Revealing druggable cryptic pockets in the Nsp1 of SARS-CoV-2 and other β-coronaviruses by simulations and crystallography
Figures
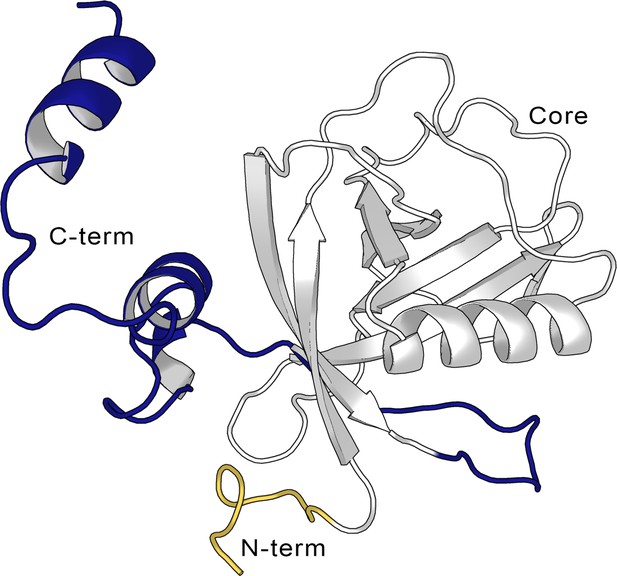
Structure of the full-length SARS-CoV-2 Nsp1.
Cartoon representation of the full-length non-structural protein 1 (Nsp1) structure from the AlphaFold (Jumper et al., 2021; Varadi et al., 2022) model, showing the N-terminus (in yellow, aa Met1-Asn9), the Nsp1N core (in gray, aa Glu10-Asn126), and the C-terminus (blue, aa Gly127-Gly180).
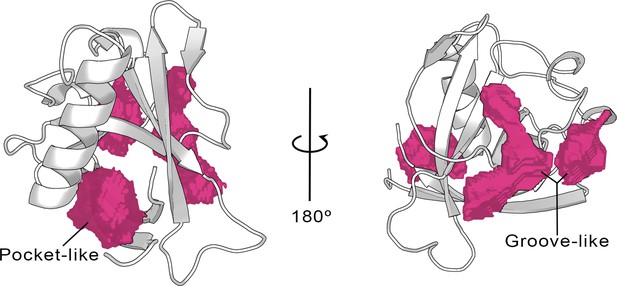
Cavities identified on the Nsp1N crystal structure (tPDB entry 7K7P) by the ProteinPlus server for the concave pocket-like structure between the β-barrel and the α-helix and the groove-like topology.
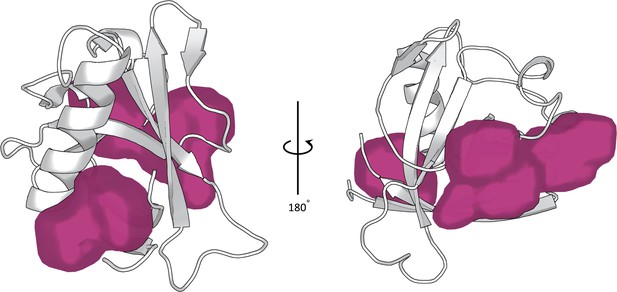
Cavities identified by the Fpocket software on the Nsp1N crystal structure (PDB entry 7K7P) (Le Guilloux et al., 2009).
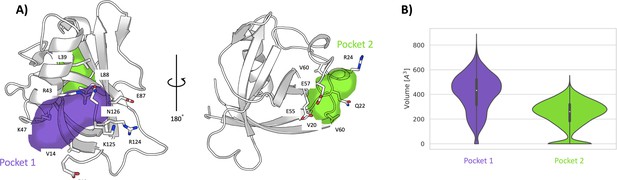
Pockets revealed from unbiased simulations.
(A) Cavities identified on Nsp1N along the 1 μs unbiased molecular dynamics (MD) simulation, namely pocket 1 (purple) and pocket 2 (green), with the main residues displayed in sticks. (B) The volume distribution of each pocket along the unbiased MD simulations.
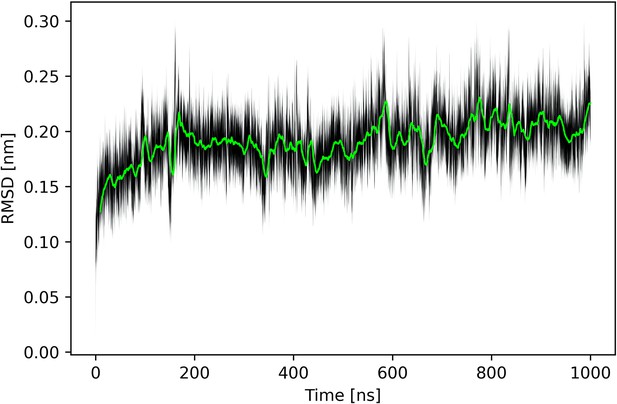
Time average of the root-mean-square deviation (RMSD) for Nsp1N.
