Microcephaly-associated protein WDR62 shuttles from the Golgi apparatus to the spindle poles in human neural progenitors
Figures
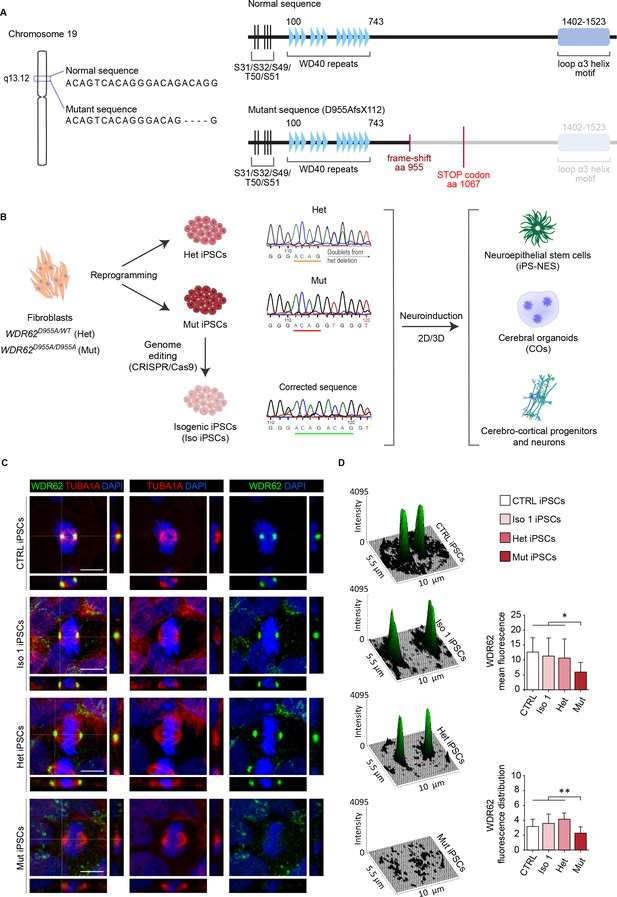
Generation of human induced Pluripotent Stem Cell (iPSC)-based 2D and 3D models to analyze WDR62 function in neurodevelopment.
(A) Schematic illustrations of the WDR62 gene on chromosome 19q13 indicating the 4 bp (ACAG) deletion in exon 23, and of WDR62 full-length and C-terminally truncated (D955AfsX112) protein structure showing the location of WD40 repeats and loop α3 helix motif. (B) Scheme of experimental design. WDR62 iPSCs harboring the heterozygous (Het; D955A/WT) or homozygous (Mut; D955A/D955A) mutation, and isogenic corrected (Iso) iPSC lines were employed. Sanger sequencing of iPSCs shows the above-described mutations from one parent and the affected offspring, and the CRISPR/Cas9 corrected sequence. A neuroinduction protocol was applied to Het, Mut, and Iso 1 and/or Iso 2 iPSCs to obtain neuroepithelial stem (iPS-NES) cells, cerebral organoids (COs), as well as cerebro-cortical progenitors and neurons. (C) Representative confocal images of WDR62 and tubulin-alpha (TUBA1A) expression in CTRL (external control), Het, Mut, and Iso 1 iPSCs during mitosis. Orthogonal projections indicate WDR62, TUBA1A, and DAPI signals. (D) Surface plots and fluorescence intensity analysis show WDR62 signal distribution. Histograms show mean fluorescence intensity (top) and fluorescence signal distribution (skewness, arbitrary unit, bottom) during metaphase. CTRL, Het, and Iso 1 iPSCs show similar WDR62 signal distribution, which is decreased in the measured area in Mut iPSCs (replicates n=3, total cells N=61, p-value <0.05, Kruskal-Wallis test, post hoc Dunn’s test in top and bottom histograms. Data are shown as mean ± SD. Scale bar = 10 μm).
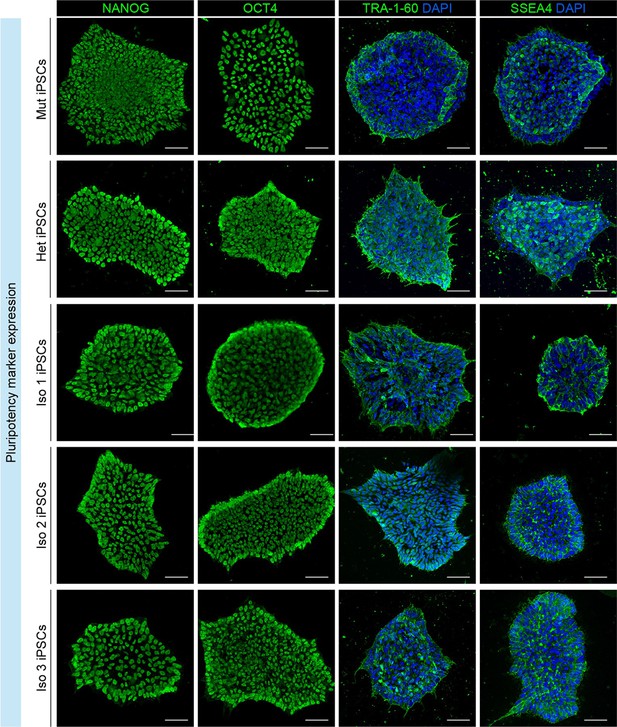
Analysis of pluripotency marker expression in patient (homozygous, Mut), parent (heterozygous, Het), and three isogenic-corrected (Iso 1, Iso 2, and Iso 3) induced Pluripotent Stem Cell (iPSC) lines.
Immunofluorescence analysis for NANOG, POU5F1 (also known as OCT4), TRA-1–60, and SSEA4 in Mut, Het, Iso 1, Iso 2, and Iso 3 iPSC lines. Scale bars = 50 μm.
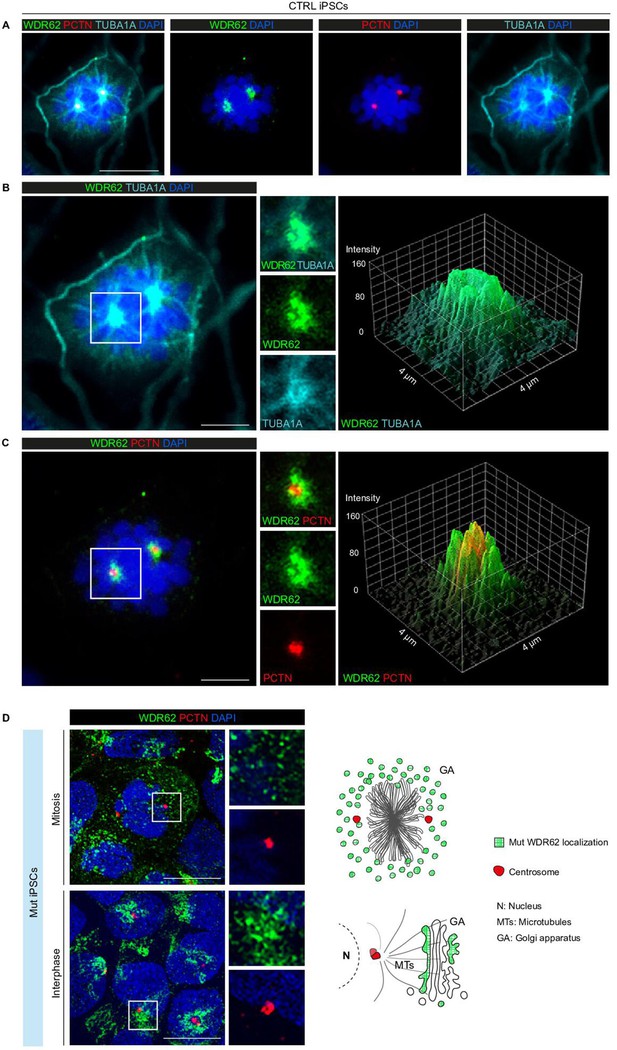
WDR62 is localized to the spindle poles in CTRL induced Pluripotent Stem Cells (iPSCs) during mitosis.
