Loss of aquaporin-4 results in glymphatic system dysfunction via brain-wide interstitial fluid stagnation
Figures
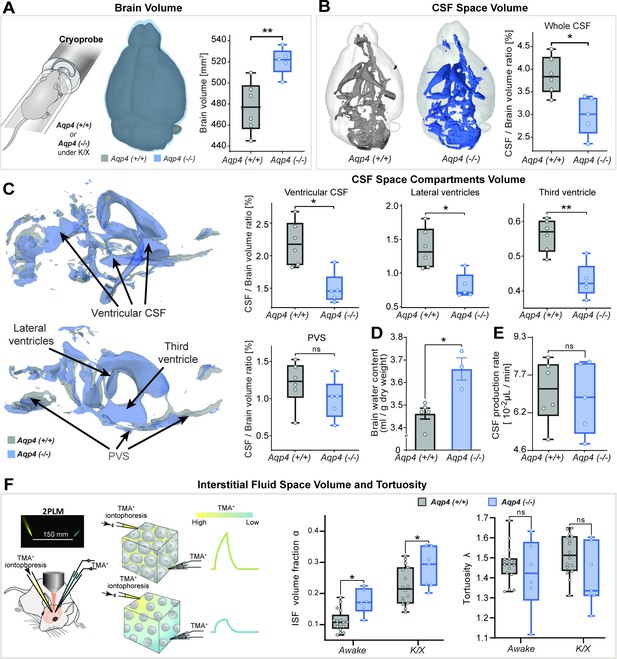
3D-CISS MRI, CSF and interstitial space volumetry in vivo.
Overlaid 3D surface images of the co-registered and averaged 3D-CISS brain volumes, and whiskers-box plots comparing (A) the brain volumes and (B) segmented CSF space volumes from 6 WT and 5 AQP4 KO mice (see Figure 1—source data 1). (C) Overlaid 3D surface reconstruction of the co-registered whole CSF spaces segmented from 3D-CISS from all WT (gray) and KO (blue) animals, along with whiskers-box plot comparisons of main CSF compartments volumes segmented: whole ventricular (left top), lateral (middle top) and third ventricular (right top), and whole perivascular space at the skull base (PVS; left bottom). (D) Whiskers-box plots comparing brain water content from 5 WT and 3 KO ex vivo (Figure 1—source data 2). (E) Whiskers-box plots comparing CSF production rates measured in 6 WT and 5 KO in vivo (Figure 1—source data 3). (F) Whiskers-box plots for the extracellular space volume and tortuosity measured using real-time iontophoresis with tetramethylammonium (TMA) in awake (17 WT, 6 KO) and K/X anesthetized (16 WT, 7 KO) animals (Figure 1—source data 4). Legend: ns-not significant, *-p<0.05, **-p<0.01; Mann-Whitney U-test (A–E), one-way ANOVA with Bonferroni’s post-hoc correction (F).
-
Figure 1—source data 1
3D-CISS CSF space volumetry in vivo - source data.
- https://cdn.elifesciences.org/articles/82232/elife-82232-fig1-data1-v3.xlsx
-
Figure 1—source data 2
Brain water content ex vivo - source data.
- https://cdn.elifesciences.org/articles/82232/elife-82232-fig1-data2-v3.xlsx
-
Figure 1—source data 3
CSF production in vivo - source data.
- https://cdn.elifesciences.org/articles/82232/elife-82232-fig1-data3-v3.xlsx
-
Figure 1—source data 4
ISF space volume estimation with TMA - source data.
- https://cdn.elifesciences.org/articles/82232/elife-82232-fig1-data4-v3.xlsx
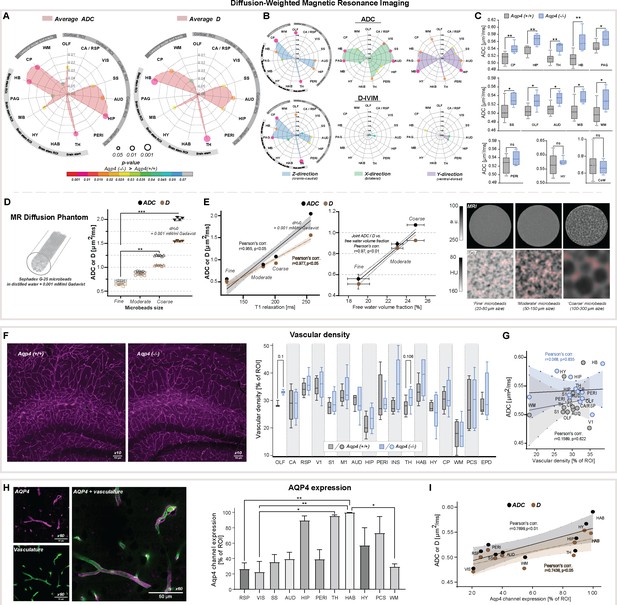
Diffusion-weighted MRI in vivo and ex vivo, vascularity, and AQP4 cellular surface expression ex vivo.
