Synergistic stabilization of microtubules by BUB-1, HCP-1, and CLS-2 controls microtubule pausing and meiotic spindle assembly
Figures
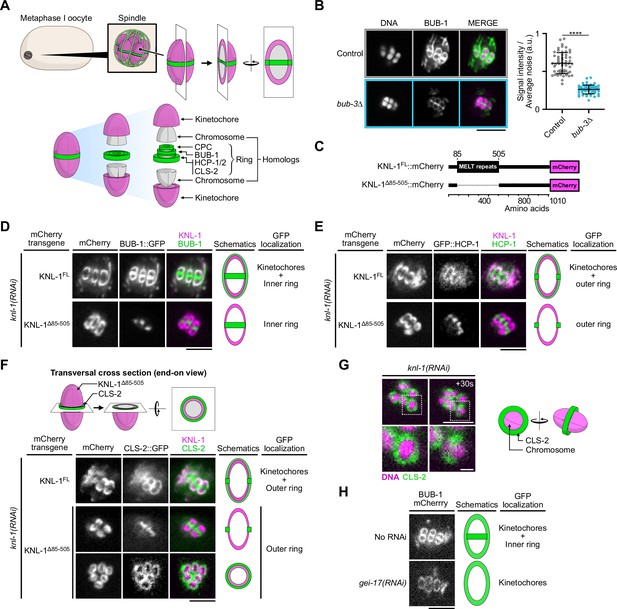
The N-terminal MELT repeats of KNL-1, but not BUB-3, are required for BHC module kinetochore targeting in oocytes.
(A) Schematic of kinetochore and ring domain protein organization around a bivalent chromosome during metaphase I in a C. elegans oocyte. CPC: Chromosome Passenger Complex. (B) Immunolocalization of BUB-1 (left) and quantification of BUB-1 signal at kinetochores (right) in bub-3(ok3437) mutants (bub3∆, n=58) compared to wild type controls (n=58). Error bars, Mean and standard deviation. Unpaired t-test, alpha = 0.05, p<0.0001. (C) Schematic of KNL-1::mCherry protein fusions. (D–F) Localization of BUB-1::GFP (D), GFP::HCP-1 (E) and CLS-2::GFP (F) in worms carrying full length or MELT-deleted KNL-1::mCherry (KNL-1FL and KNL-1∆85-505 respectively, n≥10). (G) Localization of CLS-2::GFP at ring domains in knl-1-depleted oocytes (left) with corresponding schematic (right). (H) Localization of BUB-1::mCherry in gei-17-depleted oocytes (n=29) compared to controls (n=25). Scale bars 5 µm, 1 µm in insets.
-
Figure 1—source data 1
Panel B source data.
Quantification of BUB-1 signal at kinetochores in bub-3(ok3437) mutants (bub-3∆) and in control N2. Integrated intensity measurements corrected by integrated background noise (Integrated signal intensity/Average noise) of immunolocalized BUB-1 at kinetochores in fixed oocytes of bub-3(ok3437) mutants (bub-3∆) compared to N2 wild type controls. Descriptive statistics and details of unpaired t-test for comparison of samples are provided.
- https://cdn.elifesciences.org/articles/82579/elife-82579-fig1-data1-v2.xlsx
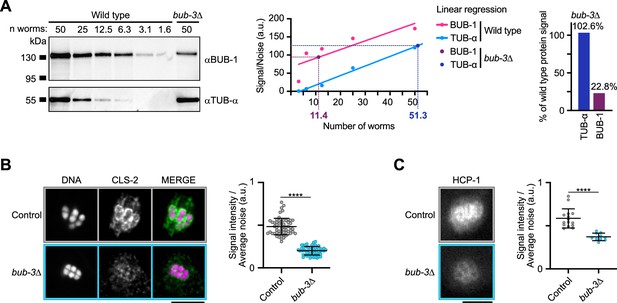
BUB-3 is not required for BHC module kinetochore targeting.
(A) Left: Western blot of full-protein extracts. BUB-1 (top) and α-tubulin (TUB-α, bottom) in wild type and bub-3(ok3437) mutants (bub-3∆). The number of worms extracted is indicated (n worms). Center: Quantification and linear regression of corrected intensities of BUB-1 and TUB-α in wild type and bub-3∆ extracts. Right: Percentage of BUB-1 and TUB-α signals in bub-3∆ relative to wild type. (B) Immunolocalization of CLS-2 (left) and quantification of CLS-2 signal at kinetochores (right) in bub-3(ok3437) mutants (bub3∆, n=58) compared to wild type controls (n=58). Note the ring localization of CLS-2, which is clearly visible on the left- and right-most chromosomes of the wild-type spindle. (C) Stills of metaphase I oocytes from live imaging of endogenously tagged GFP::HCP-1 (left), and quantification of GFP signal/average noise in the whole spindle, measured at the frame preceding spindle rotation at the cortex (right), in bub-3(ok3437) mutants (bub3∆, n=10) compared to controls (n=15). Error bars, Mean and standard deviation. Unpaired t-test, alpha = 0.05, p<0.0001. Scale bars, 5 µm.
