Antigenic mapping and functional characterization of human New World hantavirus neutralizing antibodies
Figures
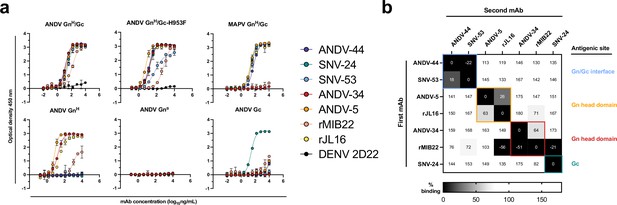
Hantavirus neutralizing antibodies target four distinct regions on the glycoprotein spike.
(a) Binding potency of mAbs to recombinant hantavirus antigens, ANDV GnH/Gc, ANDV GnH/Gc_H953F, MAPV GnH/Gc, ANDV GnH, ANDV GnB, ANDV Gc, expressed in S2 cells. Binding curves were obtained using non-linear fit analysis, with the bottom of curve constrained to 0, using Prism software. The data shown are representative curves from 3 independent experiments. Mean ± SD of technical duplicates from one experiment are shown. (b) Competition binding analysis of neutralizing antibodies to ANDV GnH/Gc recombinant protein measured using BLI. % Competition is designated by the heatmap, where black boxes indicate complete competition, gray boxes indicate intermediate competition, and white boxes indicate no competition. The data are shown are representative from two independent experiments.
Escape mutant generation and mutagenesis mapping indicate critical binding residues for hantavirus mAbs.
(a) Results from viral escape selection for indicated antibodies. Real-time cellular analysis escape mutant mapping shows the number of replicates with escape over the total number of replicates for each selection mAb against the indicated selection virus. Mutations from serial passaging were identified for each mAb, and escape was confirmed in the presence of saturating mAb concentrations. VSV/ANDV or VSV/SNV were used for escape selections. (b) Side and top view of escape mutants mapped to the ANDV Gn/Gc spike (PDB: 6ZJM). The colored spheres designate escape mutants for the indicated antibody. Gn is shown in white, and Gc is shown in grey. (c) Heatmap of mAb binding in the presence of ANDV mutant constructs. Dark blue boxes indicate loss of binding. The black boxes designate escape mutants for the indicated antibody. The percent binding (% WT) of each mAb to the mutant constructs was compared to the WT SNV or ANDV control. The data are shown as average values from three to four independent experiments. All numbering for ANDV sequences was based on GenBank AF291703.2 and SNV sequences were based on GenBank KF537002.1. (d) Heatmap of SNV mutant constructs as described in c. All numbering for SNV sequences were based on GenBank KF537002.1.
-
Figure 2—source data 1
Percent mAb binding in the presence of ANDV mutants.
- https://cdn.elifesciences.org/articles/81743/elife-81743-fig2-data1-v2.docx
-
Figure 2—source data 2
Percent mAb binding the presence of SNV mutants.
- https://cdn.elifesciences.org/articles/81743/elife-81743-fig2-data2-v2.docx
Mutagenesis expression levels and gating strategy.
(a) Neutralization of escape mutant viruses to antibodies at a saturating concentration (10 µg/mL). The data shown are averages ± SD from three experiments, n=9. (b) Expression levels of Gn/Gc point mutants were measured based on binding of a positive control oligoclonal mix of antibodies targeting multiple epitopes on the Gn/Gc antigens. The value for % PE + cells was determined by gating on untransfected Expi293F cells (mock). The data are shown as average values from two to three independent experiments. (c) Gating strategy for antibody binding to Expi293F cells expressing ANDV M-segment. Cells were first gated by forward and side scatter and dead cells were excluded using a viability dye (DAPI). Binding of the positive control sample (oligoclonal mix of hantavirus-reactive antibodies), shown in green, was detected with a PE-conjugated goat anti-human IgG secondary. The gate for the glycoprotein-specific subset (PE+) was placed based on staining of untransfected Expi293F cells, shown in red. (d) Gating strategy for antibody binding to Expi293F cells expressing SNV M-segment, as described above in (c).
Amino acid alignment of hantavirus species.
