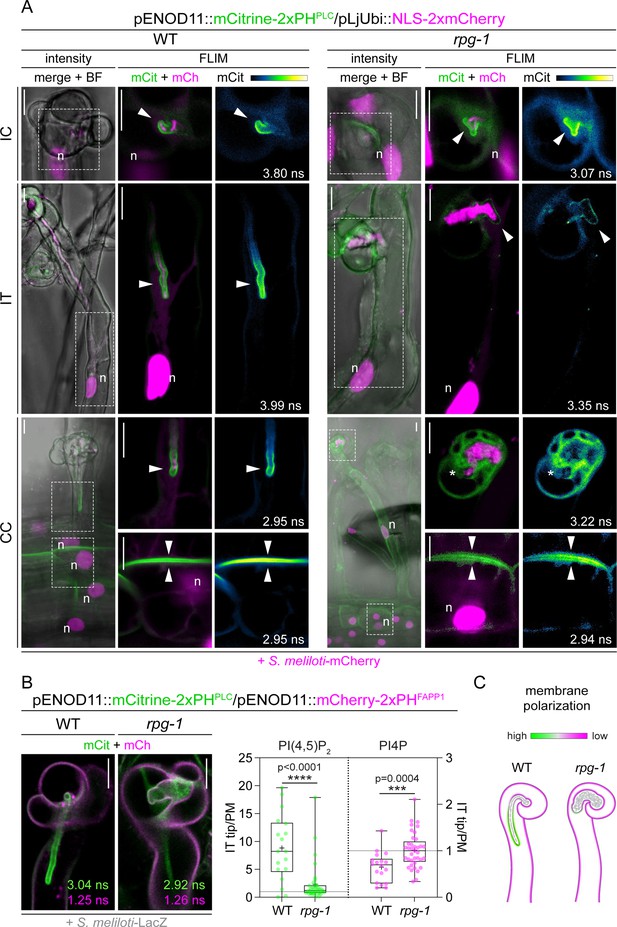
Rhizobium-directed polar growth (RPG) is required to maintain strong membrane polarization at the infection thread (IT) tip.
(A) Live-cell confocal images of root hairs from transgenic WT and rpg-1 roots expressing the PI(4,5)P2 biosensor mCitrine-2xPHPLC at 4–8 dpi with S. meliloti-mCherry. The distribution of mCitrine-2xPHPLC in WT and rpg-1 transgenic roots are compared during the emergence of rhizobia from the infection chamber (IC), in root hairs hosting infection threads (IT) and in cortical cells underlying epidermal infection events (CC). Membrane domains where differential enrichment of PI(4,5)P2 occurred are indicated with arrowheads. Cytoplasmic signal of the biosensor in rpg-1 infected root hairs is indicated with an asterisk. Intensity-based images are merges of maximum intensity projections of mCitrine (green) and mCherry (magenta) channels overlaid with the bright field (BF) channel. The dashed white line box in intensity images indicates the region shown in Fluorescent Lifetime Imaging Microscopy (FLIM) images. FLIM images are single focal planes; the mCitrine component is shown in green when merged with the mCherry component (magenta) and in Green Fire Blue when isolated, with yellow indicating the maximum intensity and blue a low level of fluorescence. Lifetime values (ns) obtained from the exponential reconvolution of decay profiles of the mCitrine channel are reported. A nuclear-localized tandem mCherry was used as a transformation marker. Each infection stage has been monitored on a minimum of three composite plants from at least three independent replicas. n=nucleus. (B) Co-visualization of PI4P and PI(4,5)P2 in root hairs harboring ITs from wild-type (WT) and rpg-1 transgenic plants co-expressing mCitrine-2xPHPLC and mCherry-2xPHFAPP1 at 4–7 dpi with S. meliloti-LacZ. Images are merges of mCitrine (green) and mCherry (magenta) components with indicated lifetime obtained from exponential reconvolution of decay profiles of the respective channels. A nuclear-localized triple mScarlet was used as a transformation marker (not visible in these images). The IT tip versus plasma membrane fluorescence ratio calculated for each biosensor in WT and rpg-1 infected root hairs is reported in the box plot, where the top and bottom of each box represents the 75th and 25th percentiles, the middle horizontal bar indicates the median and whiskers represent the range of minimum and maximum values. Crosses represent sample means. Horizontal gray lines are positioned at y=1. Asterisks indicate statistical significance based on a Mann-Whitney test with p-values <0.05 (*), <0.01 (**), <0.001 (***), and <0.0001 (****). Data are from three independent experiments with 13 (WT) and 14 (rpg-1) composite plants analyzed. n=18 (WT) and 37 (rpg-1) infected root hairs. (C) Schematic representation of the differential enrichment of PI(4,5)P2 (green) and PI4P (magenta) on membranes of WT and rpg-1 infected root hairs, indicative of a high and low membrane polarization characterizing the tip of WT and rpg-1 ITs, respectively. Bacteria within infection threads are colored in gray. Scale bars in (A) and (B) = 10 µm.