The black shaded area delimits the RMSD values within two standard deviations from the time average.
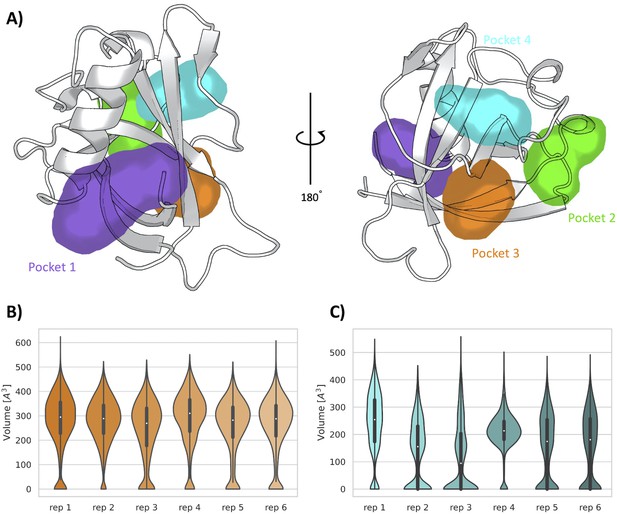
Pockets revealed from SWISH simulations.
(A) Pockets sampled during the 500 ns of replica-exchange SWISH (sampling water interfaces through scaled Hamiltonians) simulations. Volume distributions of the cryptic binding sites pocket 3 (B) and pocket 4 (C) along the six replicas of the SWISH simulations.
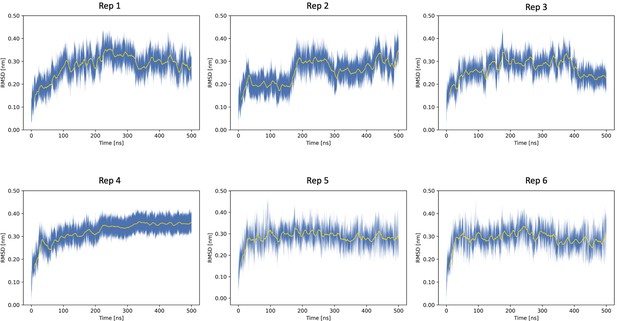
Time averages of the root-mean-square deviation (RMSD) for Nsp1N across the six SWISH (sampling water interfaces through scaled Hamiltonians) replicas.
The blue shaded area delimits the RMSD values within two standard deviations from the time average.
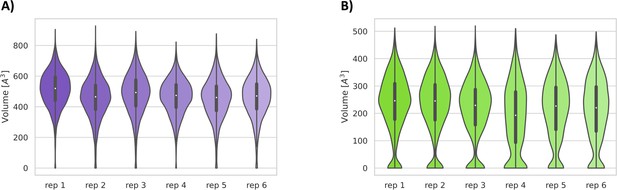
Violin plots of the pocket volumes.
Volume distributions of the cryptic binding sites pocket 1 (A) and pocket 2 (B) along the six replicas of the SWISH (sampling water interfaces through scaled Hamiltonians) simulation of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Nsp1N.
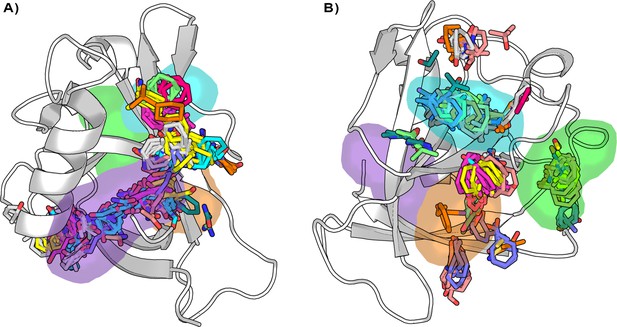
Distribution of the binding hotspots on the Nsp1N surface around (A) pocket 1 and (B) pockets 2, 3, and 4.
Multiple consensus clusters are shown in sticks. Each consensus cluster is represented in different colors.
-
Figure 5—source data 1
Consensus clusters information as obtained from the FTMap program for the 11 selected Nsp1N structures.