(A) Representative immunofluorescence assay for WDR62, PCTN (centrosome marker), and TUBA1A (mitotic spindle marker) showing WDR62 signal surrounding the centrosome. (B) Magnified view (left) of the mitotic cell in (A) showing the WDR62 and TUBA1A signals in the boxed area overlapping at the spindle poles (middle). The 3D surface plot of fluorescence intensity shows similar WDR62 and TUBA1A signal distribution at the spindle poles (right). (C) Magnified view (left) of the mitotic cell in (A) showing the WDR62 signal surrounding the PCTN signal in the boxed area (middle). The 3D surface plot of fluorescence intensity shows non-overlapping WDR62 and PCTN signal distribution at the spindle poles (right). (D) Endogenous WDR62 signal in Mut iPSCs. Representative immunofluorescence assay for WDR62 and PCTN during mitosis and interphase. Magnified view of the boxed areas showing subcellular localization of mutant WDR62 protein. Schematic representation of mutant WDR62 signal (green) distribution during mitosis (diffuse) and interphase (in the Golgi apparatus). Scale bars = 10 μm in (A) and 5 μm in (B, C), and 20 μm in (D).
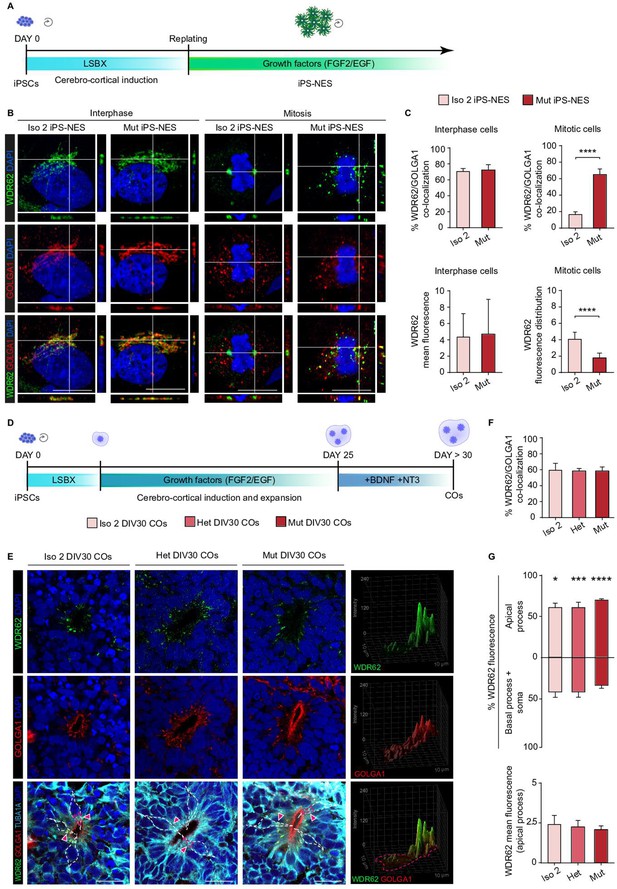
WDR62 is associated with the Golgi apparatus in neural progenitors during interphase.
(A) Schematic illustration of Neuroepithelial Stem Cell derivation protocol from induced Pluripotent Stem Cells (iPSCs) (iPS-NES). (B) Immunofluorescence analysis of WDR62 and GOLGA1 in interphase and metaphase iPS-NES cells. During interphase, the WDR62 subcellular localization is comparable in Iso 2 and Mut iPS-NES cells. In metaphase, the WDR62 signal is localized to the spindle poles in Iso 2 iPS-NES cells, but remains diffuse and co-localized with GOLGA1 in Mut iPS-NES cells. Orthogonal projections indicate WDR62, GOLGA1, and DAPI signals. (C) Quantification of WDR62 and GOLGA1 signal co-localization during interphase (top left histogram) and mitosis (top right histogram); WDR62 mean fluorescence in interphase (bottom left histogram) shows no differences in Iso 2 versus Mut iPS-NES cells. Fluorescence distribution in mitotic cells (bottom right histogram) (skewness, arbitrary units) show differences in Iso 2 versus Mut cells (top left histogram: replicates n=3, total cells N=60, p-value >0.05, unpaired Student’s t-test; top right histogram: replicates n=3, total cells N=33, p-value <0.0001, Kolmogorov-Smirnov test; bottom left histogram: replicates n=3, total cells N=164, p-value >0.05, unpaired Student’s t-test; bottom right histogram: replicates n=3, total cells N=21, p-value <0.0001, unpaired Student’s t-test). (D) Schematic illustration of cerebral organoid (CO) derivation protocol from iPSCs. (E) Immunofluorescence analysis of WDR62, GOLGA1, and TUBA1A in Iso 2, Het, and Mut COs at 30 days in vitro (DIV30). WDR62 and GOLGA1A signals co-localize to the apical domain of neural progenitors in COs during interphase. 3D surface plots show WDR62 and GOLGA1 signal distribution within the apical process. (F) Quantification of WDR62 and GOLGA1 signal co-localization in neural progenitors of Iso 2, Het, and Mut COs at DIV30 (COs n=6, total cell N=193, p-value >0.05, Kruskal-Wallis test, post hoc Dunn’s test). (G) Quantification of WDR62 fluorescence within the apical process versus the basal process and the soma of neural rosettes of Iso 2, Het, and Mut COs at DIV30 (COs n=6, total cell N=54, p-value <0.05, p-value <0.001, and p-value <0.0001, two-way ANOVA, post hoc Tukey’s test top histogram; p-value >0.05, one-way ANOVA, post hoc Tukey’s test bottom histogram). Data are shown as mean ± SD. Scale bar = 5 μm in (B) and 20 μm (E).
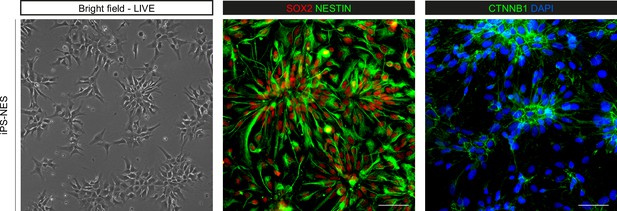
iPS-NES cells are polarized progenitors.
Morphology of Neuroepithelial Stem cells derived from induced Pluripotent Stem cells (iPS-NES cells), live imaging and representative immunofluorescence assay for SOX2, nestin (progenitor markers), and CTNNB1 (beta catenin, apical domain marker) showing iPS-NES cells at early passages self-organizing into neural rosettes. Scale bars = 50 μm.
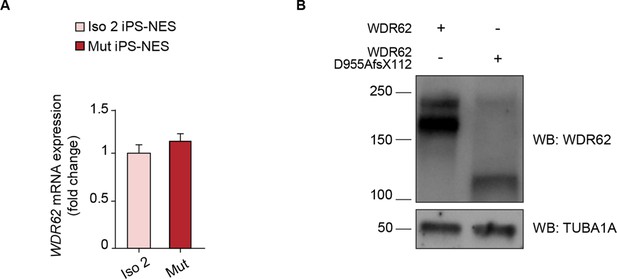
iPS-NES cells express mutant WDR62.
(A) WDR62 mRNA expression in Neuroepithelial Stem cells derived from induced Pluripotent Stem cells (iPS-NES cells). Quantitative analysis of mRNA expression levels detected by RT-qPCR reveals no differences in Iso 2 and Mut iPS-NES cells. Data are shown as fold change in mRNA expression of WDR62 relative to GAPDH, a housekeeping gene, according to the 2−ΔΔCT method. (B) Western blot showing full-length (165 KDa) and mutant (118 KDa) WDR62 protein expression. CTRL iPS-NES cells were transfected with WDR62 or WDR62 D955AfsX112 expression plasmids. Transfected cell lysates were immunoblotted with antibodies against WDR62 and TUBA1A.
-
Figure 2—figure supplement 2—source data 1
Uncropped images of western blots.