Multiple sequence alignment of the M-segment from six representative Orthohantaviruses. Domains are colored as followed, Gn domain A: purple, domain B: dark purple, β-ribbon domain: light purple, insertions in white, capping loop: pink; Gn domain C in grey. Gc: domain I: red, domain II: yellow, domain III: dark blue, fusion loop: orange. Strictly conserved residues are highlighted in a red background, and similar residues are colored red. Escape mutations are indicated by colored spheres; SNV-53: light blue, SNV-24: green, ANDV-44: indigo, ANDV-5: orange, rJL16: yellow, ANDV-34: red, rMIB22: salmon. Contact residues for ANDV-5 and ANDV-34 are indicated by orange or red asterisks, respectively. Sequences were aligned from Andes virus (ANDV, NC_003467.2), Sin Nombre virus (SNV, L37903.1), Puumala virus, (PUUV, KJ994777.1), Hantaan virus (HTNV, JQ083394.1), Dobrava-Belgrade virus (DOBV, JF920149.1), and Seoul virus (SEOV, NC_005237.1). Figure was generated using ESprit 3.0 (Robert and Gouet, 2014).
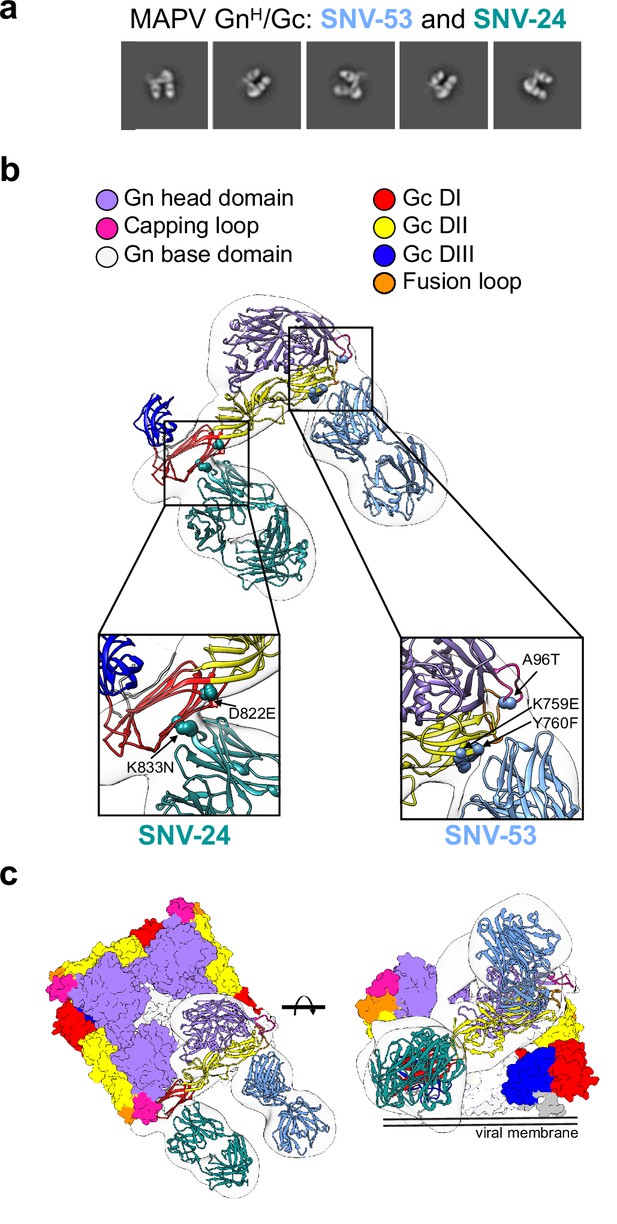
BnAbs SNV-53 and SNV-24 target two sites on the GnH/Gc heterodimer.
(a) Representative nsEM 2D-class averages of SNV-53 and SNV-24 Fabs in complex with MAPV GnH/Gc heterodimer. (b) Surface representations (light grey) of SNV-53 (blue) and SNV-24 (green) in complex with MAPV GnH/Gc. Escape mutations are indicated by the colored spheres. GnH is colored in purple, GnB is colored in light grey, and the capping loop is colored in pink. Domain I, II, and III of Gc are colored in red, yellow, and blue, respectively, and the fusion loop is colored in orange. (c) Model of bnAbs in complex with the (Gn-Gc)4 spike as colored in b, top and side view are shown.