- https://cdn.elifesciences.org/articles/81167/elife-81167-fig5-data1-v1.docx
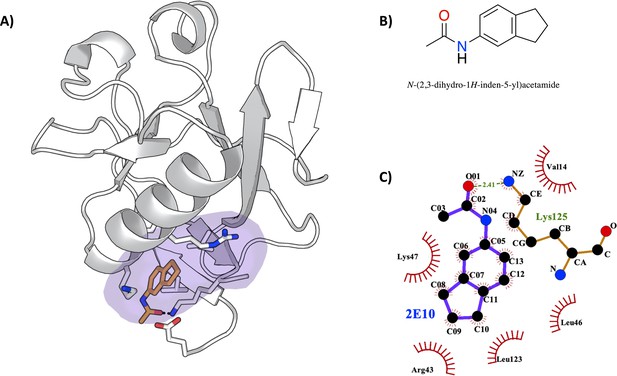
Characterization of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) non-structural protein 1 (Nsp1)-2E10 complex.
(A) Binding pose of the fragment hit obtained by crystal soaking and structure determination methods. The fragment is located in pocket 1. (B) Chemical structure and name of fragment hit. (C) Magnification of the Nsp1N –2E10 binding pocket showing the interactions the fragment establishes with residues of Nsp1. Hydrophobic interactions are shown by red half-moons and the hydrogen bond interaction is displayed with a dotted green line.
-
Figure 6—source data 1
Crystallographic data and model refinement statistics for the SARS CoV-2 Nsp1N-2E10 complex.
- https://cdn.elifesciences.org/articles/81167/elife-81167-fig6-data1-v1.docx
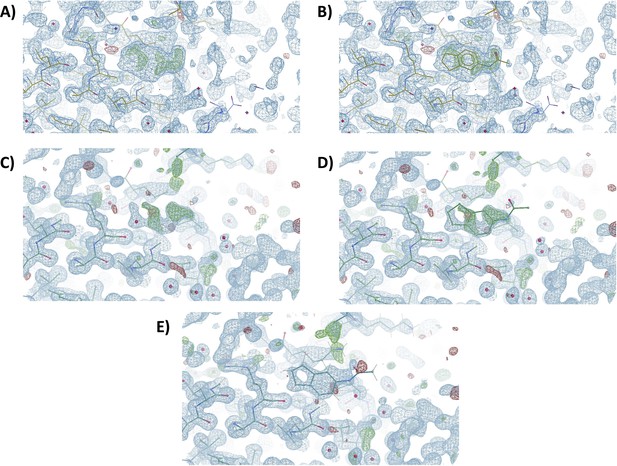
Pan-Dataset Density Analysis (PanDDA) event map and standard single dataset map focused on the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Nsp1N binding site of fragment hit 2E10.
PanDDA event map (A) without ligand and (B) in the presence of 2E10. Standard single dataset (D) map with and (C) without ligand 2E10. (E) Refined standard single dataset map with ligand 2E10. The sigma level for both the standard single dataset map and PanDDA maps is 1.
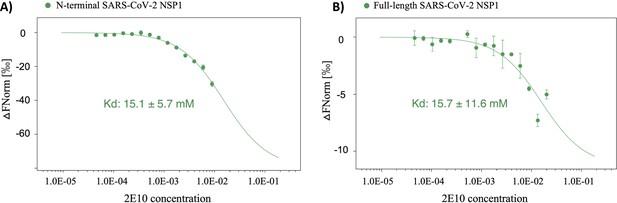
Microscale thermophoresis (MST) measurements of the binding affinity between fragment hit 2E10 and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) non-structural protein 1 (Nsp1).
(A) Concentration-response plot of fragment hit 2E10 and SARS-CoV-2 Nsp1N domain. (B) Concentrations-response curve of 2E10 and SARS-CoV-2 Nsp1 FL. All measurements were conducted at least in triplicate.