- https://cdn.elifesciences.org/articles/81716/elife-81716-fig2-figsupp2-data1-v1.zip
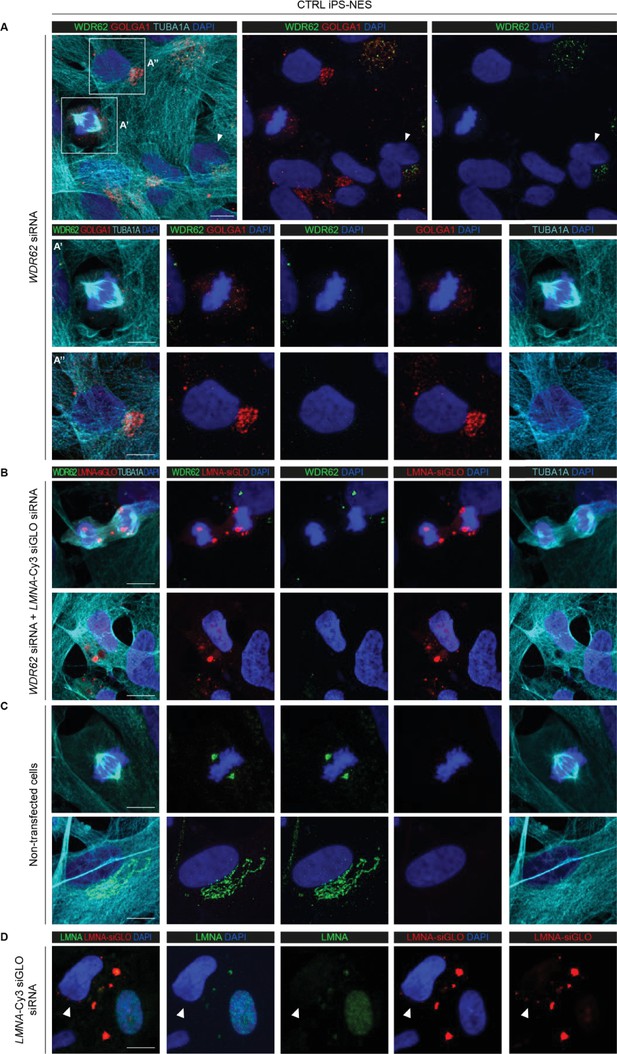
WDR62 knockdown supports localization to the Golgi apparatus.
(A) siRNA-mediated knockdown of WDR62 leads to loss of endogenous signal. Immunofluorescence assay for WDR62, GOLGA1, and TUBA1A shows no WDR62 signal in CTRL Neuroepithelial Stem cells derived from induced Pluripotent Stem cells (iPS-NES cells) transfected with WDR62-siRNA. In non-transfected cells (arrowhead), the WDR62 signal is present at the Golgi apparatus. Magnification of WDR62-siRNA transfected cells in mitosis (A’) and interphase (A’’). (B) Immunofluorescence assay for WDR62 and TUBA1A in CTRL iPS-NES cells co-transfected with WDR62- and LMNA-Cy3 siGLO (positive control for transfection) siRNAs. No WDR62 signal is detected in co-transfected cells (Cy3 fluorescence). (C) Immunofluorescence assay for WDR62, LMNA-Cy3, and TUBA1A in non-transfected CTRL iPS-NES cells. Representative confocal images of WDR62 signal in both mitotic and interphase cells. (D) Representative confocal images of LMNA and LMNA-Cy3 in CTRL iPS-NES cells transfected with LMNA-Cy3 siGLO siRNA. Contrary to non-transfected cells, transfected cells show no LMNA signal (arrowhead). Scale bars = 10 μm.
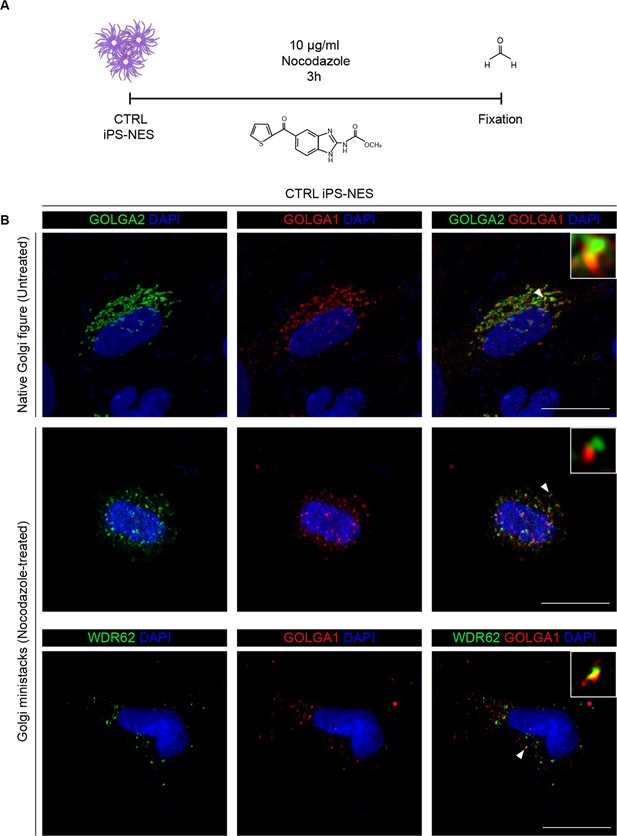
Supporting evidence for WDR62 localization to the Golgi apparatus via Golgi fragmentation.
(A) Schematic illustration of the protocol for Golgi apparatus fragmentation in CTRL Neuroepithelial Stem cells derived from induced Pluripotent Stem cells (iPS-NES cells) through nocodazole treatment and formation of Golgi ministacks. (B) Representative confocal images of GOLGA2 and GOLGA1 in untreated (top), and of WDR62 and GOLGA1 in nocodazole-treated cells showing Golgi apparatus fragmentation. In nocodazole-treated cells, WDR62 follows the same pattern as GOLGA1, indicating WDR62 association with the Golgi ministacks. Arrowheads indicate the areas magnified in the insets. Replicates n=4, total cell N=8. Scale bars = 20 μm.
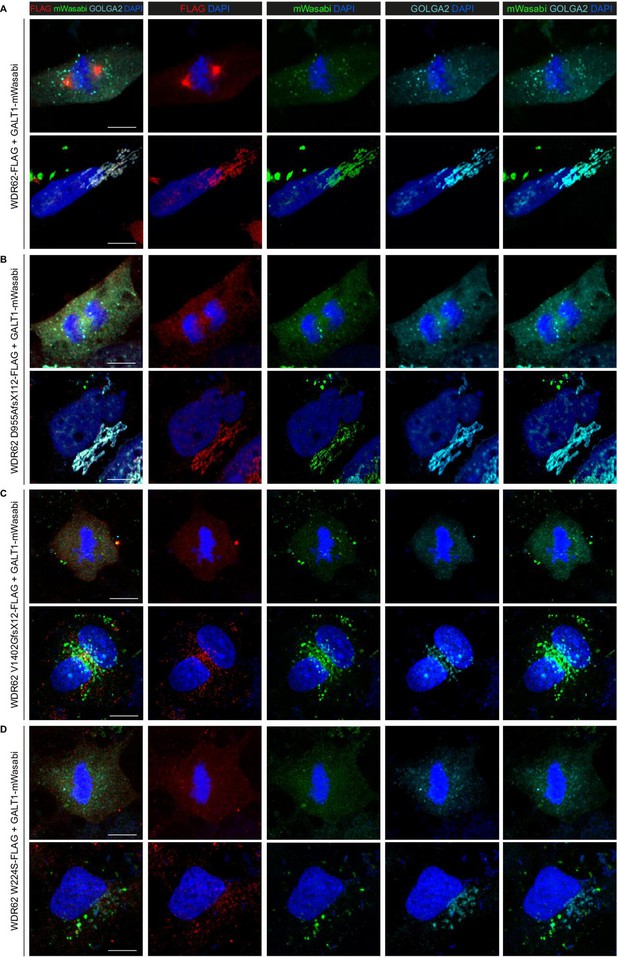
Localization pattern of full-length and mutant WDR62 in iPS-NES cells.