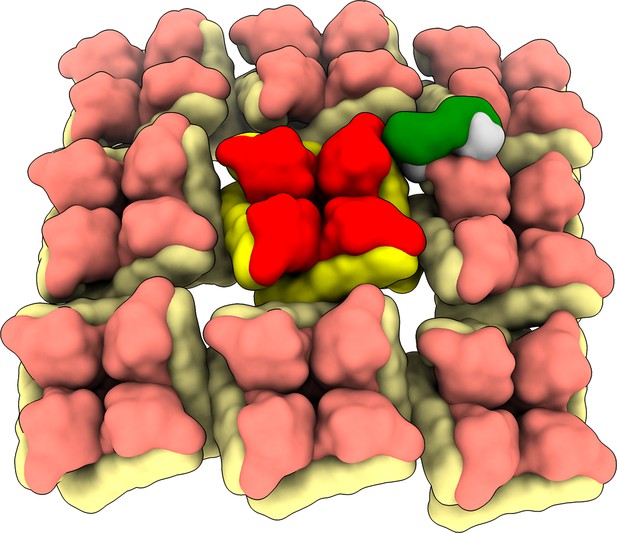
SNV-53 Fab docked to the hantavirus surface glycoprotein lattice (EMD-11236).
Model is colored as follows, Gn: red, Gc: yellow, SNV-53 Fab: green/blue.
Hydrogen-deuterium exchange mass spectrometry analysis of hantavirus antibodies in complex with ANDV GnH/Gc.
Relative fractional deuterium uptake difference at the 5000 s time point was mapped onto the cryo-EM reconstruction of the glycoprotein complex (PDB: 6ZJM). The relative fractional uptake difference (%) is derived from the relative fractional uptake of the complex (Fab +ANDV GnH/Gc) subtracted by the antigen only (ANDV GnH/Gc); percentages are shaded from blue to red. The color scale corresponds to the highest observed difference and is adjusted to –10% to 10% for ANDV-44, SNV-24, ANDV-34, and SNV-53 (different from color scale in the heat map: –50% to 50%) and –50% to 50% for ANDV-5 for visualization purpose.
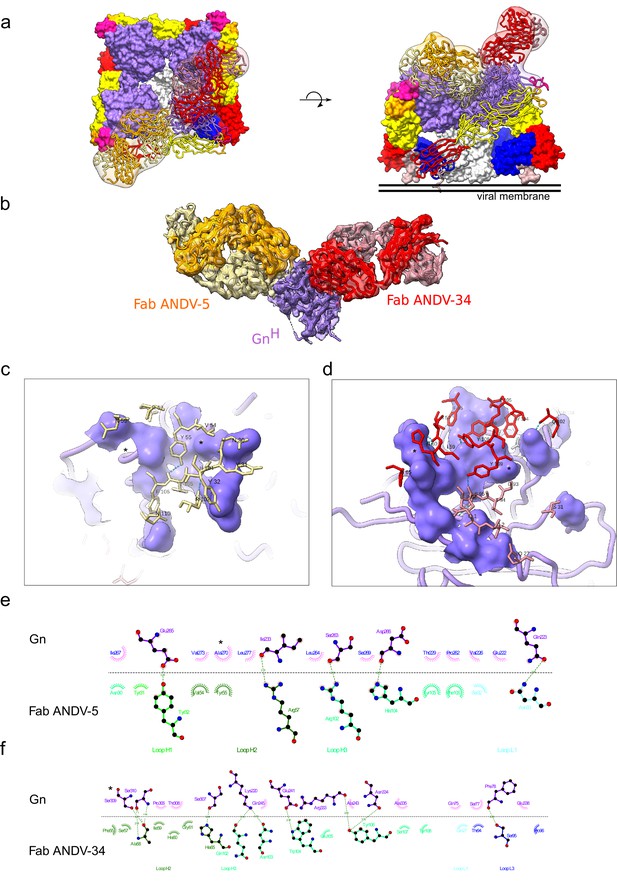
Cryo-EM structure of neutralizing antibodies ANDV-5 and ANDV-34 in complex with ANDV GnH.