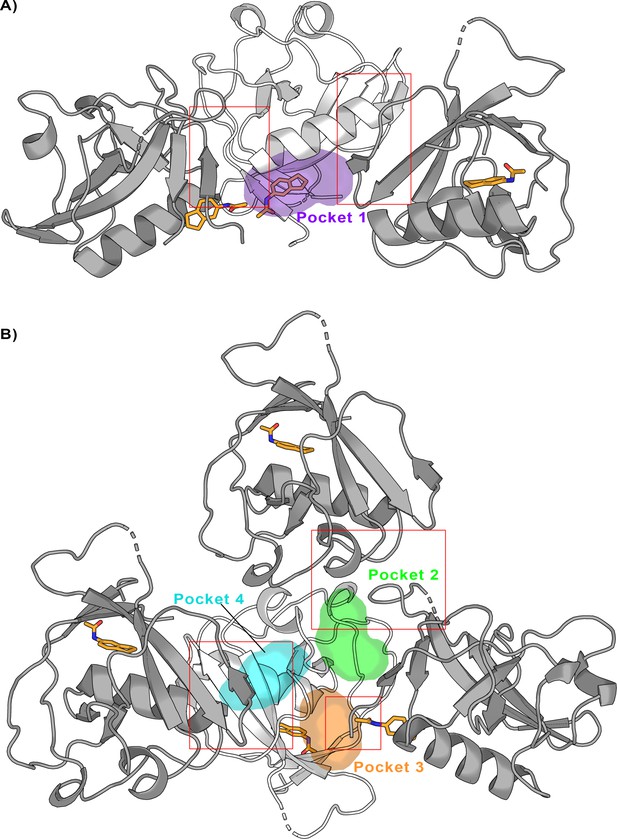
Crystal packing of the Nsp1N-fragment complex (PDB entry 8A4Y) was obtained from X-ray soaking experiments.
The direct crystal contacts around (A) pocket 1 and (B) pockets 2, 3, and 4 are highlighted with squares.
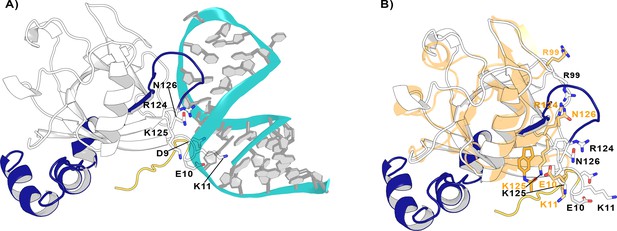
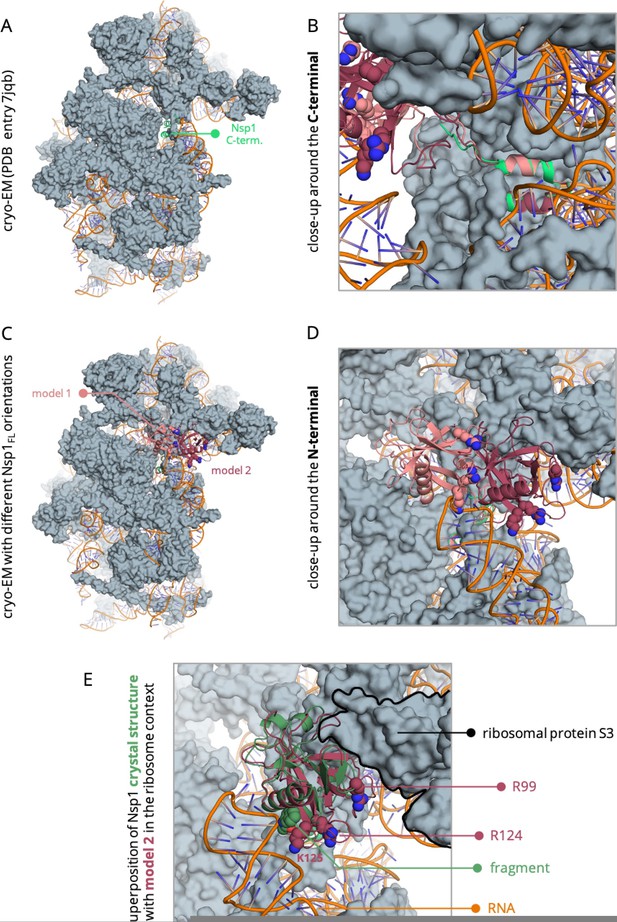
Nsp1FL-RNA complex obtained from the HADDOCK program.
(A) Residues in the proximity of pocket 1 involved in crucial Nsp1FL-SL1 contacts are displayed in sticks. (B) Crystal structure from soaking experiments (orange transparent cartoon) is superimposed over the Nsp1FL of the Nsp1FL-RNA model. The most important residues for the Nsp1FL-RNA interaction are highlighted in white (Nsp1FL model) and orange (PDB entry 8A4Y from our soaking experiments) sticks. Figures obtained from model A.
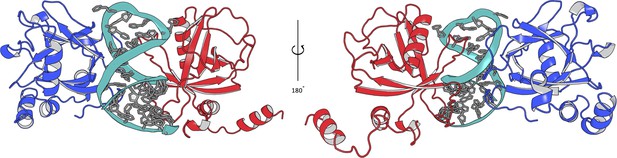
Different Nsp1FL-RNA models proposed in this work.
The Nsp1FL from model A (blue) and B (red) are represented.