-
Figure 1—figure supplement 1—source data 1
Panel A source data 1.
Folder containing raw images and uncropped annotated images of Western blot of BUB-1 and tubulin in full-protein extracts of wild type and bub-3(ok3437) (bub-3∆) mutant worms.
- https://cdn.elifesciences.org/articles/82579/elife-82579-fig1-figsupp1-data1-v2.zip
-
Figure 1—figure supplement 1—source data 2
Panel A source data 2.
Signal measurements on western blots, performed in Fiji. Simple linear regression was performed using GraphPad Prism.
- https://cdn.elifesciences.org/articles/82579/elife-82579-fig1-figsupp1-data2-v2.xlsx
-
Figure 1—figure supplement 1—source data 3
Panel B source data.
Quantification of CLS-2 signal at kinetochores in bub-3(ok3437) mutants (bub-3∆) compared to wild type N2 controls. Signals were measured in Fiji from immunolocalized proteins. Details of statistical analyses are provided.
- https://cdn.elifesciences.org/articles/82579/elife-82579-fig1-figsupp1-data3-v2.xlsx
-
Figure 1—figure supplement 1—source data 4
Panel C source data.
Quantification of HCP-1 signal at kinetochores in bub-3(ok3437) mutants (bub-3∆) compared to controls. Signals were measured in Fiji from live imaging of GFP::HCP-1. Details of statistical analyses are provided.
- https://cdn.elifesciences.org/articles/82579/elife-82579-fig1-figsupp1-data4-v2.xlsx
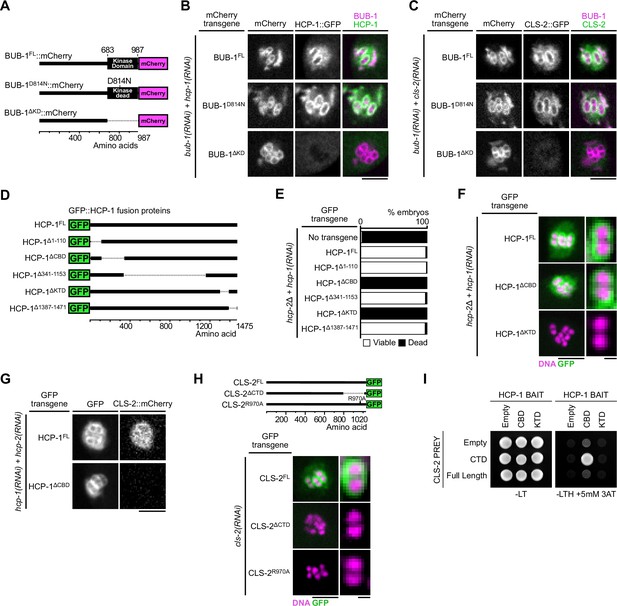
Molecular determinants of BHC module assembly.
(A) Schematic of BUB-1::mCherry protein fusions. (B,C) Localization of indicated RNAi-resistant BUB-1::mCherry and GFP::HCP-1 (B) or CLS-2::GFP (C) upon depletion of corresponding endogenous gene products (n≥10). (D) Schematic of truncated GFP::HCP-1 fusions. (E) Embryonic viability assay in indicated transgenic hcp-2(ijm6) (hcp-2∆) mutants upon depletion of endogenous hcp-1. (F) Localization of indicated GFP::HCP-1 fusions in hcp-2∆ worms depleted of endogenous hcp-1 (n≥9). (G) Localization of CLS-2::mCherry in indicated conditions (n≥13). (H) Localization of schematized (top) RNAi-resistant CLS-2::GFP fusions upon depletion of endogenous cls-2 (bottom, n≥10). (I) Yeast-two-hybrid interaction assay between HCP-1 domains (baits) and CLS-2 CTD (prey). Scale bars, metaphase plate 5 µm, single chromosome details 1 µm.
-
Figure 2—source data 1
Panel E source data.