-
Figure 5—source data 1
FLIM fit values, IT tip/PM values, and statistical analysis.
- https://cdn.elifesciences.org/articles/80741/elife-80741-fig5-data1-v2.zip
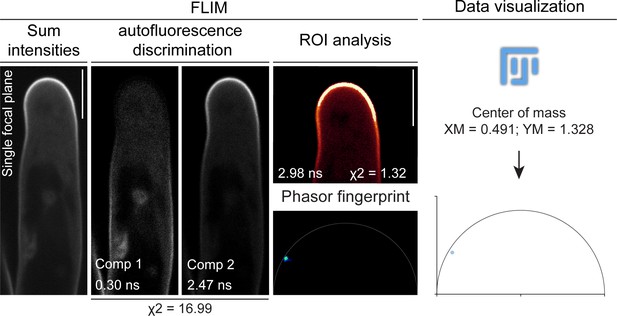
A Fluorescent Lifetime Imaging Microscopy (FLIM)-based systematic approach was adopted to visualize PI(4,5)P2 membrane enrichment.
The sequential steps used for visualization and analysis of PI(4,5)P2 distribution patterns are shown in a confocal image of a root hair from wild-type (WT) transgenic roots expressing mCitrine-PHPLC as an example. FLIM-based confocal imaging initially yields an image (sum intensities) where the intensities of the different fluorescent components present in the scanned area are summed up. By applying n-exponential reconvolution of the fluorescence intensity decay profile of the whole image, the major fluorescent components are separated and a lifetime value is assigned to each of them. The number of components is determined according to a reduced chi-squared (χ2) criterion (Lakowicz, 2006). The components exhibiting lifetime values ≤1.1 ns are considered as autofluorescence and are subtracted from the image. To determine with higher accuracy the enrichment of mCitrine-PHPLC at the apical membrane domain, the decay profile at a such region of interest (ROI) is analyzed. The lifetime is calculated via mathematical fitting, yielding a more precise estimation, as shown by the chi-squared (χ2) reaching values closer to one. A lifetime value of ~3 ns for mCitrine is consistent with previous reports (Söhnel et al., 2016). In addition, Phasor fingerprints of fluorescence emissions at the ROIs are generated using the fit-free Phasor approach, which assigns a position to each pixel in the ROI according to its composition in terms of fluorescent species (Malacrida et al., 2021). Enrichment of the mCitrine-PHPLC biosensor at the apical membrane domain results in pixels forming a cloud positioned on or close to the semicircle in the left part of the plot, where long mono-exponential decays are located. The Phasor fingerprint is further converted to a point in an XY plot by measuring the center of mass of the pixel population in the Phasor image using ImageJ/(Fiji) (Schindelin et al., 2012). In this way, data can be visualized on a graph. Scale bars = 10 µm.
-
Figure 5—figure supplement 1—source data 1
FLIM fit values.
- https://cdn.elifesciences.org/articles/80741/elife-80741-fig5-figsupp1-data1-v2.zip
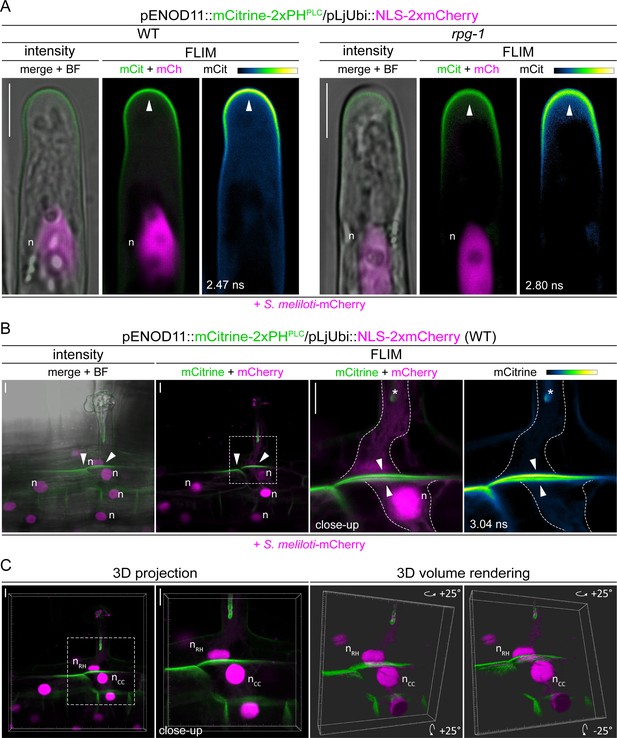
PI(4,5)P2 patterning in transgenic roots of wild-type (WT) and rpg-1.