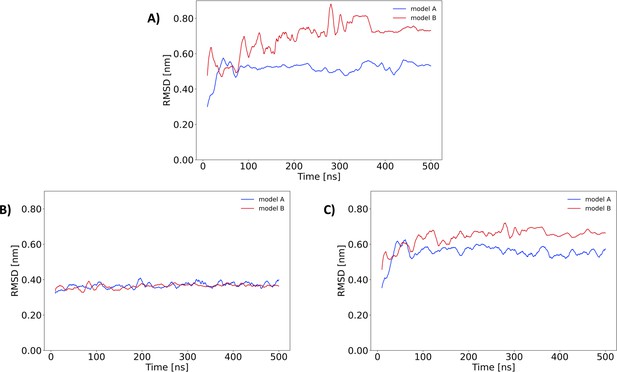
RMSD for model A and B of the Nsp1-RNA complex.
Root-mean-square deviation (RMSD, nm) along the 500 ns of unbiased molecular dynamics (MD) simulations considering (A) the backbone atoms of the Nsp1FL protein from the Nsp1FL-RNA complex, (B) only the RNA atoms of the stem loop 1 (SL1) from the complex and (C) the backbone atoms of Nsp1FL and all the RNA atoms of the full Nsp1FL-RNA complex.
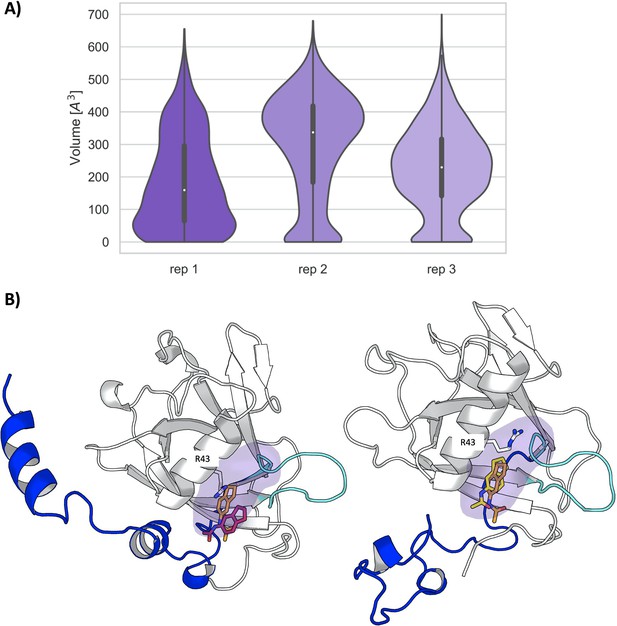
Ligand accesibility to pocket 1 in Nsp1FL.
(A) Volume distribution of pocket 1 along the three unbiased molecular dynamics (MD) simulations of the Nsp1FL. (B) Docked poses of the fragment to the representative structure from the most populated cluster in replica 3 (left) and to the second most populated cluster in replica 2 (right). The original crystallographic pose is depicted in orange, whereas the docked poses in pink and yellow. The flexible loop comprising aa Arg124-Gly137 is displayed is cyan. N- and the C-terminal are depicted in purple and blue, respectively.
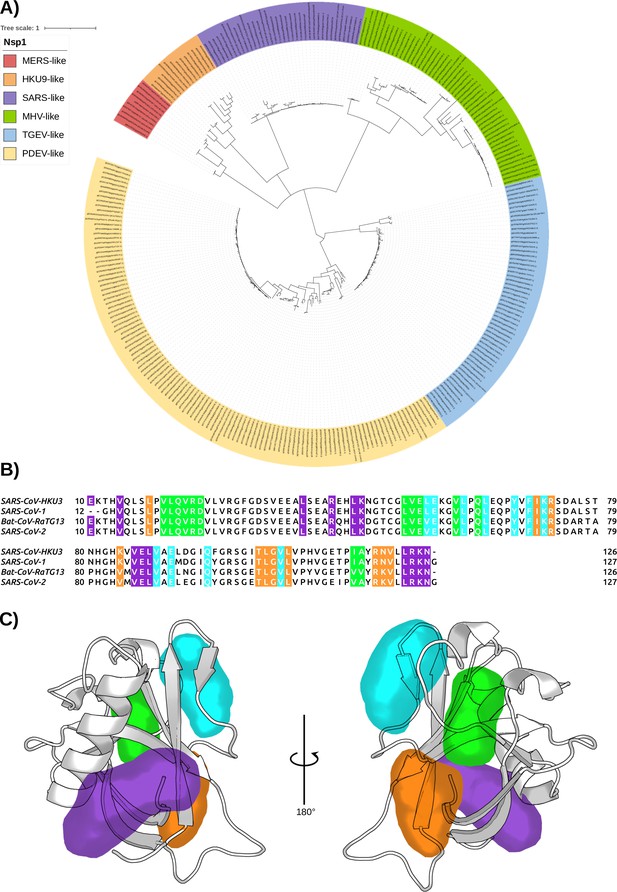
The conservation of Nsp1 sequence.