(a) Top view (right) and side view (left) of the heterotetramer Gn/Gc (Serris et al., 2020) (PDB: 6ZJM) with low resolution map and model of Gn(H) ANDV-5 and ANDV-34 (purple, orange and red, respectively) complex superimpose with GnH. (b) Cryo-EM map and model of the Gn-Fabs complex. Right, transparent EM map color by chain with Gn purple, ANDV-5 heavy chain in yellow, ANDV-5 light chain in orange, ANDV-34 heavy chain in red and ANDV-34 light chain in pink. (c) Zoom-in on the paratope/epitope interface of the ANDV-5:GnH. Fab ANDV-5 residues that is in close contact with Gn. Heavy chain residues (yellow stick), label with single letter and residue number. Gn is shown in purple with the contact residues shown in a surface representation. Asterisks correspond to residues found in the escape mutant viruses. Blue dashed line, H-bond. (d) Zoom-in on the paratope/epitope interface of the ANDV-34:GnH. Fab ANDV-34 residues that is in close contact with Gn. Heavy residues (red stick), light chain residues (pink stick), label with single letter and residue number. Gn is shown in purple with the contact residues shown in a surface representation. Asterisks correspond to residues found in the escape mutant viruses. Blue dashed line, H-bond. (e) Fab ANDV-5 paratope and epitope residues involved in hydrogen bonding (dashed lines) and hydrophobic interactions. Hydrophobic interactions residues are shown as curved lines with rays. Atoms shown as circles, with oxygen red, carbon black, and nitrogen blue. Interacting residues that belong to CDR loops are colored in different shade. Image was made with Ligplot+49 (Laskowski and Swindells, 2011). (f) Fab ANDV-34 paratope and epitope residues involved in hydrogen bonding (dashed lines) and hydrophobic interactions. Hydrophobic interactions residues are shown as curved lines with rays. Atoms shown as circles, with oxygen red, carbon black, and nitrogen blue. Interacting residues that belong to CDR loops are colored in different shade. Image was made with Ligplot+49.
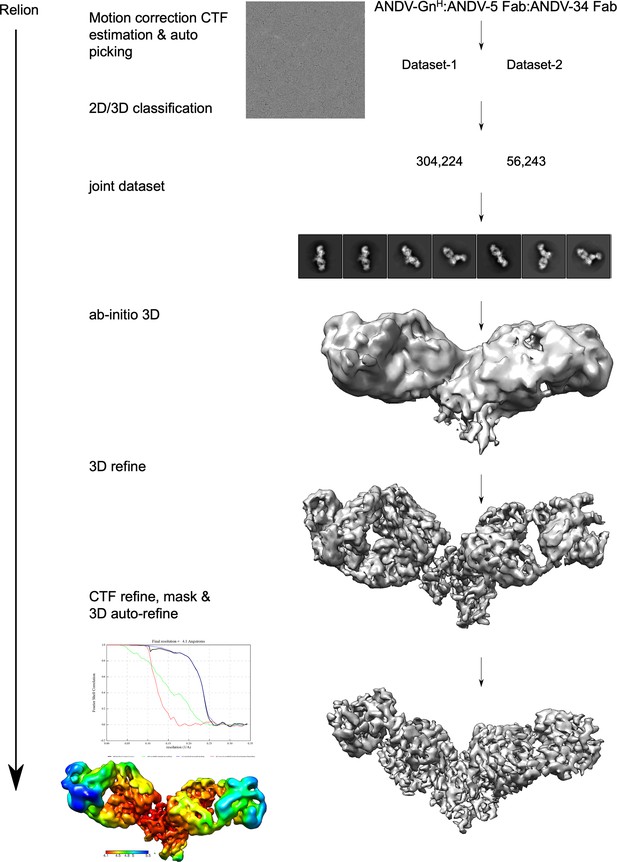
Workflow of cryo-electron microscopy processing for model of ANDV-5 and ANDV-34 Fabs in complex with ANDV GnH.
Potently neutralizing hantavirus mAbs inhibit viral entry through viral attachment blocking and/or fusion inhibition.
(a) Neutralization curves of IgG1 and Fab forms of broad mAbs (SNV-53, ANDV-44 or SNV-24) to VSV/SNV and VSV/ANDV determined through real-time cellular analysis. The data shown are representative curves from three independent experiments. Mean ± SD of technical duplicates from one experiment are shown.(b) Neutralization curves of IgG1 and Fab forms of ANDV-nAbs (ANDV-5, ANDV-34, rJL16, or rMIB22) to VSV/SNV and VSV/ANDV determined through real-time cellular analysis. The data shown are representative curves from three independent experiments. Mean ± SD of technical duplicates from one experiment are shown. (c) sEC1 or sEC1-EC2 blocking activity of neutralizing antibodies determined through a flow cytometric assay, in which mAbs were added at saturating concentration before the addition of the each soluble PCDH-1 domain labeled with Alexa Fluor 647 dye. The data shown are averages ± SD from three experiments, n=9. One-way ANOVA with Dunnett’s test, ****p<0.0001; ns, non-significant. (d) FFWO assay testing VSV/ANDV or VSV/SNV post-attachment antibody neutralization in a permissive (pH 5.5) conditions at 30 µg/mL. The data shown are averages ± SD from three experiments, n=9. One-way ANOVA with Dunnett’s test, ****p<0.0001. (e) Representative curves of dose-dependent VSV/ANDV fusion inhibition. The data shown are average values for technical replicates ± SDs. The experiments were performed three times independently with similar results.