Live-cell confocal images of WT and rpg-1 transgenic roots expressing the PI(4,5)P2 biosensor mCitrine-2xPHPLC at 4–8 dpi with S. meliloti-mCherry. (A) Accumulation of PI(4,5)P2 (arrowhead) at the apical membrane of growing root hairs occurring in both WT and rpg-1 transgenic roots. (B–C) PI(4,5)P2 enrichment (arrowheads) at the basal domain of infected root hairs and at the apical membrane of underlying cortical cells visualized in WT transgenic roots (enlarged panels of Figure 4A -CC). mCherry-derived cytoplasmic signals occurring due to leakage of the nuclear-localized transformation marker highlight the presence of a cytoplasmic column (dashed freehand line) connecting the tip of the IT (asterisk) to the base of the root hair and surrounding the nucleus (n) in the underlying cortical cells. A 3D projection of the image in (B) (left panel) is shown in (C), together with a 3D volume rendering of its close-up, obtained by rotating to 25 degrees around the X and Y tilting axis using the Imaris software (Bitplane AG, Switzerland). The dashed white line box indicates the region shown in close-ups. Intensity-based images are merges of maximum intensity projections of mCitrine (green) and mCherry (magenta) channels overlaid with the bright field (BF) channel. Fluorescent Lifetime Imaging Microscopy (FLIM) images are single focal planes; the mCitrine component is shown in green when merged with the mCherry component (magenta) and in Green Fire Blue when isolated, with yellow indicating the maximum intensity and blue a low level of fluorescence. Lifetime values (ns) obtained from the exponential reconvolution of decay profiles of the mCitrine channel are reported. n=nucleus. Scale bars = 10µm.
-
Figure 5—figure supplement 2—source data 1
FLIM fit values.
- https://cdn.elifesciences.org/articles/80741/elife-80741-fig5-figsupp2-data1-v2.zip
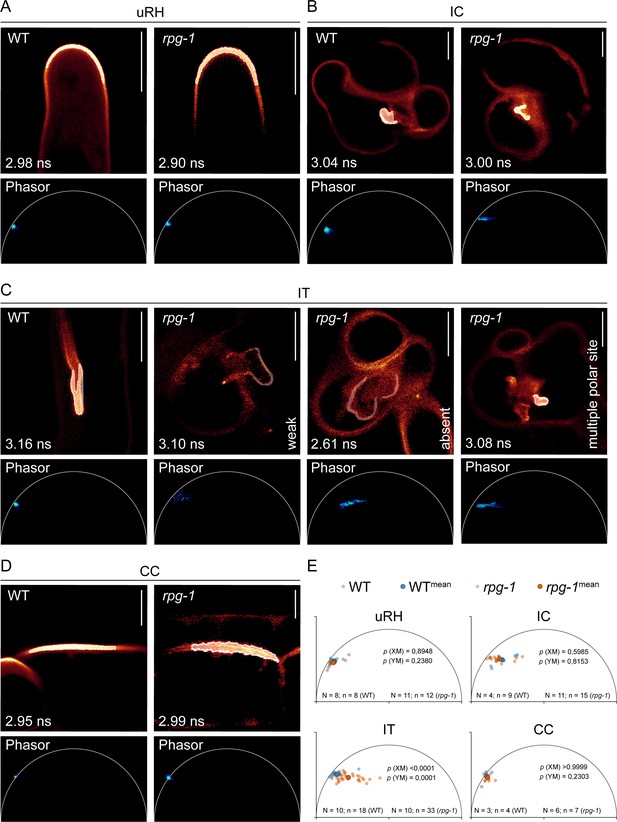
Phasor analysis statistically supports loss of infection thread (IT) membrane polarization occurring in rpg-1.