(A) Phylogenetic tree based on 283 sequences from distinct α- and β-coronaviruses (CoVs) of different subgenera. The sequences were obtained from the Conserved Domain Database (CDD) with accession number cl41742. The scale bar indicates the number of substitutions per site in the amino acid sequence. Six different Nsp1 domain models can be identified, two for the α-CoVs (transmissible gastroenteritis virus [TGEV]-like and PDEV-like), and four for the β-CoVs genus (MERS-like, HKU9-like, SARS-like, and murine hepatitis virus [MHV]-like). (B) Multi-sequence alignment of the four homologues selected. The alignment was performed with the MUSCLE algorithm (Edgar, 2004). The residues of the different pockets are highlighted in the corresponding color, namely pocket 1 in purple, pocket 2 in green, pocket 3 in orange, and pocket 4 in cyan. (C) Representation of the pockets found in the severe acute respiratory syndrome coronavirus 1 (SARS-CoV-1) Nsp1N variant.
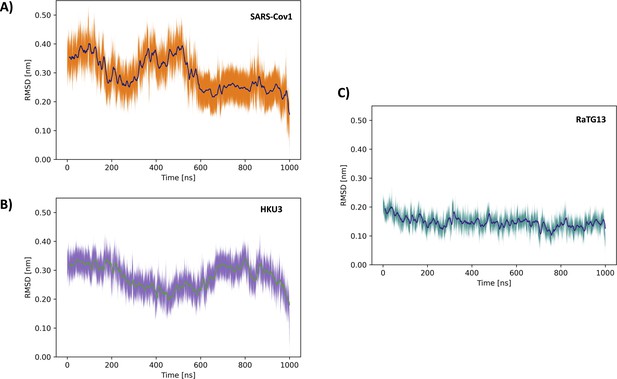
Time averages of the backbone root-mean-square deviation (RMSD) along the 1 μs unbiased molecular dynamics (MD) simulation for Nsp1N of (A) severe acute respiratory syndrome coronavirus 1 (SARS-CoV-1), (B) SARS-HKU3, and (C) Bat-CoV-RatG13.
The shaded area delimits the RMSD values within two standard deviations from the time average.
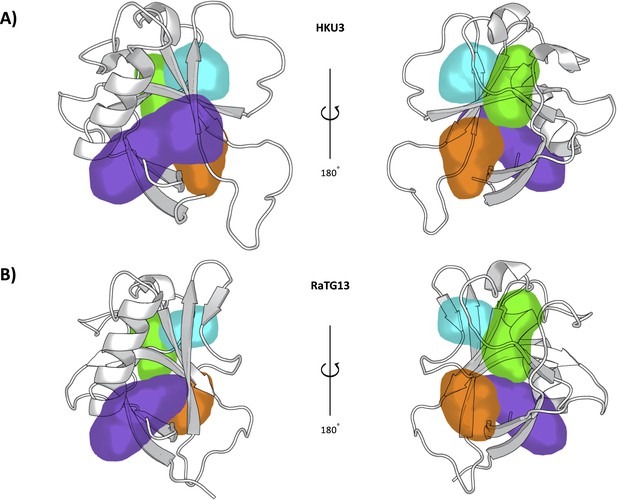
Representation of the pockets found in the Bat-CoV-HKU3 (A) and Bat-Cov-RaTG13 (B) Nsp1N variants.
Tables
SMILES and identification number of the 59 tested fragments and corresponding predicted binding.
The predicted pose for each fragment can be found at https://github.com/Gervasiolab/Gervasio-Protein-Dynamics/tree/master/nsp1/virtual_screening (Gervasiolab, 2022;copy archived at swh:1:rev:936e929db11aff39faed53e5fbe6f902f1456a6d). The above reported crystal hit (lig_1427) is highlighted in bold.