Reactivity and potency of germline revertant forms of NWH antibodies.
(a) Representative binding curves for all germline reverted forms of SNV-53 and ANDV-44 bnAbs to Expi293F cells transfected with ANDV, SNV, PUUV, DOBV, HTNV, or SEOV Gn/Gc. The value for % PE+ cells was determined by gating on cells stained only with secondary antibodies. Data shown are average values for technical replicates ± S.D. The experiment was performed 3 times independently with similar results; one experiment is shown. (b) Representative neutralization curves for all bnAbs to VSVs bearing ANDV or SNV glycoproteins Gn/Gc. % Neutralization was measured using real-time cellular analysis and calculated by comparing CPE in the treatment wells with a cells only control well. Data shown are average values for technical replicates ± S.D. The experiment was performed 3 times independently with similar results. (c) Representative binding curves for all ANDV-nAbs, including germline reverted forms, to Expi293F cells transfected with ANDV, as described above in a. (d) Representative neutralization curves for all ANDV-nAbs to VSVs bearing ANDV, as described above in a. (e) Alignment of MIB22, ANDV-34, and ANDV-5 CDHR2 sequences to the human germline IGHV1-69 gene. Somatically mutated residues are indicated in red. Alignment was generated using IMGT/DomainGapAlign.
SNV-53 protects hamsters when given before or after HTNV inoculation.
(a) 8-week-old Syrian hamsters (n=8 per treatment group) were administered 5 mg/kg of the indicated Ab treatment and then inoculated 1 day later with 200 PFU of HTNV i.m. All animals were sacrificed at 28 dpi and organs were harvested. (b) Ab detection in the serum at day 0 by pseudovirion neutralization assay (PsVNA) and at day 28 by nucleoprotein ELISA. Dotted line indicated limit of detection. (c) qRT-PCR detection of HTNV genome in the lungs and kidneys. Kruskal-Wallis test with multiple comparisons of each group to vehicle, * p<0.01, ** p<0.001, *** p<0.001, **** p<0.0001. ns, not significant. Dashed line indicates the limit of detection, which was 50 copies of HTNV S-RNA per 100 ng/100 ng RNA.(d) In-situ hybridization detection of HTNV genome in the lungs and kidneys. One-way ANOVA with multiple comparisons of each group to vehicle, * p<0.01, ** p<0.001, *** p<0.001, **** p<0.0001. ns, not significant. (e) Eight-week-old Syrian hamsters (n=6 per treatment group) were inoculated with 10 PFU of HTNV i.m., and 5–10 mg/kg of indicated Ab was administered via i.p. route at 3 dpi. All animals were sacrificed at 28 dpi and organs were harvested. (f) Serum was analyzed as indicated in b. (g) Lung and kidneys were assayed as described in c.(h) Lung and kidneys were assayed as described in d.
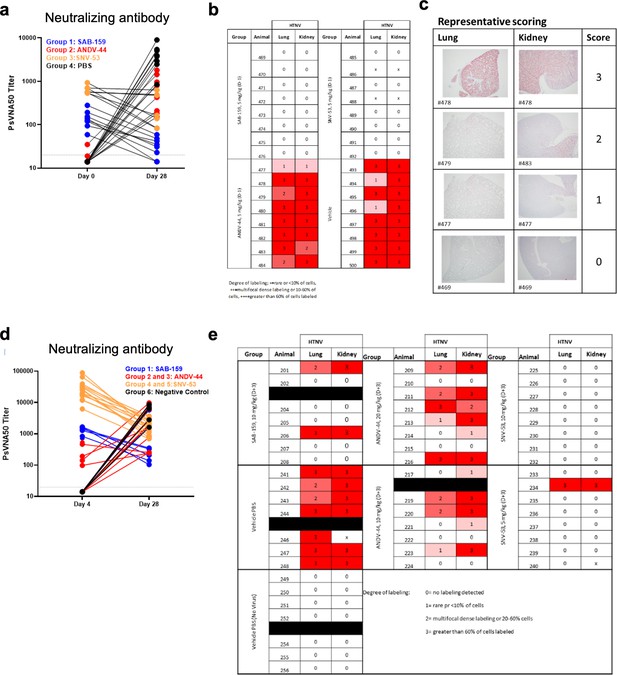
Pre- and post-exposure HTNV serum PsVNA50 data and pathology staining.