Radar plots showing statistical significances for the differences between the slow diffusion measures among 15 parenchymal ROI assessed, for average ADC (A) and D, and in (B) 3 diffusion-encoding direction separately; (C) Whiskers-box plots for the mean of the average ADC values among 6 AQP4 KO and 6 WT animals analyzed, including: the regions showing the most significant differences (top); exemplary regions showing significant differences (middle); exemplary regions showing no differences (bottom), by means of the Mann-Whitney U-test (Figure 2—source data 1); (D) mean and 95% confidence intervals of ADC and D calculated in a water phantom (+0.001 mM/ml gadobutrol) and 3 water phantoms filled with Sephadex-G25 microbeads of fine, moderate and coarse sizes; (E) Correlation plots of the calculated mean ± SD of ADC and D to the T1 relaxation values and free fluid volumes obtained from the phantoms using MRI (left), and micro-computed tomography (µCT; middle) (Figure 2—source data 2). Single-slices of turbo-spin echo (RARE) MR images from respective phantoms (right, upper) along with µCT images of the central portion of the respective phantoms (right, lower) filled with 1:1 solution of Ominpaque 350 contrast agent and 0.9% NaCl. Semi-transparent red area marks the free fluid space considering only voxels above 75th percentile of Hounsfield units (HU) intensity distribution in each µCT image; (F) Exemplary immunohistochemistry images (Olympus UplanXApo 10 x/numerical aperture 0.40, ∞/compatible cover glass thickness 0.17 mm/field number 26.5 mm, no immersion liquid) (left) from the hippocampal area from WT and KO animal (magenta vascular labeling) along with whiskers-box plot for the comparison of the mean vascular density among 17 regions analyzed (right) (Figure 2—source data 3); (G) Region-wise correlation plot of calculated ADC to vascular density among 12 regions analyzed with both methods, in WT and KO animals; (H) Exemplary image for vasculature (green, AlexaFluor 488) and AQP4 (magenta) immunohistochemistry staining (Olympus UplanXApo 60 x/numerical aperture 1.42, ∞/compatible cover glass thickness 0.17 mm/field number 26.5 mm, oil immersion) (left), bar-plot comparison of the mean AQP4 channel expression among 11 ROI assessed from 4 WT mice (right) (Figure 2—source data 4); (I) ROI-wise correlation plot comparing average ADC and D with the mean AQP4 channel expression among 10 regions assessed with both methods. Legend: OLF-olfactory, CA / RSP-cingulate / retrosplenial, VIS (V1)-visual, SS (S1)-somatosensory, M1-motorcortex, AUD-auditory, HIP-hippocampus, PERI-perirhinal, INS-insular, TH-thalamus, HAB-habenula, HY-hypothalamus, MB-midbrain, PAG-periaqueductal gray, HB-hindbrain; CP-caudate putamen, WM-white matter; PCS-pericisternal space, CoW-Circle of Willis, EPD-ependymal layer around lateral ventricles; SD-standard deviation; ns-not significant, *-p<0.05, **-p<0.01, by means of Mann-Whitney U-test (A–C, F), Kruskal-Wallis one-way ANOVA with Dunn’s correction (D, H). All correlation plots show respective regression lines along with semi-transparent areas marking 95% confidence intervals of the fitting. The highest obtained Pearson’s linear or Spearman’s range correlation scores are reported and considered significant if correlation value >0.5 with p<0.05, and non-zero regression slope.
-
Figure 2—source data 1
DWI and IVIM-DWI estimates in vivo - source data.
- https://cdn.elifesciences.org/articles/82232/elife-82232-fig2-data1-v3.xlsx
-
Figure 2—source data 2
DWI, IVIM-DWI, and T1 estimates in the phantom - source data.
- https://cdn.elifesciences.org/articles/82232/elife-82232-fig2-data2-v3.xlsx
-
Figure 2—source data 3
Immunohistochemistry: vascular labeling - source data.
- https://cdn.elifesciences.org/articles/82232/elife-82232-fig2-data3-v3.xlsx
-
Figure 2—source data 4
Immunohistochemistry: AQP4 channel labeling - source data.
- https://cdn.elifesciences.org/articles/82232/elife-82232-fig2-data4-v3.xlsx
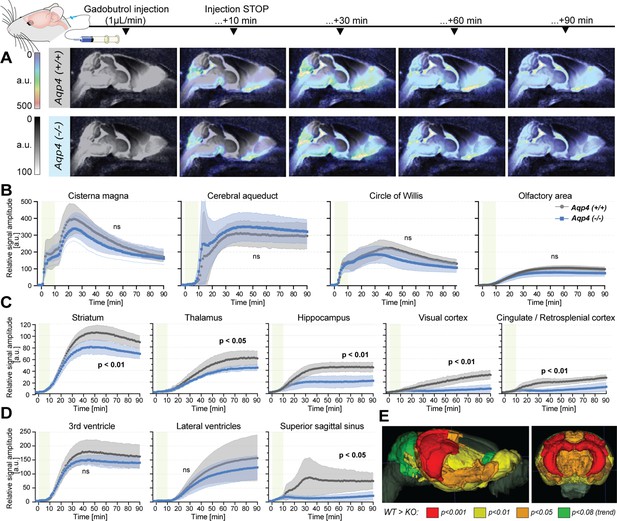
Dynamic contrast-enhanced MRI in vivo (Figure 3—source data 1).