Embryonic viability assay of hcp-2(ijm6) mutant worms carrying gfp::hcp-1 transgene variants, upon depletion of endogenous hcp-1.
- https://cdn.elifesciences.org/articles/82579/elife-82579-fig2-data1-v2.xlsx
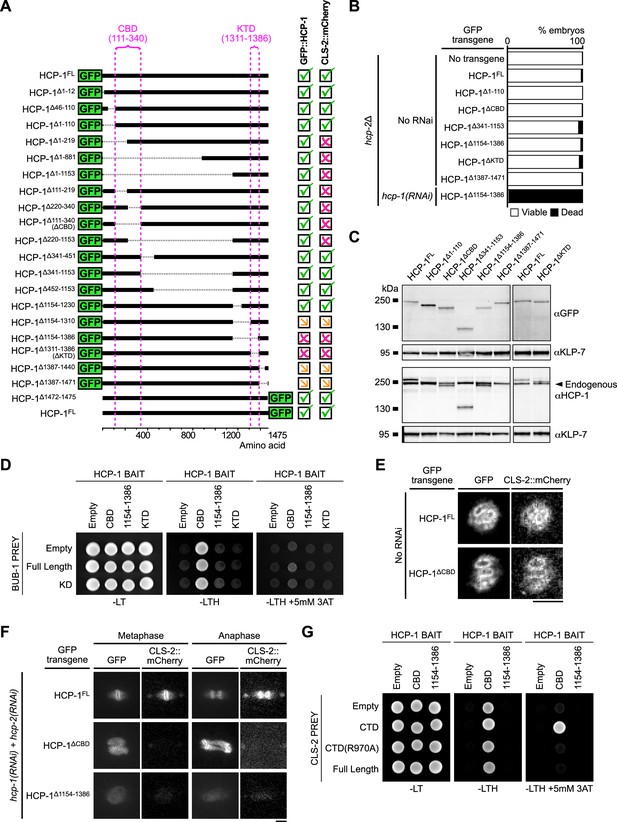
Protein domains essential for BHC module assembly.
(A) Schematics of truncated GFP::HCP-1 fusions used to identify the CBD and KTD regions. Indicated on the right, presence (green tick), absence (magenta cross) or decrease (orange arrow) of GFP::HCP-1 and CLS-2::mCherry signals in oocytes and zygotes. (B) Embryonic viability assay of hcp-2(ijm6) (hcp-2∆) mutants in indicated conditions. (C) Western blot of full-protein extracts (50 worms per lane). Arrowhead shows endogenous HCP-1. (D) Yeast two hybrid interaction assay between HCP-1 (bait) and BUB-1 (prey) domains. (E,F) Localization of CLS-2::mCherry in meiosis I (E) and during the first mitotic division of the zygote (F) in indicated conditions (n≥10). Scale bars, 5 µm. (G) Yeast-two-hybrid interaction assay between HCP-1 domains (bait) and CLS-2 (prey).
-
Figure 2—figure supplement 1—source data 1
Panel B source data.
Embryonic viability assay of hcp-2(ijm6) mutant worms carrying gfp::hcp-1 transgene variants, when endogenous hcp-1 is not depleted.
- https://cdn.elifesciences.org/articles/82579/elife-82579-fig2-figsupp1-data1-v2.xlsx
-
Figure 2—figure supplement 1—source data 2
Panel C source data.
Raw images and uncropped annotated images of western blots of GFP::HCP-1 fusion protein variants in full-protein worm extracts.
- https://cdn.elifesciences.org/articles/82579/elife-82579-fig2-figsupp1-data2-v2.zip
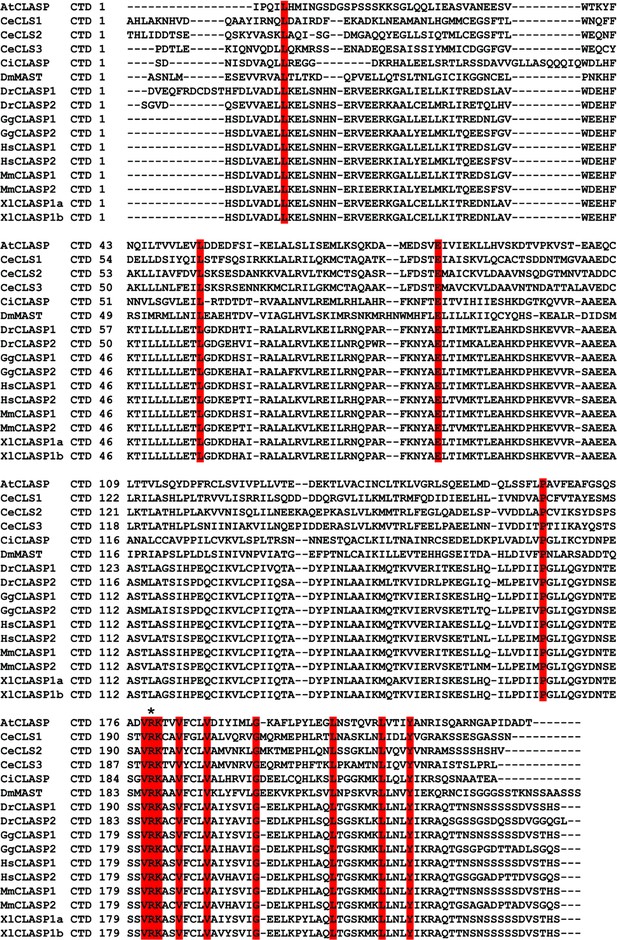
Protein sequence alignment of indicated eukaryotic CLASP CTDs.