(A) CTRL Neuroepithelial Stem cells derived from induced Pluripotent Stem cells (iPS-NES cells) were co-transfected with WDR62-FLAG and GALT1-mWasabi expression plasmids. Representative confocal images for FLAG, mWasabi (a green fluorescent protein), and GOLGA2. Representative magnified views of a metaphase cell (top row) and of an interphase cell (bottom row) showing WDR62 localization to the mitotic spindle poles and to the Golgi apparatus during interphase. (B – D) Cells were co-transfected with microcephaly (MCPH)-associated WDR62 D955AfsX112-FLAG (B), WDR62 V1402GfsX12-FLAG (C), or WDR62 W224S-FLAG (D) and GALT1-mWasabi expression plasmids. Representative confocal images for FLAG, mWasabi, and GOLGA2. Magnified views of mitotic (top rows) and interphase cells (bottom rows) showing that mutant WDR62 do not localize to the mitotic spindle poles and that the FLAG and mWasabi signals overlap in the Golgi apparatus in interphase cells in a manner similar to the endogenous WDR62/GOLGA1 signals. Scale bars = 10 μm.
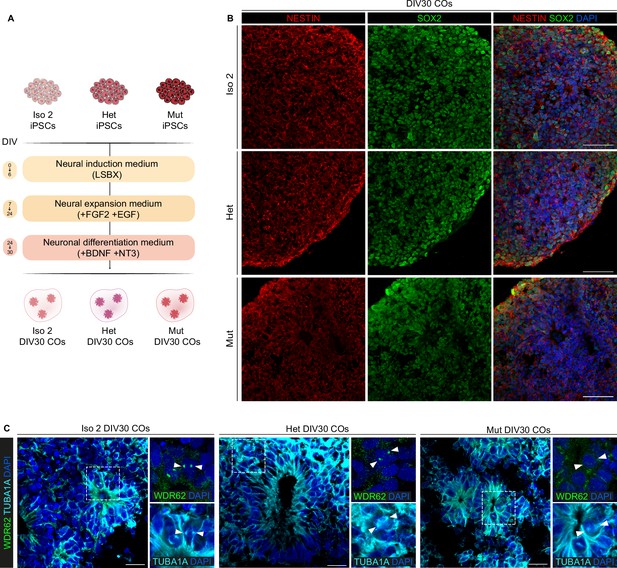
Characterization of cerebral organoids (COs).
(A) Schematic representation of CO generation (LSBX) via inhibitors of SMAD (LDN193189, SB431542) and WNT (XAV939) from Iso 2, Het, and Mut induced Pluripotent Stem Cell (iPSC) lines. (B) Immunofluorescence assay for nestin and SOX2 in sections of Iso 2, Het, and Mut COs at 30 days in vitro (DIV30) and representative confocal images. (C) Immunofluorescence assay for WDR62 and TUBA1A in Iso 2, Het, and Mut COs at DIV30 and representative confocal images showing the formation of neural rosettes. Boxed areas indicate the magnification in the insets, and white arrowheads indicate mitotic cells. Scale bars = 50 μm in (B) and 20 μm in (C).
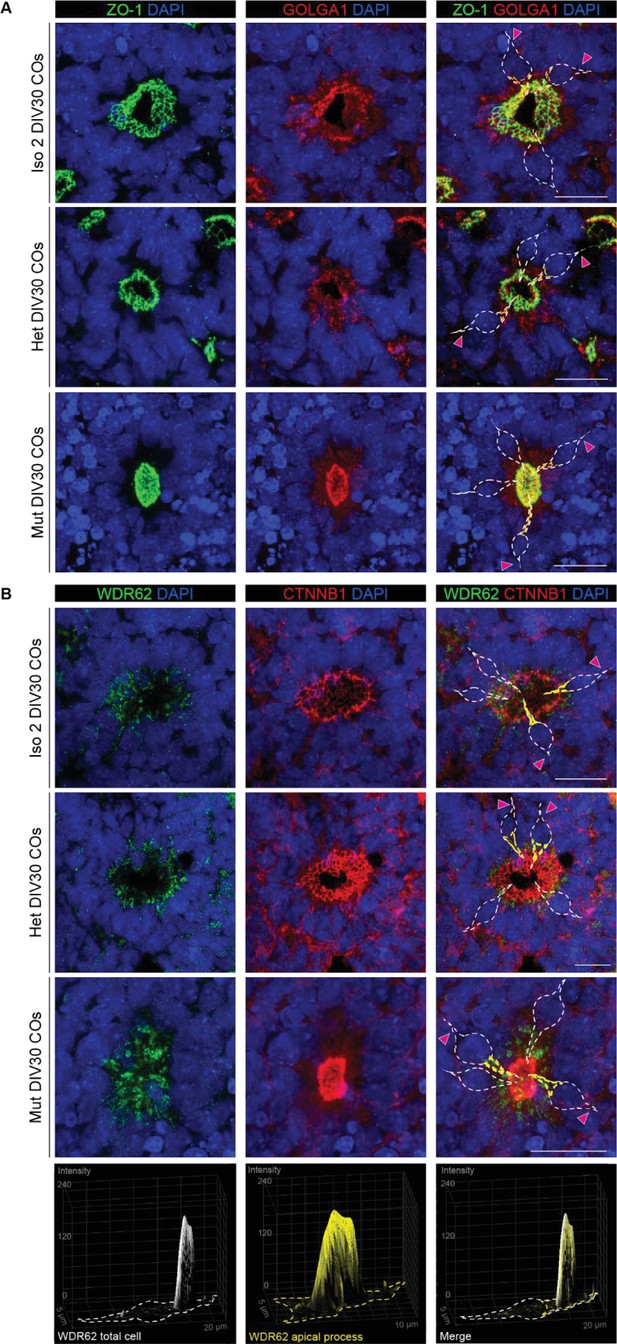
WDR62 and GOLGA1 localize to the apical domain of radial glia (RG)-like progenitor cells in cerebral organoids (COs).
(A) Immunofluorescence analysis for ZO-1 and GOLGA1 in Iso 2, Het, and Mut COs at 30 days in vitro (DIV30) identifies the position of the Golgi apparatus within the apical process of RG-like cells in neural rosettes. (B) Immunofluorescence analysis for WDR62 and CTNNB1 showing apical localization of WDR62 in rosettes of Iso 2, Het, and Mut COs at DIV30. White dotted lines outline individual cells lining the rosette lumen; yellow lines outline the apical process of RG-like cells. Pink arrowheads indicate the basal process of RG-like cells. 3D surface plots show fluorescence distribution of WDR62 in the apical process of Iso 2, Het, and Mut COs. Scale bars = 20 μm.
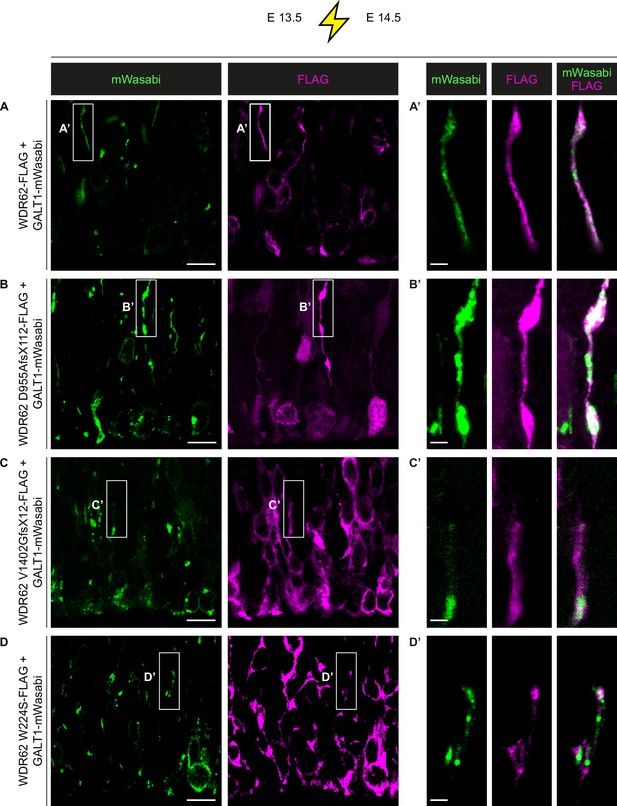
Full-length and mutant WDR62 overexpression in mouse neural progenitors supports localization to the Golgi apparatus.