Estimation of PI(4,5)P2 membrane enrichment in wild-type (WT) and rpg-1 transgenic roots expressing the PI(4,5)P2 biosensor mCitrine-PHPLC via analysis of fluorescence emission at regions of interest (ROI) corresponding to different membrane domains. (A–D) Representative Fluorescent Lifetime Imaging Microscopy (FLIM) images of analyzed membrane domains showing the Phasor fingerprint (Phasor) associated with the ROI selected on the membrane (white outlines) and the lifetime value (ns) calculated via fitting of the decay profile of the ROI. Phasor fingerprints of ROIs selected at the apex of uninfected root hairs (uRH) (A), at the site of symbiont emergence from the infection chamber (IC) (B), and on the membrane domain on the trajectory of IT propagation in cortical cells (CC) (D) in cells of WT and rpg-1 transgenic root show similar positioning on the left half of the Phasor plot close to the semicircle, indicative of similar PI(4,5)P2 enrichment. (C) The Phasor signature associated with the membrane of ITs in rpg-1 markedly differs from the signature of WT ITs: a pixel cloud with a similar Phasor position but composed of few scattered pixels is indicative of weak signal of the mCitrine-PHPLC biosensor, while a consistent shift of the pixel cloud away from the semicircle towards the right part of the plot where short lifetimes are located is indicative of the dominance of autofluorescent components over mCitrine-PHPLC, whose signal appears mostly absent. The Phasor fingerprint associated to the membrane surrounding the multiple polar sites emerging from the enlarged infection threads in rpg-1 is similar to the one recorded at the IC of rpg-1 in (B). (E) XY plots representing the center of mass calculated from Phasor fingerprints of different membrane domains analyzed in multiple cells of different plants for each genotype. A Mann-Whitney test was applied to detect statistical differences between XM and YM values. Calculated p values are shown. N=number of plants; n=number of cells. Five independent replicas were conducted. Scale bars = 10 µm.
-
Figure 5—figure supplement 3—source data 1
FLIM fit values and Phasor analysis values.
- https://cdn.elifesciences.org/articles/80741/elife-80741-fig5-figsupp3-data1-v2.zip
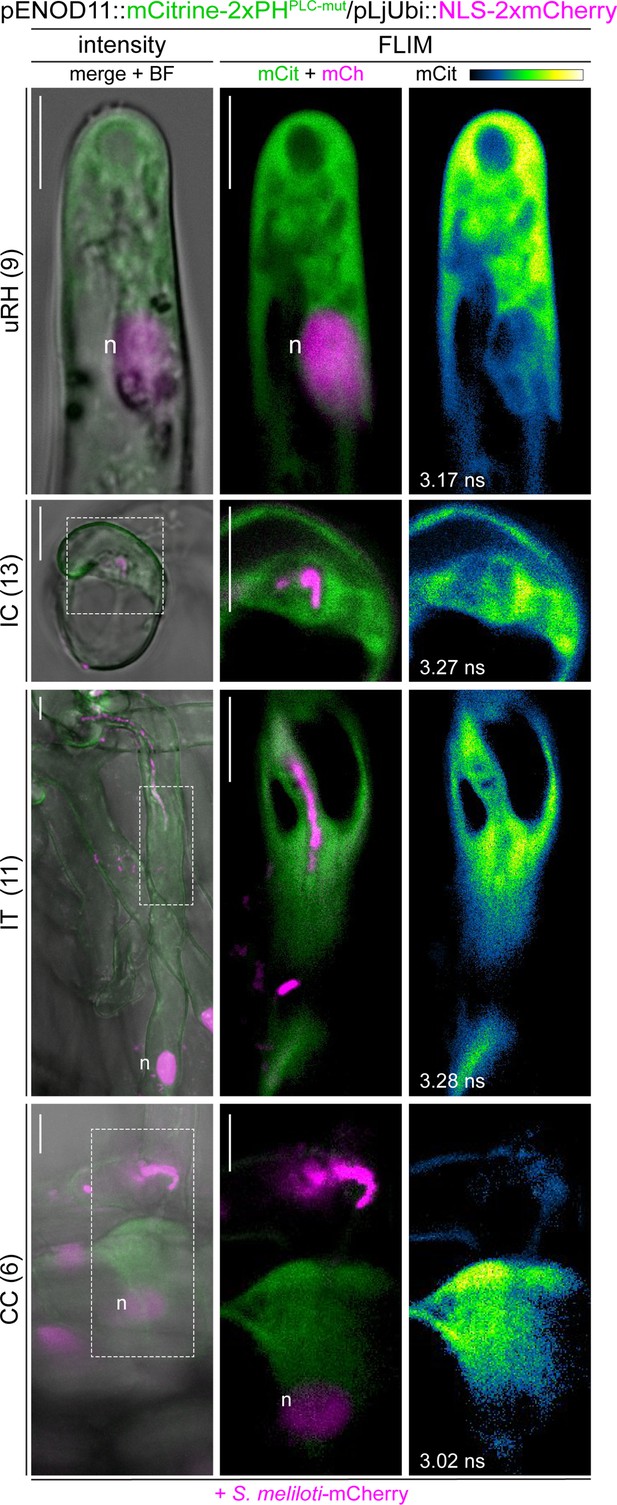
A mutant PI(4,5)P2 biosensor does not accumulate at membrane domains during rhizobial infection.