(A) 3D multiplanar reconstructions of dynamic-contract enhanced (DCE) MRI – sagittal slices from mean images of 6 WT (top) and 5 AQP4 KO (bottom) using 3D-FISP, after applying gadobutrol injection via cisterna magna (CM); mean ± SD DCE time-curves from WT (gray) and AQP4 KO (blue): (B) main CSF compartments ventrally and caudo-cranially from CM, (C) parenchymal regions where significant differences between WT and KO were found, (D) ventricular and CSF efflux regions. (E) 3D multiplanar reconstruction of DCE-MRI from mean 3D FISP images of 6 WT animals, along with Allen Brain Atlas-based segmentation maps color-coded according to p-significance value from nonparametric Two-way ANOVA with post-hoc, showing WT vs. KO differences in the CSF tracer dynamics at 60th minute after gadobutrol injection start. Legend: ns-not significant, *-p<0.05, **-p<0.01, by means of nonparametric Two-way Anova with Bonferroni’s post-hoc.
-
Figure 3—source data 1
DCE-MRI in vivo - source data.
- https://cdn.elifesciences.org/articles/82232/elife-82232-fig3-data1-v3.xlsx
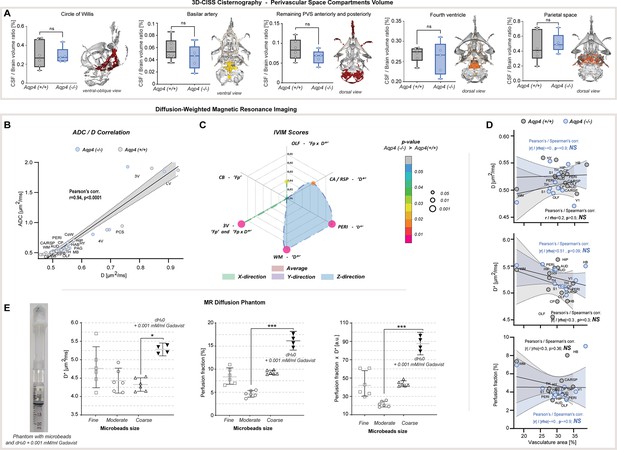
Supplementary results for MR CSF space volumetry and MR diffusion evaluation.
(A) Whiskers-box plot comparison of segmented CSF regions, where no statistical difference between KO and WT was found, along with overlaid 3D surface images of separate compartments (colored) from exemplary WT animal: CSF space at the skull base - Circle of Willis; perivascular space (PVS) around the basilar artery and its branches; PVS anteriorly and posteriorly, remaining after separating the ventricular and the space at the skull base; CSF space in the fourth ventricle; parietal PVS and cisterns (Figure 1—source data 1). (B) Correlation plot for ADC and D slow diffusion obtained from 6 AQP4 KO and 6 WT animals analyzed. (C) Radar plot showing statistical significances for the differences between IVIM scores, found among 5 parenchymal and 1 CSF space ROI assessed, for average and in 3 diffusion-encoding directions jointly (Figure 2—source data 1). (D) Correlation plots for ROI-wise comparison between IVIM measures and vascular densities from 12 parenchymal ROI analyzed with both methods in KO and WT animals, along with calculated linear and range correlation scores. (E) Fast diffusion IVIM measures calculated in a distilled water phantom (+0.001 mM/ml gadobutrol) and water phantoms filled with Sephadex-G25 microbeads of fine, moderate, and coarse sizes (Figure 2—source data 2). Legend: OLF-olfactory, CA / RSP-cingulate / retrosplenial, VIS (V1)-visual, SS (S1)-somatosensory, AUD-auditory, HIP-hippocampus, PERI-perirhinal, TH-thalamus, HAB-habenula, HY-hypothalamus, MB-midbrain, PAG-periaqueductal gray, HB-hindbrain; CP-caudate putamen, WM-white matter; 3V-third ventricle, LV-lateral ventricle, 4V-fourth ventricle, PCS-pericisternal space, CB-cerebellum, D*-pseudodiffusion (fast diffusion), Fp-perfusion fraction; ns-not significant, *-p<0.05, **-p<0.01, by means of Mann-Whitney U-test (A, C), Kruskal-Wallis one-way ANOVA with Dunn’s correction (E). All correlation plots show respective regression lines along with semi-transparent areas marking 95% confidence intervals of the fitting. The highest obtained Pearson’s linear or Spearman’s range correlation scores reported and considered significant if correlation value >0.5 with P<0.05, and non-zero regression slope.