(A–D). Immunofluorescence analysis of E14.5 mouse brains for FLAG and mWasabi following in utero electroporation (IUE) at E13.5 with GALT-mWasabi and WDR62-FLAG (A), WDR62 D955AfsX112-FLAG (B), WDR62 V1402GfsX12-FLAG (C), or WDR62 W224S-FLAG (D) expressing plasmids. (A’–D’) Representative confocal images and magnified views of the boxed areas (left) showing overlapping mWasabi (green) and FLAG (magenta) signals in radial processes. Scale bars = 10 μm in (A–D) and 2 μm in (A’–D’).
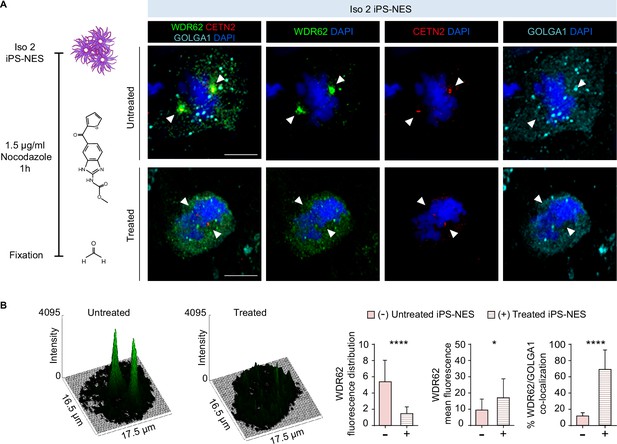
WDR62 shuttling from the Golgi apparatus to the mitotic spindle poles depends on microtubules.
(A) Schematics of the experiment and relative immunofluorescence analysis of WDR62, GOLGA1, and CETN2 in Iso 2 Neuroepithelial Stem cells derived from induced Pluripotent Stem cells (iPS-NES cells) before and after microtubule depolymerization with 1.5 μg/ml nocodazole for 1 hr. In untreated metaphase cells, the WDR62 signal is pericentrosomal. In nocodazole-treated cells, WDR62 remains associated with the Golgi apparatus (marked by GOLGA1). White arrowheads indicate the centrosome (marked by CETN2). (B) Surface plots and analysis of WDR62 fluorescence signal distribution in mitotic untreated and nocodazole-treated Iso 2 iPS-NES cells. Histograms show fluorescence signal distribution (skewness, arbitrary units) (left), mean fluorescence intensity (center), and co-localization with GOLGA1 (right) in untreated and nocodazole-treated Iso 2 iPS-NES cells during mitosis. Treated mitotic cells show different WDR62 signal distribution, increased mean fluorescence values, and higher WDR62/GOLGA1 co-localization percentage in the measured area (see also Figure 2B and C for comparison with Mut iPS-NES cells). Replicates n=3, total cells N=42, p-value <0.0001, Kolmogorov-Smirnov test, left histogram; replicates n=3, total cells N=42, p-value <0.05, Kolmogorov-Smirnov test, center histogram; replicates n=3, total cells N=32, p-value <0.0001, unpaired Student’s t-test, right histogram. Data are shown as mean ± SD. Scale bar = 10 μm.
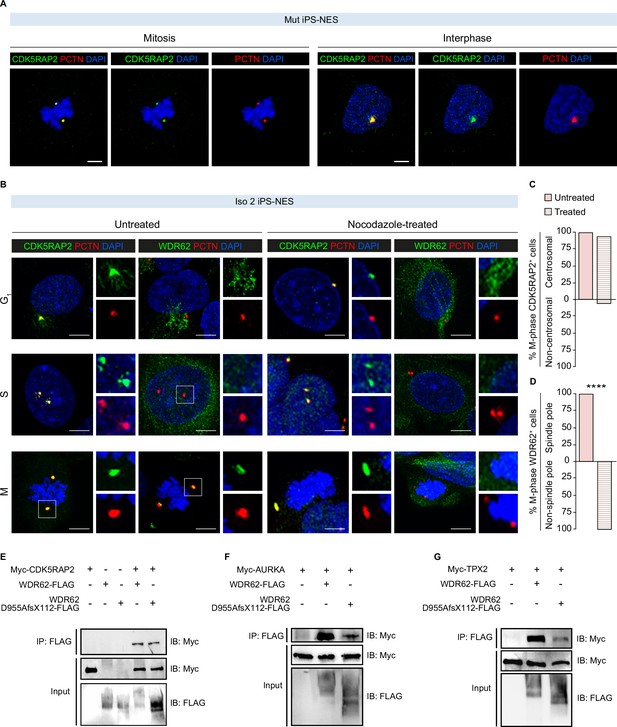
Impact of the D955AfsX112 mutation on WDR62 interactions with other proteins.
(A) Immunofluorescence assay for CDK5RAP2 and PCTN in Mut Neuroepithelial Stem cells derived from induced Pluripotent Stem cells (iPS-NES cells) during mitosis and interphase. Representative confocal images show correct localization of CDK5RAP2 to the centrosome. (B) Immunofluorescence assay for WDR62, CDK5RAP2, and PCTN in untreated and nocodazole-treated (1.5 μg/ml for 1 hr) Iso 2 iPS-NES cells during G1, S, and mitosis (M) phases. Representative confocal images of CDK5RAP2 and PCTN signal localization. (C) Quantitative analysis of CDK5RAP2 centrosomal localization during mitosis, which is unaltered in nocodazole-treated cells. (D) Quantitative analysis of WDR62 spindle pole localization during mitosis; nocodazole treatment results in a dispersed perinuclear signal. Data are shown as %, p-value >0.05 (Chi-square test) and %, p-value <0.05 (Fisher’s exact test), respectively (replicates n=3, total cells N=46). (E – G) The interactions of WDR62 with CDK5RAP2 (E), AURKA (F), and TPX2 (G) are not disrupted by the mutation. HEK293T cells were transfected with Myc-CDK5RAP2, Myc-AURKA, or Myc-TPX2 and WDR62-FLAG or WDR62 D955AfsX112-FLAG expression plasmids, as indicated. Cell lysates were immunoprecipitated with an antibody against Myc and immunoblotted with an antibody against FLAG. Interactions of WT and mutant WDR62 are detectable with all three partners. Scale bars = 5 μm in (A) and 10 μm in (B).
-
Figure 3—figure supplement 1—source data 1
Uncropped images of western blots.
- https://cdn.elifesciences.org/articles/81716/elife-81716-fig3-figsupp1-data1-v1.zip
-
Figure 3—figure supplement 1—source data 2
Uncropped images of western blots.
- https://cdn.elifesciences.org/articles/81716/elife-81716-fig3-figsupp1-data2-v1.zip
-
Figure 3—figure supplement 1—source data 3
Uncropped images of western blots.