| Identification Number | SMILES | Predicted pocket(s) |
|---|---|---|
| lig_30 | C9H7NO2 | 3, 4 |
| lig_83 | C12H13NO2 | 1, 3, 4 |
| lig_113 | C12H13ClF3NO | 1, 3, 4 |
| lig_168 | C13H10O3 | 1, 3, 4 |
| lig_171 | C11H10O3 | 1, 4 |
| lig_194 | C8H6F2O3 | 1, 3, 4 |
| lig_212 | C11H12O2 | 1, 3, 4 |
| lig_243 | C14H17NO2S | 1 |
| lig_262 | C15H16N2O | 1, 3, 4 |
| lig_286 | C13H13NO2S | 1, 3, 4 |
| lig_329 | C9H8F3NOS | 3, 4 |
| lig_335 | C13H11NO2S | 1, 3 |
| lig_349 | C11H9NO2 | 1, 4 |
| lig_355 | C13H13NO2S | 4 |
| lig_369 | C11H12O2 | 1, 3, 4 |
| lig_377 | C12H10O2S | 1, 4 |
| lig_394 | C16H13NO2 | 1, 3, 4 |
| lig_400 | C14H12O2 | 1, 3, 4 |
| lig_422 | C14H9NO2 | 1, 3, 4 |
| lig_490 | C8H5NO2S2 | 1, 4 |
| lig_502 | C12H10O3 | 1, 3, 4 |
| lig_507 | C12H12N2O | 1, 4 |
| lig_552 | C12H15NO2 | 1, 3, 4 |
| lig_570 | C12H9NO2 | 1, 4 |
| lig_575 | C10H8O3 | 1, 4 |
| lig_579 | C12H9NO2 | 1, 3, 4 |
| lig_685 | C14H12O3 | 1, 3, 4 |
| lig_706 | C12H14N2O | 1, 4 |
| lig_752 | C9H7ClFN3S2 | 1, 3, 4 |
| lig_783 | C12H11FO2 | 1, 3, 4 |
| lig_806 | C14H13NO2 | 1, 3 |
| lig_812 | C13H11ClN2O | 1 |
| lig_864 | C15H12O3S | 1, 3, 4 |
| lig_892 | C7H6N2OS | 1, 3 |
| lig_897 | C8H6FNO2S | 1, 3, 4 |
| lig_907 | C10H11NOS2 | 1, 4 |
| lig_910 | C10H6F3NOS | 1, 3, 4 |
| lig_924 | C14H12O3S2 | 1 |
| lig_1009 | C11H10ClNO2 | 1, 4 |
| lig_1037 | C10H7ClO2S | 1, 4 |
| lig_1054 | C12H13NO2 | 1, 3, 4 |
| lig_1057 | C12H16N2O | 1, 3, 4 |
| lig_1064 | C13H12O2 | 1, 3, 4 |
| lig_1149 | C11H10F3NO | 1, 3, 4 |
| lig_1157 | C13H13NO2 | 1, 3, 4 |
| lig_1195 | C11H10N2O | 1, 3, 4 |
| lig_1209 | C9H6F3N3S2 | 1, 3, 4 |
| lig_1216 | C12H11F2NO2 | 1, 3, 4 |
| lig_1220 | C11H12O2S | 3, 4 |
| lig_1223 | C8H9ClN2OS | 1, 3 |
| lig_1228 | C15H13FN2OS | 1, 3, 4 |
| lig_1281 | C9H7F3O3 | 1, 3, 4 |
| lig_1310 | C12H13NO2 | 1, 4 |
| lig_1315 | C13H11F3O2 | 1, 3, 4 |
| lig_1381 | C9H6F3NO | 3, 4 |
| lig_1382 | C12H8F3NO2S | 1, 3, 4 |
| lig_1410 | C9H11NO | 3, 4 |
| lig_1427 | C11H13NO | 1, 4 |
| lig_1428 | C11H11NO2 | 1, 4 |
Thermal shift assay (TSA) results for 59 potential fragment hits from computational screening using severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Nsp1N.
Fragments showing atypical curves are labelled as atypical curve in the table.