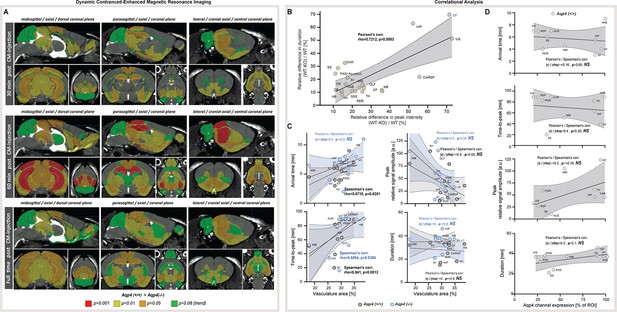
Supplementary results for dynamic contrast-enhanced MRI in vivo and correlational analysis.
(A) 3D multiplanar reconstructions of dynamic-contract enhanced (DCE) MRI – midsagittal, parasagittal, and lateral along with orthogonal axial and coronal slices from mean 3D FISP images from 5 AQP4 KO and 6 WT along with segmentation maps color-coded according to p-significance value from nonparametric Two-way ANOVA with post-hoc, for 3 time points: 30 minutes (top), 60 minutes (middle) and 90 minutes (full time, bottom) after applying gadobutrol injection via cisterna magna (CM) (Figure 3—source data 1). (B) Correlation plot for the relative duration and peak intensity differences between AQP4 KO and WT animals analyzed. (C) Correlation plots between DCE-derived scores and the vascular densities from 12 parenchymal ROI analyzed with both methods in AQP4 KO and WT animals. (D) Correlation plots between DCE-derived scores and AQP4 expression from 10 parenchymal ROI analyzed with both methods in AQP4 KO and WT animals. Legend: OLF-olfactory, CA / RSP-cingulate / retrosplenial, VIS (V1)-visual, SS (S1)-somatosensory, AUD-auditory, HIP-hippocampus, PERI-perirhinal, TH-thalamus, HAB-habenula, HY-hypothalamus, MB-midbrain, PAG-periaqueductal gray, HB-hindbrain; CP-caudate putamen, WM-white matter; 3V-third ventricle, LV-lateral ventricle, CoW-Circle of Willis, CM-cisterna magna, SSS- superior sagittal sinus; NS-not significant. All correlation plots show respective regression lines along with semi-transparent areas marking 95% confidence intervals of the fitting. The highest obtained Pearson’s linear or Spearman’s range correlation scores reported and considered significant if correlation value >0.5 with P<0.05, and non-zero regression slope.
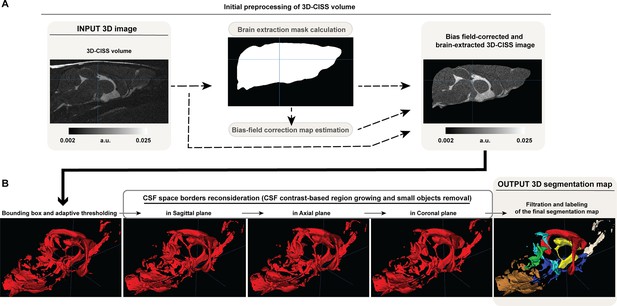
Schematic flow chart of the algorithm for the 3D-CISS-based CSF space volumetry and cisternography in Aqp4(-/-) (KO) and Aqp4(+/+) (WT) mice, including initial preprocessing (bias field correction and brain extraction) of the computed 3D-CISS volume (A) and subsequent automatic CSF space segmentation (B), based on representative 3D-CISS volume from a single animal.

Inverse Laplace transform (ILT) performed using INSPECT algorithm assuming presence of 2 spectral components.
Presences of fast diffusivities (>10 um2/ms, perfusion-like) components was confirmed in aggregated ADC spectra from 3 regions analyzed (A-C, see ‘Distribution diffusion-encoding direction-wise’). Although contribution of fast diffusivities is minimal to the mean spectrum from all diffusion-encoding directions jointly (‘Mean of aggregated distribution’), its presence confirms applicability of IVIM method for separating the perfusion-related effects from the spectrum of slow diffusivities. Two distinctive compartments in the range of restricted and low range of free water diffusivities were found for all cortical (A) and parenchymal (B) ROI (see ‘Mean of aggregated distribution’ and ‘Averaged distribution’) but not the 3rd ventricular CSF space ROI where an additional peak was visible in the fast diffusivity range (C). As further requested, we have provided comparison for ratios of depicted two diffusivity peaks fractions for analyzed ROI (D). Legend: Frestricted – fraction of restricted compartment, Ffree – fraction of free water compartment, ns-not significant (Mann-Whitney U-test).