- https://cdn.elifesciences.org/articles/81716/elife-81716-fig3-figsupp1-data3-v1.zip
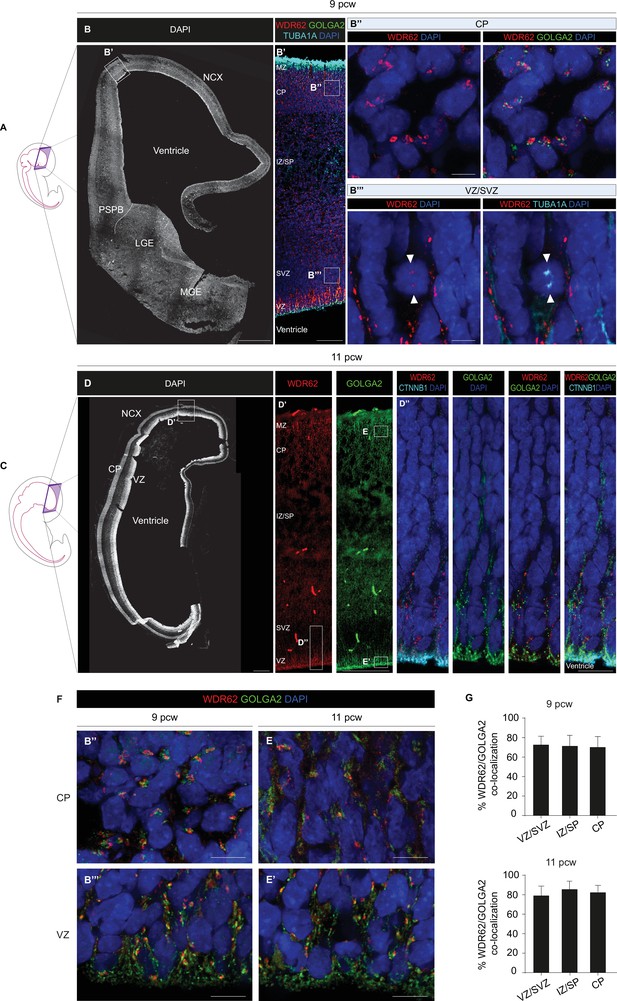
WDR62 is associated with the Golgi apparatus in human fetal telencephalic cells.
(A) Schematic illustration of a human fetal specimen at 9 post-conceptional weeks (pcw) showing the developing central nervous system (CNS). (B) DAPI staining of a coronal hemisection of the human telencephalon at 9 pcw. (B’) Magnification of the boxed area in (B) showing WDR62/GOLGA2/TUBA1A expression in the developing NCX. (B’’, B’’’) Magnified views of the boxed areas in (B’) showing WDR62/GOLGA2 expression in CP (B’’) and WDR62/TUBA1A expression in VZ/SVZ (B’’’). White arrowheads indicate WDR62 signal and TUBA1A+ spindle poles in a mitotic cell. (C) Schematic illustration of a human fetal specimen at 11 pcw showing the developing CNS. (D) DAPI staining of a coronal hemisection of the human telencephalon at 11 pcw. (D’) Magnification of the boxed area in (D) showing WDR62 and GOLGA2 expression in the developing NCX. (D’’) Magnified views of the boxed areas in (D’) showing WDR62/CTNNB1 and WDR62/GOLGA2 expression at the apical domain of VZ progenitors. (F) Representative magnified views of the boxed areas in (B’) and (D’) showing WDR62 and GOLGA2 signal distribution during interphase at the CP and VZ at 9 and 11 pcw. WDR62 and GOLGA2 signals overlap in post-mitotic neuroblasts at the CP (B’’, E) and in neural progenitor cells (NPCs) at the VZ (B’’’, E’). (G) Analysis of WDR62 and GOLGA2 co-localization at the VZ/SVZ, IZ/SP, and CP at 9 and 11 pcw (sections n=2, total Golgi objects N=7087 in 9 pcw sections; sections n=2, total Golgi objects N=5103 in 11 pcw sections). Data are shown as mean ± SD. Scale bar = 500 μm in (B, D), 100 μm in (B’, D’), 5 μm in (B’’, B’’’, E, and E’). NCX (neocortex); PSPB (pallial-subpallial boundary); LGE (lateral ganglionic eminence); MGE (medial ganglionic eminence); MZ (marginal zone); CP (cortical plate); IZ/SP (intermediate zone/subplate); SVZ (subventricular zone); VZ (ventricular zone).
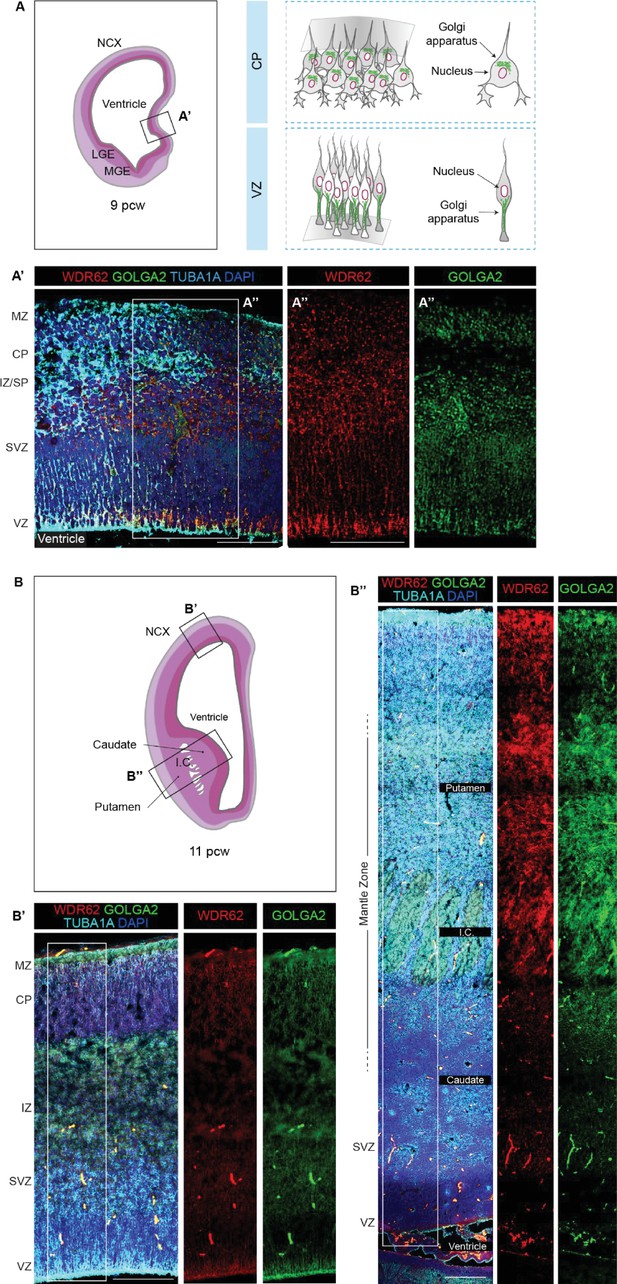
Expression pattern and subcellular localization of WDR62 in human fetal telencephalon.
(A) Schematic illustration of a coronal hemisection of human telencephalon at 9 post-conceptional weeks (pcw) (left), and schematic illustration of Golgi apparatus patterns in neural progenitor cells (NPCs) at the VZ and in post-mitotic neurons at the CP (right). (A’, A”) Immunofluorescence assay for WDR62, GOLGA2, and TUBA1A (medial pallium) (A’) and magnified view of WDR62 and GOLGA2 signal distribution (A’’) in the CP and VZ at 9 pcw. In early post-mitotic neurons at the CP, WDR62 shows a discrete perinuclear pattern and follows a diffuse-to-punctate pattern along NPC processes at the VZ, overlapping with GOLGA2. (B) Schematic illustration of a coronal hemisection of human telencephalon at 11 pcw. (B’, B”) Immunofluorescence assay for WDR62, GOLGA2, and TUBA1A in the developing neocortex (B’) and basal ganglia (B’’); boxed areas show WDR62 and GOLGA2 signal distribution in the NCX (B’) and basal ganglia (B”). Scale bars = 100 μm. NCX (neocortex); MZ (marginal zone); LGE (lateral ganglionic eminence); MGE (medial ganglionic eminence); CP (cortical plate); IZ (intermediate zone); SP (subplate); SVZ (subventricular Zone); VZ (ventricular zone); IC (internal capsule).