Live-cell confocal images of wild-type (WT) transgenic roots expressing the mCitrine-2xPHPLC-mut biosensor at 4–7 dpi with S. meliloti-mCherry. The signal from the mutated version of the biosensor appeared diffused in the cytoplasm of uninfected (uRH) and infected (IC-IT) root hairs, and in cortical cells (CC) on the infection thread trajectory. A nuclear-localized tandem mCherry was used as a transformation marker. Intensity-based images are merges of mCitrine (green) and mCherry (magenta) channels overlaid with the bright field (BF) channel. FLIM images are single focal planes; the mCitrine component is shown in green when merged with the mCherry component (magenta) and in Green Fire Blue when isolated, with yellow indicating the maximum intensity and blue a low level of fluorescence. Lifetime values (ns) obtained from the exponential reconvolution of decay profiles of the mCitrine channel are reported. The number of cells analyzed at each infection stage is reported at the left side of each panel. A minimum of five composite plants from two independent replicas were analyzed. n=nucleus. Scale bars = 10 µm.
-
Figure 5—figure supplement 4—source data 1
FLIM fit values.
- https://cdn.elifesciences.org/articles/80741/elife-80741-fig5-figsupp4-data1-v2.zip
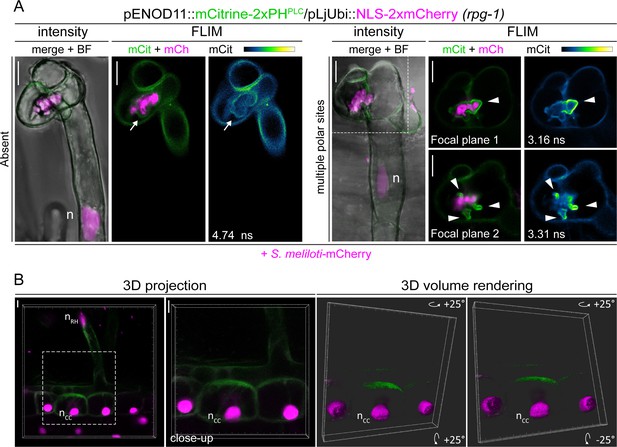
PI(4,5)P2 patterning in transgenic roots of rpg-1.
(A) Representative images of infected root hairs on rpg-1 transgenic roots showing an almost absent signal of the PI(4,5)P2 biosensor on the membrane (arrow, left panel) or its presence at multiple polar sites (arrowheads, right panel) on enlarged infection structures. The dashed white line indicates the region shown in focal planes 1 and 2. Intensity-based images are merges of maximum intensity projections of mCitrine (green) and mCherry (magenta) channels overlaid with the bright field (BF) channel. Fluorescent Lifetime Imaging Microscopy (FLIM) images are single focal planes; the mCitrine component is shown in green when merged with the mCherry component (magenta) and in Green Fire Blue when isolated, with yellow indicating the maximum intensity and blue a low level of fluorescence. Lifetime values (ns) obtained from the exponential reconvolution of decay profiles of the mCitrine channel are reported. (B) 3D projection of the same sample shown in Figure 5A -CC (different z-stack) and a 3D volume rendering (right panel) of its close-up, obtained by rotating to 25 degrees around the X and Y tilting axis using the Imaris software (Bitplane AG, Switzerland). The dashed white line box indicates the region shown in the close-up. n=nucleus. Scale bars = 10 µm.