3D-CISS-based regional brain morphometry in Aqp4 (+/+) and Aqp4 (-/-) mice brains.
Legend: OLF-olfactory, CA-cingulate, VIS (V1)-visual, SS (S1)-somatosensory, M-motor; AUD-auditory, HIP-hippocampus, PERI-perirhinal, TH-thalamus, HAB-habenula, HY-hypothalamus, MB-midbrain, PAG-periaqueductal gray, HB-hindbrain; CP-caudate putamen, WM-white matter; 3V-third ventricle, LV-lateral ventricle, CoW-Circle of Willis, CM-cisterna magna, SSS- superior sagittal sinus; NS-not significant. Minimal p-value (p=0.1385) found for the cingulate area.
Tables
Summary of demographic characteristics of the animals used in 8 experiments performed, and (B) details of MRI protocols and acquisition parameters for evaluation of the brain CSF and ISF spaces, employed in the current study.
Legend: CryoProbe – cryogenically-cooled MR coil; TA - time of acquisition; Tx/Rx - transmit/receive; TR - time to repetition; TE - time to echo; FA - flip angle; FOV - field of view; Av. / Rep. – averages for image formation or independently repeated acquisitions; FISP - steady-state free precession sequence; TrueFISP - true balanced steady-state free precession sequence; EPI - echo-planar imaging sequence; VTR – variable TR; VFA – variable flip angle; CM – cisterna magna; p - statistical p-value from Mann-Whitney U-test; N/A – data not available; NS - statistically not significant finding (p>>0.05).
| A | No.of animals | Age [weeks] | Body weight [g] | Respiration [bpm] | |||||
|---|---|---|---|---|---|---|---|---|---|
| KO | WT | Male [%] | Overall (mean ±SD) | p | Overall (mean ±SD) | p | Overall (mean ±SD) | p | |
| 3D-CISS volumetry | 5 | 6 | 72.7 | 13.8±1.8 | NS | 27.3±2.1 | NS | 176±30 | NS |
| MR-DWI | 6 | 6 | 41.7 | 10.4±0.7 | NS | 22.4±3.0 | NS | 175±11 | NS |
| DCE-MRI | 5 | 6 | 45.5 | 13.4±1.6 | NS | 27.3±2.2 | NS | 182±14 | NS |
| AQP4 expression | - | 4 | 50 | 10.0±0.0 | NS | 25–30 | N/A | - | - |
| Vascular density | 6 | 6 | 0 | 14.6±1.0 | NS | 24.3±4.6 | NS | - | - |
| CSF production | 5 | 6 | 45.5 | 15.4±0.5 | NS | 28.7±4.2 | NS | - | - |
| ISF space volume estimation using TMA | 8 | 20 | 55 | 10.0±0.0 | NS | 25–30 | N/A | - | - |
| Brain water content | 3 | 5 | 62.5 | 12.0±1.0 | NS | 25–30 | N/A | - | - |
| B | MR sequences and acquisition parameters | ||||||||
| Sequence (Tx/Rx coil; slice orientation) | TR [ms] | TE [ms] | FA [deg] | Av./Rep. | Voxel size [mm3](interpolation) | Bandwidth [Hz/pix] | Matrix size | TA | |
| MR CSF space volumetry | |||||||||
| 3D-TrueFISP (CryoProbe; sagittal) | 5.2 | 2.6 | 50 | 2 | 0.033×0.033×0.033 (2.0×1.6×1.0) | 260 | 19.2×12.8×12.8 | 27 min | |
| MR-DWI (in vivo and ex vivo) (δ=3 and Δ=10ms for gradient duration and separation times) *(b-values (Av.>1)): 40, 50, 59, 70, 92, 113, 165, 197, 238, 342, 445, 649, 854, 1057 (2), 1564 (2), 2071 (2), 3081 (2) s2/mm | |||||||||
| 2D-EPI (volumetric; axial) | 3600 | 30 | 90 | 3 | 0.15×0.15×0.5 (0.2 mm gap, 16 slices) | 3307 | 16.2×14.4×11.2 | 20–30 min (respiratory-gated) | |
| DCE-MRI via CM-injection | |||||||||
| 3D-FISP (CryoProbe; sagittal) | 3.26 | 1.63 | 15 | 1 | 0.1×0.1×0.1 | 781 | 19.2×12.8×12.8 | 90 min | |
| Microbeads phantom ex vivo - T1 mapping (VTR: 12000, 9000, 6500, 4000, 2000, 1000, 800, 500, 300, 100, 80, 50, 15ms) (VFA: 45° and 90°) | |||||||||
| 2D-RARE (volumetric; axial) | VTR | 3.1 | 90 | 2 | 0.1×0.1×3.0 | 671 | 16.2×16.2×1 | 3 h 20 min | |
| 2D-RARE (volumetric; axial) | 12000 | 3.1 | VFA | 3 | 0.1×0.1×3.0 | 671 | 16.2×16.2×1 | 1 hr 40 min | |
-
*
Presented averages of measured values from 3 diffusion encoding directions are slightly higher than the set-up, due to gradient preparation time.