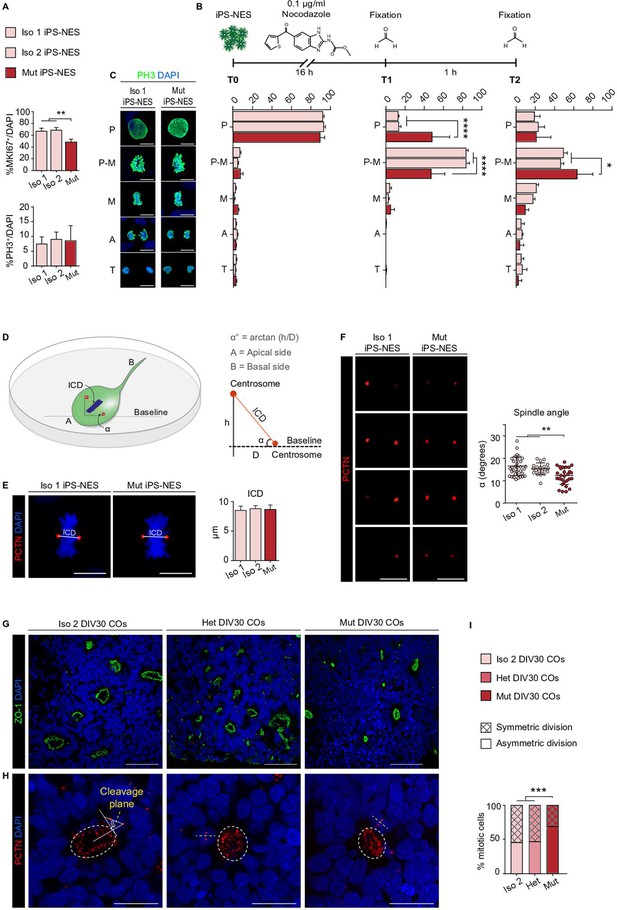
WDR62 regulates mitotic progression and asymmetric versus symmetric divisions in neural progenitors.
(A) Quantitative analysis for MKI67 and PH3 in Iso 1, Iso 2, and Mut Neuroepithelial Stem cells derived from induced Pluripotent Stem cells (iPS-NES cells) reveals a decrease in G1-M (MKI67+) but not in mitotically active (PH3+) cells (MKI67+, replicates n=4, total cell N=3315, one-way ANOVA, post hoc Tukey’s test, p-value <0.001; PH3+, replicates n=4, total cell N=12684, one-way ANOVA, post hoc Tukey’s test, p-value >0.05). (B) Scheme of synchronization experiment in iPS-NES cells. (C) Representative confocal images and quantification of mitotic figure distribution in iPS-NES cells. The distribution of mitotic figures (quantified as % of mitotic figures at each phase) is not significantly different in unsynchronized (T0) Iso 1, Iso 2, and Mut iPS-NES cell cultures (replicates n=4, total cell N=12684, p-value <0.05, two-way ANOVA, post hoc Dunnett’s test). Quantitative analysis after 16 hr of treatment (T1) shows increased mitotic cell fraction in P-M. The percentage of cells arrested in P-M is higher in Iso 1 and Iso 2 than in Mut iPS-NES cell cultures. Fewer Mut iPS-NES cells proceed to M following nocodazole treatment and subsequent release in standard culture conditions (T2) compared to Iso 1 and Iso 2 iPS-NES cells (replicates n=4, total cell N=8764, P-value <0.0001, two-way ANOVA, post hoc Dunnett’s test and replicates n=4, total cell N=9751, p-value <0.05, two-way ANOVA, post hoc Dunnett’s test). P (prophase); P-M (prometaphase); M (metaphase); A (anaphase), and T (telophase). (D) Schematic 3D representation of inter-centrosomal distance (ICD) and spindle angle α measurements in iPS-NES cell cultures. α was measured as the angle of the spindle plane relative to the surface of the culture dish (baseline). (E) Representative confocal images of PCTN and DAPI staining in Iso 1 and Mut iPS-NES cells in metaphase and example of ICD measurements. Quantitative analysis shows no differences in ICD (replicates n=6, total cell N=81, p-value >0.05, one-way ANOVA, post hoc Sidak’s test). (F) Confocal Z-stack images of PCTN in Iso 1 and Mut iPS-NES cell lines during metaphase. Quantitative analysis shows smaller α in Mut iPS-NES cell compared with Iso 1 and Iso 2 iPS-NES cells (replicates n=6, total cell N=81, p-value <0.01, one-way ANOVA, post hoc Dunnett’s test). (G) Representative confocal images of ZO-1 in COs of Iso 2, Het, and Mut at 30 days in vitro (DIV30) showing the lumen of neural rosettes. (H) Confocal images of PCTN in Iso 2, Het, and Mut COs at DIV30. (I) Analysis of the cleavage plane angle θ in neural progenitors (% mitotic cells) showing a higher percentage of asymmetric (horizontal) divisions in Mut COs compared with Iso 2 and Het COs (COs n=6, total cell N=184, data are shown as %, p-value >0.001, Chi-square test). Data are shown as mean ± SD. Scale bar = 5 μm in (C), 10 μm in (E, F), and 20 μm (G, H).
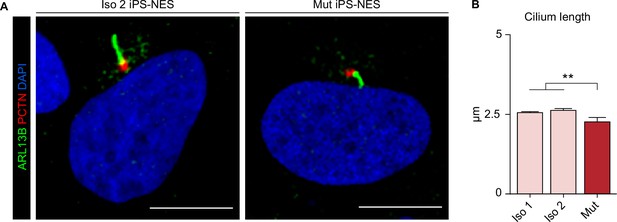
Mut iPS-NES cells harbor shorter cilia.
(A) Immunofluorescence analysis of PCTN and ARL13B in Iso 2 and Mut Neuroepithelial Stem cells derived from induced Pluripotent Stem cells (iPS-NES cells) during interphase. (B) Quantitative analysis of ciliary length (measured from the tip on the base of the cilium) in Iso 1, Iso 2, and Mut iPS-NES cells shows shorter primary cilia in the latter (replicates n=3, total cell N=850, p-value <0.01, one-way ANOVA, post hoc Dunnett’s test). Data are shown as mean ± SD. Scale bars = 10 μm.
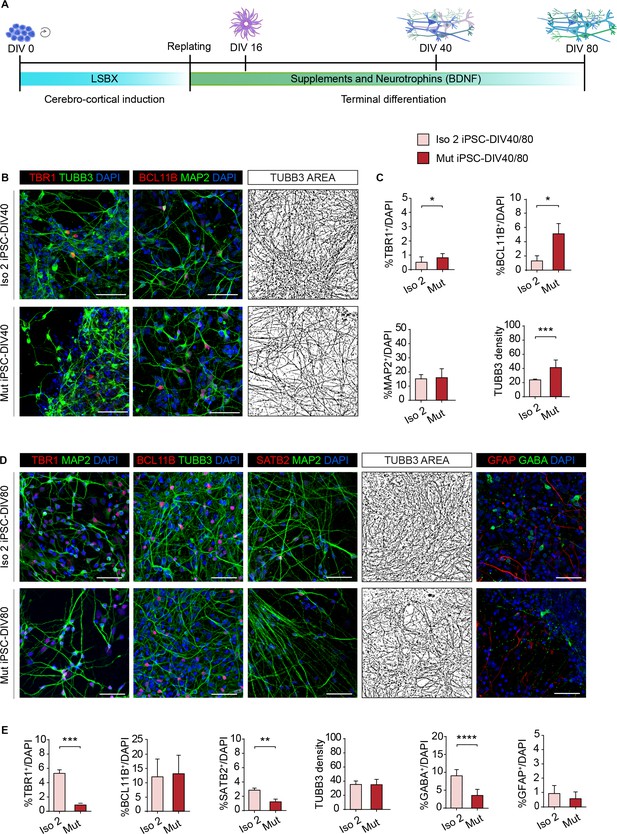
WDR62 regulates neural progenitor cell fate.