-
Figure 5—figure supplement 5—source data 1
FLIM fit values.
- https://cdn.elifesciences.org/articles/80741/elife-80741-fig5-figsupp5-data1-v2.zip
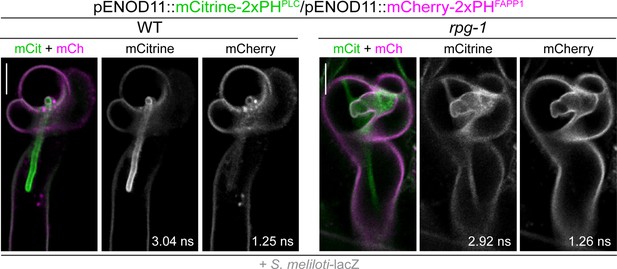
Co-visualization of PI4P and PI(4,5)P2 in wild-type (WT) and rpg-1 infected root hairs.
Fluorescent Lifetime Imaging Microscopy (FLIM) based confocal images of infected root hairs shown in Figure 4B from WT and rpg-1 transgenic plants co-expressing mCitrine-2xPHPLC and mCherry-2xPHFAPP1 at 4–7 dpi with S. meliloti-LacZ. Images are single focal planes with individual channels shown in gray. Lifetime values (ns) obtained from the exponential reconvolution of decay profiles of mCitrine and mCherry channels are reported. When components are merged, mCitrine is shown in green and mCherry in magenta. Scale bars = 10 µm.
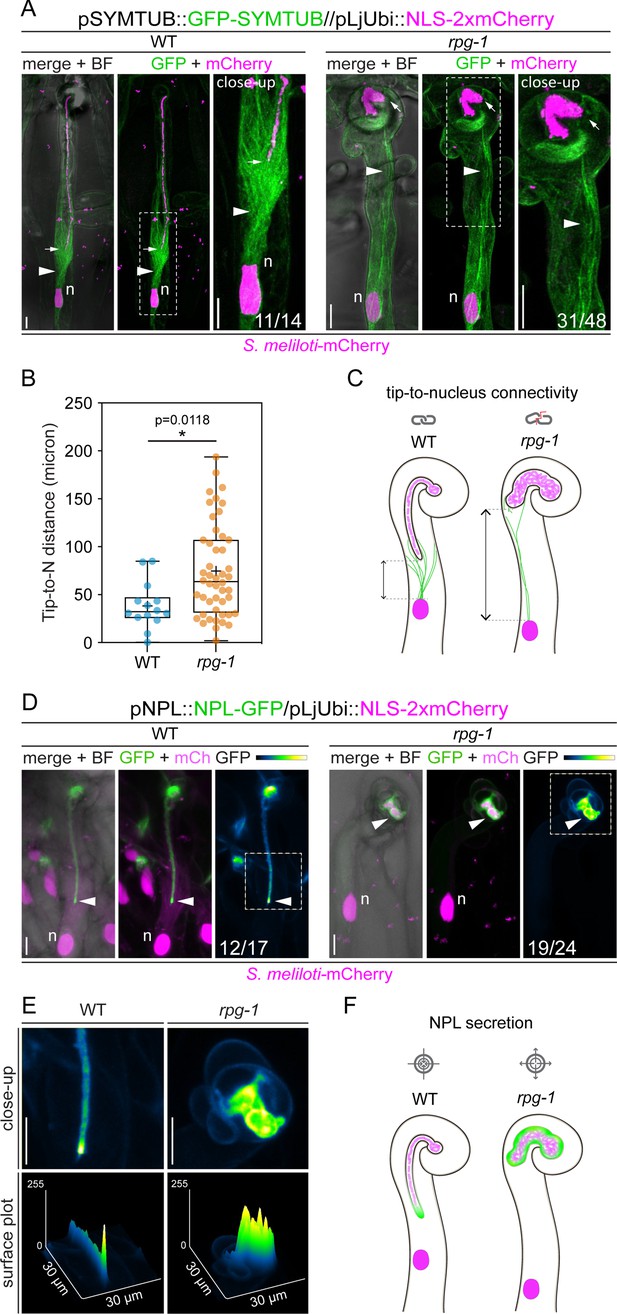
The tip-to-nucleus microtubule connectivity and polar secretion of Nodule pectate lyase (NPL) are affected in rpg-1.