Summary of findings for (A) average ADC and D-IVIM, (B) direction-wise MR diffusion and pseudodiffusion among 21 ROI assessed, along with statistical scoring.
Asterisks reflect ‘p’ significance values from the nonparametric Mann-Whitney test comparing diffusion measures between KO vs. WT animals ROI-wise (total n=21, balanced groups), along with the magnitude of the difference expressed by the inequality sign. Legend: OLF-olfactory, CA / RSP-cingulate / retrosplenial, VIS-visual (V1), SS-somatosensory (S1), AUD-auditory, HIP-hippocampus, PERI-perirhinal, TH-thalamus, HAB-habenula, HY-hypothalamus, MB-midbrain, PAG-periaqueductal gray, HB-hindbrain; CP-caudate putamen, WM-white matter; 3V-third ventricle, LV-lateral ventricle, 4V-fourth ventricle, PCS-pericisternal space, CoW-Circle of Willis, CB-cerebellum. NS- no significant difference, *-p<0.05, **-p<0.01, by means of Mann-Whitney U-test.
| A | ROI | Average ADC | Average D-IVIM | IVIM | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Finding | Significance | Finding | Significance | D*, Fp,Fp x D* | |||||||||
| Cerebral cortex | OLF | KO>WT | * | - | NS | - | |||||||
| CA / RSP | - | NS | - | NS | - | ||||||||
| VIS | - | NS | - | NS | - | ||||||||
| SS | KO>WT | * | KO>WT | * | - | ||||||||
| AUD | KO>WT | * | KO>WT | * | - | ||||||||
| HIP | KO>WT | ** | KO>WT | * | - | ||||||||
| PERI | - | NS | - | NS | - | ||||||||
| Brain stem | TH | KO>WT | ** | KO>WT | ** | - | |||||||
| HAB | - | NS | - | NS | - | ||||||||
| HY | - | NS | - | NS | - | ||||||||
| MB | KO>WT | * | KO>WT | * | - | ||||||||
| PAG | KO>WT | * | - | NS | - | ||||||||
| HB | KO>WT | ** | KO>WT | ** | - | ||||||||
| Cerebral nuclei and tracts | CP | KO>WT | ** | KO>WT | * | - | |||||||
| WM | KO>WT | * | KO>WT | * | - | ||||||||
| CSF space | 3V | - | NS | - | NS | - | |||||||
| LV | - | NS | - | NS | - | ||||||||
| 4V | - | NS | - | NS | - | ||||||||
| PCS | - | NS | - | NS | - | ||||||||
| CoW | - | NS | - | NS | - | ||||||||
| Cerebellum | CB | - | NS | - | NS | KO>WT Fp = 0.0649 | |||||||
| B | ROI | IVIM | ADC / D | ||||||||||
| Direction Z(cranio-caudal) | Direction X(bilateral) | Direction Y(ventral-dorsal) | Direction Z (cranio‐caudal) | Direction X (bilateral) | Direction Y (ventral‐dorsal) | ||||||||
| Finding | Signif. | Finding | Signif. | Finding | Signif. | Finding | Signif. | Finding | Signif. | Finding | Signif. | ||
| Cerebral cortex | OLF | - | NS | Fp x D* KO>WT | * | - | NS | KO>WT | **/* | - | NS | - | NS |
| CA / RSP | D* KO>WT | * | - | NS | - | NS | - | NS | - | NS | - | NS | |
| VIS | - | NS | - | NS | - | NS | - | NS | KO>WT | */- | KO>WT | */- | |
| SS | - | NS | - | NS | - | NS | KO>WT | -/* | KO>WT | **/ P=0.056 | KO>WT | */- | |
| AUD | - | NS | - | NS | - | NS | KO>WT | */* | KO>WT | **/* | KO>WT | */* | |
| HIP | - | NS | - | NS | - | NS | KO>WT | */* | KO>WT | */- | KO>WT | **/* | |
| PERI | D* KO>WT | ** | - | NS | - | NS | - | NS | KO>WT | */- | - | NS | |
| Brain stem | TH | - | NS | - | NS | - | NS | KO>WT | **/** | KO>WT | */- | KO>WT | */- |
| HAB | - | NS | - | NS | - | NS | - | NS | - | NS | - | NS | |
| HY | - | NS | - | NS | - | NS | - | NS | - | NS | - | NS | |
| MB | - | NS | - | NS | - | NS | KO>WT | -/* | KO>WT | */- | KO>WT | **/- | |
| PAG | - | NS | - | NS | - | NS | KO>WT | */** | KO>WT | **/- | KO>WT | */- | |
| HB | - | NS | - | NS | - | NS | KO>WT | */* | KO>WT | **/- | KO >WT | */* | |
| Cerebral nuclei and tracts | CP | - | NS | - | NS | - | NS | KO>WT | **/** | KO>WT | */- | - | NS |
| WM | D* KO>WT | ** | - | NS | - | NS | - | NS | - | NS | KO>WT | */- | |
| CSF space | 3V | - | NS | Fp / Fp x D* KO>WT | ** / * | - | NS | - | NS | - | NS | - | NS |
| LV | - | NS | - | NS | - | NS | - | NS | - | NS | - | NS | |
| 4V | - | NS | - | NS | - | NS | - | NS | - | NS | - | NS | |
| PCS | - | NS | - | NS | - | NS | - | NS | - | NS | - | NS | |
| CoW | - | NS | - | NS | - | NS | - | NS | - | NS | - | NS | |
| Cerebellum | CB | - | NS | - | NS | - | NS | - | NS | - | NS | - | NS |
DCE-derived parameters from 21 ROI (matching those analyzed by means of MR-DWI) in 5 KO and 6 WT littermate mice, along with associated nonparametric pair-wise statistics using a two-tailed Wilcoxon signed-rank test and median ± standard deviation (SD) values strain-wise.