(A) Schematic illustration of the cerebro-cortical differentiation protocol (LSBX) via inhibitors of SMAD (LDN193189, SB431542) and WNT (XAV939). (B) Representative confocal images of TBR1, BCL11B, MAP2, and TUBB3 in Iso 2 and Mut progeny at 40 days in vitro (DIV40). (C) TBR1+ and BCL11B+ neurons are significantly enriched in Mut compared with Iso 2 induced Pluripotent Stem Cell (iPSC)-DIV40 cultures. Quantification shows no differences in MAP2+ cells but significant differences in TUBB3+ density (total TUBB3+ area (μm2)) normalized to total cell number (DAPI+) between Iso 2 and Mut iPSC-DIV40 cultures (replicates n=3), total cells N=38412, p-value <0.05, unpaired Student’s t-test for TBR1; replicates n=3, total cells N=40866, p-value <0.05, unpaired Student’s t-test for BCL11B; replicates n=6, total cells N=39595, p-value >0.05, unpaired Student’s t-test for MAP2; replicates n=6, total cells N=21013, p-value <0.001, unpaired Student’s t-test for TUBB3 density. (D) Representative confocal images for TBR1, BCL11B, SATB2, TUBB3, GABA, and GFAP staining in Iso 2 and Mut progeny at DIV80. (E) Quantification shows a decrease in TBR1+ and SATB2+ cells, but no differences in BCL11B+ cells and in TUBB3 density in Mut iPSC-derived neurons, and a decrease in GABA+, but not in GFAP+, cells (replicates n=3, total cells N=23396, p-value <0.05, unpaired Student’s t-test for TBR1; replicates n=3, total cells N=29448, p-value >0.05, unpaired Student’s t-test for BCL11B; replicates n=3, total cells N=21843, p-value <0.01, unpaired Student’s t-test for SATB2; replicates n=3, total cells N=7292, p-value >0.05, Kolmogorov-Smirnov test for TUBB3 density; replicates n=3, total cells N=20375, p-value <0.0001, unpaired Student’s t-test for GABA; replicates n=3, total cells N=20370, p-value >0.05, unpaired Student’s t-test for GFAP). Data are shown as mean ± SD. Scale bar = 50 μm in (B) and (D).
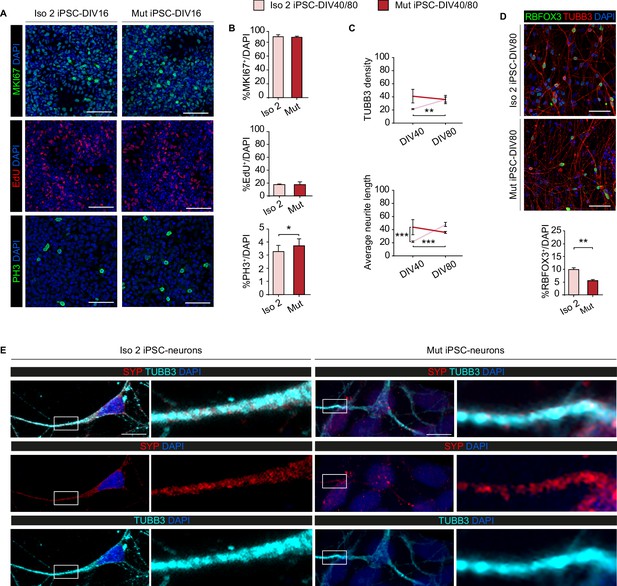
Lineage specification defects in Mut induced Pluripotent Stem Cell (iPSC)-derived cerebro-cortical neurons.
(A) Immunofluorescence analysis for MKI67, PH3, and EdU in Iso 2 and Mut iPSC-neural progeny at 16 days in vitro (DIV16). (B) Quantitative analysis shows no differences in the number of proliferating (MKI67+) or S-phase (EdU+) cells, but a significant increase in mitotic index (PH3+) in Mut iPSC neural progeny (replicates n=3, total cells N=5222, p-value >0.05, unpaired Student’s t-test for MKI67; replicates n=3, total cells N=58313, p-value <0.05, unpaired Student’s t-test for PH3; replicates n=4, total cells N=3395, p-value >0.05, unpaired Student’s t-test for EdU >50%). (C) Cross-comparison of TUBB3 density (TUBB3+ surface area normalized to total cell number [DAPI]) between DIV40 and DIV80 in Iso 2 and Mut iPSC-derived neurons shows increase during differentiation (from DIV40 to DIV80) in Iso 2 iPSC-neuroderivatives (top) (p-value <0.01, two-way ANOVA for TUBB3 density in Iso 2 iPSC-DIV40 vs. -DIV80; p-value >0.05, two-way ANOVA, post hoc Sidak’s test for TUBB3 density in Mut iPSC-DIV40 vs. -DIV80). Average neurite length cross-comparison between DIV40 and DIV80 in Iso 2 and Mut iPSC-derived neurons showing significant difference between Iso 2 and Mut neuroderivatives at DIV40 (p-value <0.001, two way-ANOVA, post hoc Tukey’s test in Iso 2 iPSC-DIV40 vs. Mut iPSC-DIV40, and Iso 2 iPSC-DIV40 vs. Iso 2 iPSC-DIV80; p-value >0.05 in Iso 2 iPSC-DIV80 vs. Mut iPSC-DIV80, and Mut iPSC-DIV40 vs. Mut iPSC-DIV80). (D) Representative immunofluorescence assay for RBFOX3 and TUBB3 in Iso 2 and Mut iPSC-DIV80 cultures. Quantitative analysis shows a decrease in RBFOX3+ cells in Mut cells (replicates n=3, total cell N=35207, p-value <0.01, unpaired Student’s t-test). (E) Representative immunofluorescence assay for SYP (pre-synaptic marker) and TUBB3 in Iso 2 and Mut iPSC-DIV80 cultures. Scale bars = 50 μm in (A), (D), and 10 μm in (E).
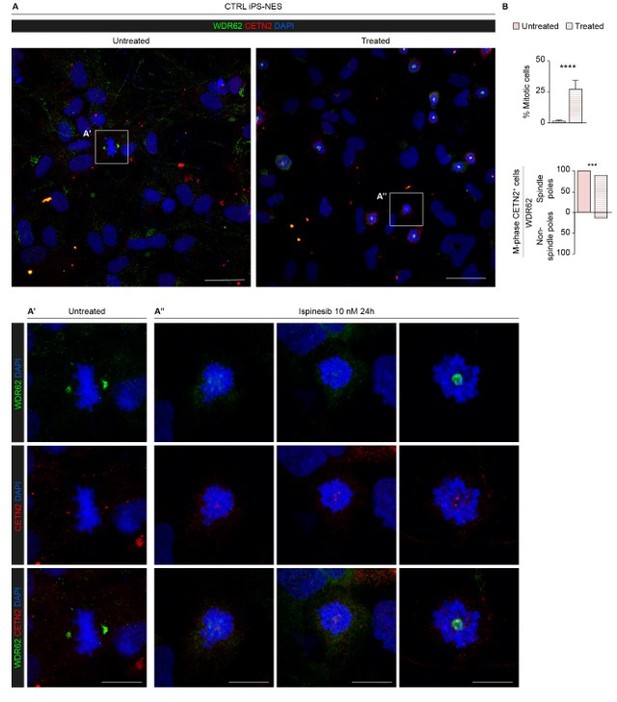
Kinesin Spindle Protein inhibition partially impairs WDR62 shuttling to the spindle poles.
(A) Representative confocal images of untreated and Ispinesib (10 nM)-treated CTRL iPS-NES cells. Treated cells were synchronized in prometaphase. (A’) Magnification of untreated mitotic iPS-NES cells showing WDR62 localized in the proximity of centrosomes (CETN2 staining). (A’’) Magnification of Ispinesib-treated mitotic iPS-NES cells showing divergent WDR62 localization/mis-localization pattern. (B) Quantitative analysis of % mitotic cells in untreated and treated conditions and of % mitotic cells with mis-localized WDR62. Mitotic cells were identified through the characteristic nuclear shape indicating chromatin compaction and CETN2 staining, highlighting duplicated mitotic centrosomes. Data are shown as mean ± SD, replicates n=3, total cells N=2085, p-value < 0.0001 (Student’s t-test, post hoc Welch’s correction), and %, replicates n=3, total cells N=120, p-value < 0.001 (Fisher’s exact test), in B top and bottom histogram, respectively. Scale bars = 20 µm in A and 5 µm in A’ and A’’.