| Identification number | Fragments | Ti ± SD [°C] | ΔTi/ °C |
|---|---|---|---|
| lig_575 | 1B9 | 52.5±0.6 | –2.7 |
| lig_490 | 1C6 | 51.5±0.4 | –3.4 |
| lig_507 | 1H6 | 50.5±0.1 | –4.1 |
| lig_1427 | 2E10 | 53.8±0.3 | –1.3 |
| lig_1428 | 2F10 | 53.5±0.4 | –1.7 |
| lig_1037 | 2F5 | 53.7±0.6 | –1.6 |
| lig_685 | 3E8 | 54.8±0.2 | –0.3 |
| lig_924 | 4B5 | 54.7±0.5 | –0.5 |
| lig_502 | 4C6 | Atypical curve | |
| lig_369 | 4E4 | 53.6±0.4 | –1.4 |
| lig_400 | 5B7 | 53.9±0.1 | –0.9 |
| lig_194 | 5D3 | Atypical curve | |
| lig_335 | 5E8 | 54.8±0.2 | –0.4 |
| lig_30 | 6E3 | Atypical curve | |
| lig_113 | 6G8 | 54.3±0.0 | –0.6 |
| lig_168 | 7A2 | 51.2±0.4 | –3.8 |
| lig_212 | 7A4 | Atypical curve | |
| lig_552 | 7B6 | 55.0±0.2 | 0.1 |
| lig_1220 | 7D10 | 54.7±0.3 | –0.2 |
| lig_171 | 7D2 | 54.6±0.2 | –0.3 |
| lig_570 | 7E6 | 51.5±0.4 | –3.1 |
| lig_579 | 7F11 | 51.8±0.2 | –3 |
| lig_1054 | 7F6 | 54.3±0.3 | –0.6 |
| lig_1310 | 8A4 | Atypical curve | |
| lig_1281 | 8A5 | Atypical curve | |
| lig_422 | 8B4 | 51.2±0.1 | –3.7 |
| lig_349 | 8C4 | Atypical curve | |
| lig_1410 | 8D3 | 53.2±0.3 | –2.4 |
| lig_1009 | 8G7 | 51.5±0.4 | –3.3 |
| lig_355 | 9A11 | 54.9±0.1 | 0 |
| lig_243 | 9C10 | 54.3±0.1 | –0.9 |
| lig_1216 | 9D8 | 54.5±0.4 | –0.7 |
| lig_812 | 9E11 | 53.1±0.1 | –1.9 |
| lig_329 | 9E2 | 54.4±0.3 | –0.5 |
| lig_262 | 9G9 | 53.2±0.3 | –1.7 |
| lig_783 | 10A6 | 54.4±0.2 | –1.2 |
| lig_910 | 10A8 | Atypical curve | |
| lig_806 | 10A9 | 54.6±0.1 | -1 |
| lig_1381 | 10B7 | 50.7±0.3 | –4.9 |
| lig_286 | 10G8 | 54.1±0.2 | –0.7 |
| lig_1209 | 11A5 | 55.8±0.1 | 0.2 |
| lig_1057 | 11B2 | Atypical curve | |
| lig_907 | 11E4 | 55.6±0.0 | 0.7 |
| lig_1064 | 11E8 | 54.6±0.1 | –0.3 |
| lig_377 | 11F10 | 54.8±0.1 | –0.8 |
| lig_1195 | 11F2 | 54.8±0.2 | –0.1 |
| lig_1149 | 11G3 | 55.4±0.5 | 0.5 |
| lig_83 | 12A11 | Atypical curve | |
| lig_1315 | 12A3 | Atypical curve | |
| lig_864 | 12B10 | 55.1±0.2 | –0.1 |
| lig_394 | 12B5 | 55.2±0.2 | 0.1 |
| lig_1382 | 12C4 | 55.5±0.4 | 0.6 |
| lig_752 | 12D5 | 54.9±0.1 | –0.3 |
| lig_1223 | 12F7 | 52.4±0.3 | –2.7 |
| lig_1157 | 12F9 | 55.0±0.2 | –0.2 |
| lig_1228 | 12G7 | 52.6±0.5 | –2.6 |
| lig_892 | 12H2 | 54.8±0.1 | –0.4 |
| lig_706 | 13D6 | 51.8±0.4 | –3.3 |
| lig_897 | 13G5 | 54.3±0.1 | –0.9 |
Pairwise identity percentages of selected non-structural protein 1 (Nsp1) sequences for representative α- and β-coronaviruses (CoVs) subfamilies.
National Center for Biotechnology Information (NCBI) accession numbers the sequences used for the analysis are as follows: transmissible gastroenteritis virus (TGEV) 6IVC_A, PDEV 5XBC_A, severe acute respiratory syndrome coronavirus 1 (SARS CoV-1) NP_828860.2, severe acute respiratory syndrome coronavirus 2 (SARS CoV-2) YP_009725297.1, HKU9 P0C6T6, MERS YP_009047229, and murine hepatitis virus (MHV) YP_209244.
| CoV-like | SARS-1 | SARS-2 | HKU9 | MERS | MHV | TGEV | PDEV |
|---|---|---|---|---|---|---|---|
| SARS-1 | 100 | ||||||
| SARS-2 | 86.1 | 100 | |||||
| HKU9 | 20.9 | 19.5 | 100 | ||||
| MERS | 20.9 | 17.1 | 24.5 | 100 | |||
| MHV | 16.9 | 16.4 | 4.1 | 18.0 | 100 | ||
| TGEV | 17.4 | 16.9 | 13.3 | 7.9 | 9.0 | 100 | |
| PDEV | 17.9 | 16.1 | 15.2 | 8.7 | 11.5 | 18.2 | 100 |