Legend: Mean AUC – mean from areas under the DCE curves along with associated p-statistical values (p-stat) from nonparametric Mann–Whitney U-test ROI-wise; Duration – duration of significantly different from the baseline signal enhancement, mimicking the parenchymal tracer accumulation; Aqp4(+/+) – wildtype control; Aqp4(-/-) – AQP4 KO mice; Δ – WT-KO difference; rel. Δ – ((WT-KO)/WT)×100% relative difference (negative ‘-‘ sign means the shorter duration of interstitial tracer accumulation in KO mice); For ROI abbreviations, see caption Figures 2 and 3, or Table 2. # –Two-way Anova with Bonferroni's post-hoc. ¶ – mean of standard deviations ROI wise. * – p<0.05, ** – p<0.01.
| #WT vs. KO time-series different? | Mean AUC [a.u.] | Arrival time [min] | Time- to-peak [min] | Peak intensity [a.u.] | Interstitial tracer accumulation time [min] | Duration [min] | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ROI | Aqp4(+/+) | Aqp4(-/-) | p-stat | Aqp4(+/+) | Aqp4(-/-) | Δ | Aqp4(+/+) | Aqp4(-/-) | Δ | Aqp4(+/+) | Aqp4(-/-) | rel. Δ [%] | Aqp4(+/+) | Aqp4(-/-) | Aqp4(+/+) | Aqp4(-/-) | rel. Δ [%] | ||||
| Start | End | Start | End | ||||||||||||||||||
| Cortical | OLF | NS | 7207 | 5619 | NS | 6 | 6 | 0 | 61 | 52 | 9 | 106 | 79 | 25.5 | 43 | 80 | 39 | 70 | 38 | 32 | –15.8 |
| CA/RSP | ** | 1662 | 616 | ** | 7 | 7 | 0 | 89 | 90 | -1 | 27 | 12 | 55.6 | 59 | 90 | 66 | 90 | 32 | 25 | –21.9 | |
| VIS | ** | 1699 | 529 | ** | 7 | 8 | -1 | 89 | 90 | -1 | 33 | 9 | 72.7 | 54 | 90 | 73 | 90 | 37 | 18 | –51.4 | |
| SS | NS | 537 | 438 | NS | 6 | 7 | -1 | 20 | 16 | 4 | 8 | 7 | 12.5 | 12 | 32 | 11 | 25 | 21 | 15 | –28.6 | |
| AUD | NS | 454 | 340 | NS | 7 | 7 | 0 | 89 | 89 | 0 | 10 | 5 | 50.0 | 67 | 90 | N/A | N/A | 24 | N/A | <-100 | |
| HIP | ** | 3258 | 1622 | * | 5.5 | 7.5 | -2 | 63 | 81 | –18 | 46 | 22 | 52.2 | 45 | 90 | 74 | 90 | 46 | 17 | –63.0 | |
| PERI | P=0.052 | 3513 | 2659 | NS | 4 | 5 | -1 | 83 | 90 | -7 | 59 | 44 | 25.4 | 53 | 90 | 57 | 90 | 38 | 34 | –10.5 | |
| Brain stem | TH | * | 3471 | 2557 | 0.052 | 2–3 | 2–3 | 0 | 82 | 90 | -8 | 61 | 45 | 26.2 | 53 | 90 | 58 | 90 | 38 | 33 | –13.2 |
| HAB | NS | 2379 | 1992 | NS | 9 | 11 | -2 | 89 | 90 | -1 | 45 | 36 | 20.0 | 54 | 90 | 58 | 90 | 37 | 33 | –10.8 | |
| HY | NS | 7908 | 6766 | NS | 4 | 4 | 0 | 39 | 34 | 5 | 123 | 105 | 14.6 | 23 | 59 | 21 | 54 | 37 | 34 | –8.1 | |
| MB | * | 6125 | 4013 | * | 3 | 4 | -1 | 66 | 83 | –17 | 87 | 56 | 35.6 | 42 | 84 | 53 | 90 | 43 | 38 | –11.6 | |
| PAG | NS | 25740 | 22474 | NS | 8 | 10 | -2 | 43 | 38 | 5 | 350 | 311 | 11.1 | 35 | 75 | 31 | 61 | 41 | 31 | –24.4 | |
| HB | NS | 9291 | 8124 | NS | 2–3 | 2–3 | 0 | 42 | 38 | 4 | 133 | 114 | 14.2 | 28 | 66 | 27 | 61 | 39 | 35 | –10.3 | |
| Nuclei / tracts | CP | NS | 1736 | 1167 | NS | 4 | 8 | -4 | 89 | 90 | -1 | 39 | 25 | 35.9 | 54 | 90 | 58 | 90 | 37 | 33 | –10.8 |
| WM | ** | 6953 | 5445 | ** | 4.5 | 6.5 | -2 | 51 | 53 | -2 | 105 | 80 | 23.8 | 37 | 73 | 38 | 69 | 37 | 32 | –13.5 | |