(A) In vivo imaging of microtubule patterning in root hairs hosting infection threads from wild-type (WT) and rpg-1 transgenic roots expressing GFP-SYMTUB at 4–8 dpi with S. meliloti-mCherry. The endoplasmic microtubule array (arrowhead) bridging the nucleus (n, magenta) and the infection thread (IT) tip (arrow) in WT root hairs appears dispersed and stretched in rpg-1 root hairs, where the IT tip does not follow the path of the migrating nucleus. A nuclear-localized tandem mCherry was used as a transformation marker. Images are merges of maximum intensity projections of GFP (green) and mCherry (magenta) channels after deconvolution of the corresponding stack series using the Huygens Professional software (Scientific Volume Imaging, The Netherlands). The bright field (BF) channel is overlaid with the merge. The dashed white line boxes indicate the region shown in the close-up. Numbers indicate frequencies of observation made on a total number of six (WT) and 14 (rpg-1) composite plants from two independent replicas. (B) Distance between the nucleus (N) and the IT tip measured in infected root hairs of WT and rpg-1 transgenic roots expressing GFP-SYMTUB. In the box plot, the top and bottom of each box represent the 75th and 25th percentiles, the middle horizontal bar indicates the median and the whiskers represent the range of minimum and maximum values. Crosses represent sample means. Asterisks indicate statistical significance based on a Mann-Whitney test with p-values <0.05 (*), <0.01 (**), <0.001 (***), and <0.0001 (****). n=14 (WT) and 48 (rpg-1) infected root hairs. (C) Schematic representation of infected root hairs showing endoplasmic microtubules (green curved lines) maintaining the nucleus (magenta oval) and the IT tip at a close distance (double-arrow line) in WT. Such tip-to-nucleus connectivity (gray icon) is altered (gray icon with red line) in rpg-1 infected root hairs, where microtubules appear rarefied and do not bridge the IT tip and the nucleus, being separated by an increased distance. Dashed light gray lines indicate the position of the nucleus and the tip. Bacteria within infection threads are colored in light magenta. (D) Live-cell confocal images showing focal accumulations of NPL-GFP at the IT tip (arrowhead) in WT root hairs in contrast to its unrestricted distribution in the apoplastic space surrounding ITs developed within root hairs of rpg-1 transgenic roots. A nuclear-localized tandem mCherry was used as a transformation marker. Images are maximum intensity projections. The GFP channel is shown in green when merged with the mCherry channel (magenta), or in Green Fire Blue when isolated, with yellow indicating the maximum intensity and blue a low level of fluorescence. The bright field (BF) channel is overlaid with the merge. Numbers indicate frequencies of observations made on a total number of 10 (WT) and nine (rpg-1) composite plants from two independent replicas. (E) Close-up and surface plot of the regions bounded by the dashed white line boxes in (D). The signal of NPL-GFP in WT ITs is described by a major intensity peak positioned on the advancing tip compared to multiple intensity peaks detected on ITs of rpg-1. (F) Schematic representation of infected root hairs showing the accumulation of NPL (green) restricted to the apoplastic space surrounding the tip of WT ITs in contrast to its accumulation in a broader apoplastic domain in IT of rpg-1, indicating that targeted secretion of this enzyme (gray icon with arrows pointing to center) is not correctly maintained in this mutant (gray icon with arrows pointing outside). Bacteria within infection threads are colored in light magenta. Scale bars = 10 µm.
-
Figure 6—source data 1
Tip-to-nucleus distance and statistical analysis.
- https://cdn.elifesciences.org/articles/80741/elife-80741-fig6-data1-v2.zip
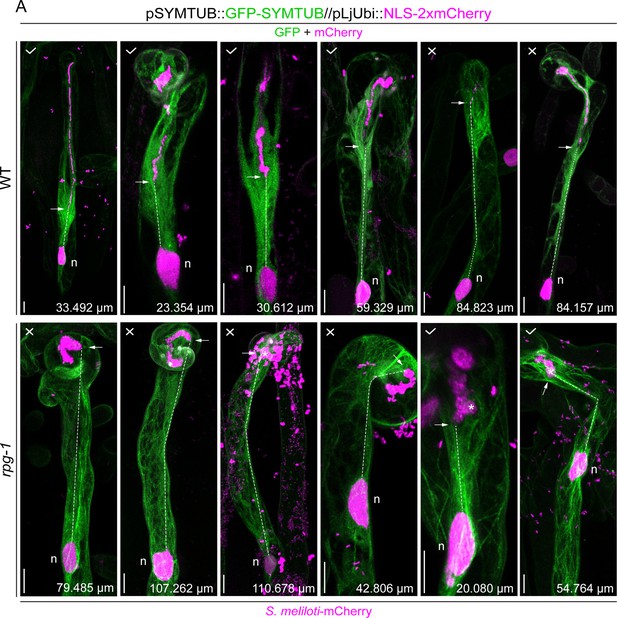
Microtubule patterning in wild-type (WT) and rpg-1 infected root hairs.