(a) Neutralizing antibody levels in serum for the four treatment groups at days 0 or 28 measured by PsVNA50 from the pre-exposure HTNV study. (b) In situ hybridization (ISH) staining scores for individual animals from the pre-exposure HTNV study. (c) Representative ISH staining in lung and kidney sections. Slides were scored by intensity ranging from 0 to 3, with median values presented. (d) Neutralizing antibody levels in serum for the four treatment groups at days 4 or 28 measured by PsVNA50 from the post-exposure HTNV study. (e) in situ hybridization (ISH) staining scores for individual animals from the post-exposure HTNV study.
ANDV-34 and ANDV-5 protect Syrian golden hamsters for lethal ANDV challenge.
(a) Eight-week-old Syrian hamsters (n=6 per treatment group) were inoculated with 200 PFU of ANDV i.m., and 10 mg/kg of indicated mAb was administered via i.p. route at 2 and 5 dpi. Animals were treated with a dengue-specific mAb (DENV 2D22) to serve as an isotype control. (b) Kaplan-Meier survival plot. Statistical analysis of survival curves was done using a log-rank (Mantel-Cox) test comparing each group to the control (DENV 2D22), **, p=0.0014.
Assay values for individual animals.
Individual animal PsVNA50 (log 10) titers were sorted highest to lowest and plotted. Day 28 ELISA titers (log 10), PCR levels (Log 10), and ISH scores (0-3) were also plotted. (A) Data from pre-exposure experiment where antibody was injected 1 day before exposure to HTNV. Sera were collected just prior to challenge with virus. (B) Data from post-exposure experiment where antibody was injected 3 days after exposure to HTNV and sera was collected one day later (Day 4). ISH = in situ hybridization, PCR = quantitative polymerase chain reaction.
Relationship between level of neutralizing antibodies in serum and protection for pre-exposure experiment.
There is a negative correlation for neutralizing antibody levels and infection for all endpoints measured. When antibody is administered 1 day pre-exposure, the level of neutralizing antibody (PsVNA50 log 10) negatively correlates with protection as measured by Day 28 anti-N ELISA, lung PCR, lung ISH, kidney PCR, or kidney ISH. Pearson r values range -0.7315 to -0.8993. An ELISA value <100 (2 log 10), qPCR less than 50 (1.69897 log 10), or an ISH score <1 were considered negative (dotted line). A PsVNA50 titer below the assay limit was given a value of 1.14 (grey shaded area). All hamsters that were negative by PsVNA were infected with HTNV as measured by positive ELISA, Lung and Kidney PCR, and Kidney ISH. There was one hamster that was negative for neutralizing antibodies that was negative by Lung ISH. There were only 3-4 of 23 hamsters positive by PsVNA with positive Lung and or Kidney PCR. Those animals were negative by ISH.
Relationship between level of neutralizing antibodies in serum and protection for post-exposure experiment.
There is a negative correlation for neutralizing antibody levels and infection for all endpoints measured. When antibody is administered 3 days post-exposure, the level of neutralizing antibody (PsVNA50 log 10) negatively correlates with protection as measured by Day 28 anti-N ELISA, lung PCR, lung ISH, kidney PCR, or kidney ISH. Pearson r values range -0.8533 to -0.9384. An ELISA value < 100 (2 log 10), qPCR less than 50 (1.69897 log 10), or an ISH score <1 were considered negative (dotted line). A PsVNA50 titer below the assay limit was given a value of 1.14 (grey shaded area). All hamsters that were negative by PsVNA were infected with HTNV as measured by positive ELISA, Lung and Kidney PCR, and Lung and Kidney ISH. There were only 2 of 30 hamsters positive by PsVNA with positive PCR in lung and/or kidney.
Additional files
-
Supplementary file 1
Summary table of electron microscopy statistics.
- https://cdn.elifesciences.org/articles/81743/elife-81743-supp1-v2.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/81743/elife-81743-mdarchecklist1-v2.docx