| CSF space | CM | NS | 23276 | 19903 | NS | 1 | 1 | 0 | 24 | 24 | 0 | 397 | 339 | 14.6 | 15 | 50 | 16 | 47 | 36 | 32 | –11.1 |
| PCS | NS | 15163 | 12498 | NS | 2–3 | 2–3 | 0 | 41 | 31 | 10 | 224 | 187 | 16.5 | 23 | 59 | 20 | 50 | 37 | 31 | –16.2 | |
| 3V | NS | 12228 | 10441 | NS | 5 | 6 | -1 | 45 | 46 | -1 | 180 | 151 | 16.1 | 34 | 76 | 33 | 61 | 43 | 29 | –32.6 | |
| LV | NS | 8288 | 6555 | NS | 6 | 11 | -5 | 90 | 90 | 0 | 157 | 123 | 21.7 | 54 | 90 | 58 | 90 | 37 | 33 | –10.8 | |
| Caudal | SSS | * | 5832 | 1430 | ** | 1 | 3 | -2 | 64 | 88 | –24 | 77 | 22 | 71.4 | 38 | 90 | 75 | 90 | 53 | 16 | –69.8 |
| CB | NS | 7369 | 6151 | NS | 6 | 7 | -1 | 47 | 59 | –12 | 97 | 80 | 17.5 | 29 | 75 | 31 | 67 | 47 | 37 | –21.3 | |
| MEDIAN ±SD | 6125±1653¶ | 4013±1306¶ | 5.0±2.2 | 6.5±2.8 | 63±23 | 81±27 | 56 | 87 | 24±19 | 37±7 | 32±7 | ||||||||||
| KO vs. WT difference (Wilcoxon signed-rank test) | WT >KO, p<0.0001 | WT <KO, p<0.01 | WT ~KO, p=0.2971 | WT >KO, p<0.0001 | WT >KO, P<0.0001 | ||||||||||||||||
Descriptive summary of findings presented in the current study.
Bold italic font highlights the regions of the largest differences found between KO and WT animals, by means of 3 MRI and 5 physiological and histological assessment methods applied.
| Magnetic resonance imaging in vivo | |||
|---|---|---|---|
| Measurement | General findings in KO compared to WT | Region of largest difference | |
| non-invasive | 3D cisternography | - 5–10% larger brain volumes - 22–29% smaller CSF space / brain volume ratio | ventricular space, 3rd ventricle |
| 2D diffusion-weighted imaging | 5–6% higher ADC and D | (difference present for average and in all diffusion directions) - thalamus, hindbrain, periaqueductal gray regions, auditory cortex and hippocampus | |
| Higher Fp and Fp × D* only in the 3rd ventricle | |||
| invasive | Dynamic CSF tracer imaging via cisterna magna` | reduced parenchymal tracer influx and evacuation | - influx: cortical ROI, hippocampus - efflux: superior sagittal sinus |
| Physiological and histological measurements | |||
| Measurement | General findings in KO compared to WT | Region of largest difference | |
| ex vivo and histology | Brain water content | ~6% larger brain water content | |
| AQP4 expression | (only WT) heterogenous AQP4 expression in the brain | largest expression in the thalamus, hippocampus, habenula | |
| Vascular density | similar vascular density to WT | trend for larger vascular density in the thalamus and olfactory area | |
| in vivo | CSF production | similar CSF production to WT | |
| Real-time ionophoresis TMA | ISF space volume larger | ||
Additional files
-
MDAR checklist
- https://cdn.elifesciences.org/articles/82232/elife-82232-mdarchecklist1-v3.docx
-
Source data 1
Aggregated data set, including all source data.
- https://cdn.elifesciences.org/articles/82232/elife-82232-data1-v3.zip