Representative images of root hairs hosting infection threads from WT and rpg-1 transgenic roots expressing GFP-SYMTUB at 4–8 dpi with S. meliloti-mCherry. A nuclear-localized tandem mCherry was used as a transformation marker. The tick and the cross symbols in white in the upper left side of each image indicate the presence or absence of a microtubule-mediated physical connection between the nucleus (n) and the infection thread (IT) tip (white arrow). White asterisks denote IT tips with the tilted direction of growth in rpg-1 infected root hairs. The dashed white lines indicate the tip-to-nucleus distance, whose value is reported in microns (μm) at the bottom right side of each image. Images are merges of maximum intensity projections of GFP (green) and mCherry (magenta) channels after deconvolution of the corresponding stack series using the Huygens Professional software (Scientific Volume Imaging, The Netherlands). The first two images of the upper and lower panel are the same presented in Figure 5A. Scale bar = 10 um.
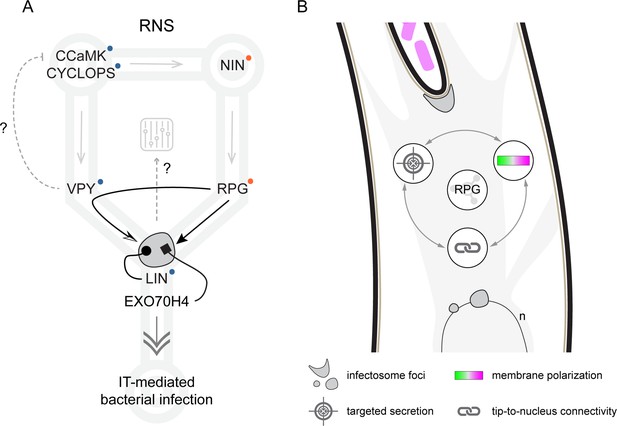
Proposed model for Rhizobium-directed polar growth (RPG) role during intracellular rhizobial infection.
(A) During Root Nodule Symbiosis (RNS), NIN-dependent transcription of RPG (gray arrow, Liu et al., 2019a; Soyano and Hayashi, 2014) leads to accumulation of the encoded protein (black arrow) within infectosome foci hosting VAPYRIN (VPY), LUMPY INFECTION (LIN), and EXO70H4 (gray irregular round shape, this study, Liu et al., 2019a). The coalescence of VPY into cytoplasmic foci is fully dependent on RPG (curved arrow, this study), while LIN is required to stabilize VPY (dotted arrow, Liu et al., 2019a, Liu et al., 2021). Since VPY and LIN belong to a conserved endosymbiotic signaling pathway including the core components CCaMK and CYCLOPS (Radhakrishnan et al., 2020, blue dots), we propose that RPG serves as a specificity determinant for infectosome-related endo- and exocytotic events (double arrow) enabling IT-mediated rhizobial infection. This is further supported by the capacity of RPG to interact with the exocyst subunit EXO70H4 (rectangular arrow). RPG-dependent segregation of VPY in infectosome foci might additionally allow to modulate the symbiotic signaling circuit (gray dashed arrow), if the putative negative regulation exerted by VPY on CCaMK (dashed gray flat arrow) during AM (Lindsay et al., 2022) would be conserved during RNS. Gray arrows indicate transcriptional activation/dependency (according to Murray et al., 2011; Singh et al., 2014; Liu et al., 2019a; Li et al., 2023). Orange dots indicate loss of the corresponding gene in non-nodulating species of the FaFaCuRo clade. (B) RPG represents a protein hub for infectosome assembly and its function is required to maintain a functional interplay between membrane polarization, tip-to-nucleus connectivity, and targeted secretion enabling infection thread (IT) polar progression.
Additional files
-
Supplementary file 1
List of constructs used in this study.
- https://cdn.elifesciences.org/articles/80741/elife-80741-supp1-v2.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/80741/elife-80741-mdarchecklist1-v2.pdf