Conserved amino acids in red. The star indicates residue R970.
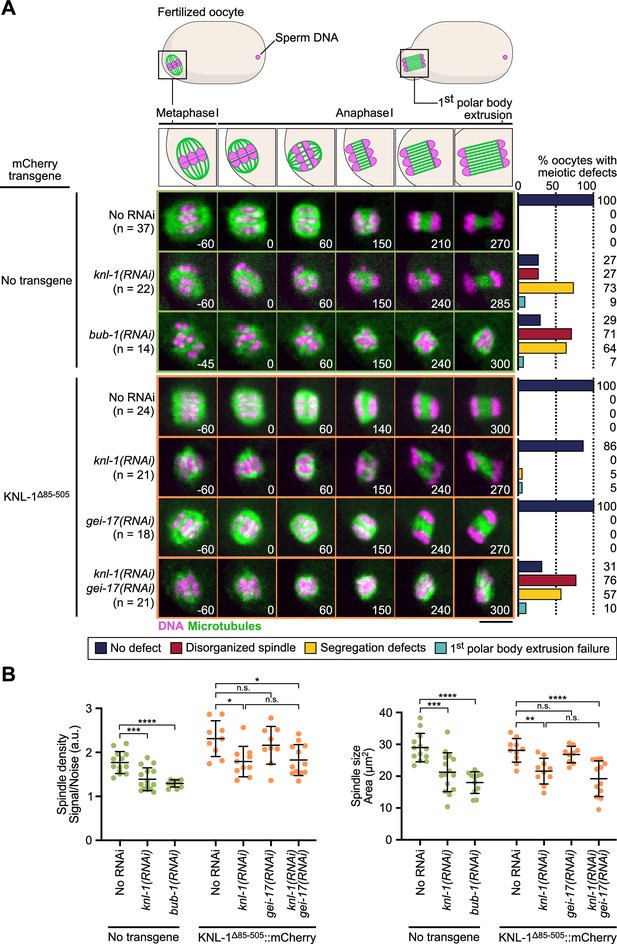
BHC module has both kinetochore-dependent and -independent functions in spindle assembly and chromosome segregation in oocytes.
(A) Schematic of the meiotic spindle during meiosis I division (top) and stills from live imaging of meiosis I in indicated conditions (bottom). Microtubules (GFP::TBA-2α -tubulin) in green, chromosomes (mCherry::HIS-11H2B) in magenta. Time in seconds relative to anaphase I onset. Scale bar 5 µm. Graphs on the right show quantifications of meiotic defects. (B) Plots of spindle density (corrected GFP intensity, left) and spindle area (right) 45 seconds before anaphase I onset. Tests, One-way ANOVA multiple comparisons alpha = 0.05, *p<0.05, **p<0.01, ***p<0,001, ****p<0,0001, n.s. not significant. Error bars, Mean and standard deviation.
-
Figure 3—source data 1
Panel A source data.
Quantifications of the meiotic defects observed in indicated conditions.
- https://cdn.elifesciences.org/articles/82579/elife-82579-fig3-data1-v2.xlsx
-
Figure 3—source data 2
Panel B source data.
Quantifications of spindle density and spindle size during meiosis I in indicated conditions. Raw quantifications and corrected intensities of GFP::TBA-2α-tubulin signal (GFP Integrated Intensity/Background Noise). Details of statistical analyses are provided.
- https://cdn.elifesciences.org/articles/82579/elife-82579-fig3-data2-v2.xlsx
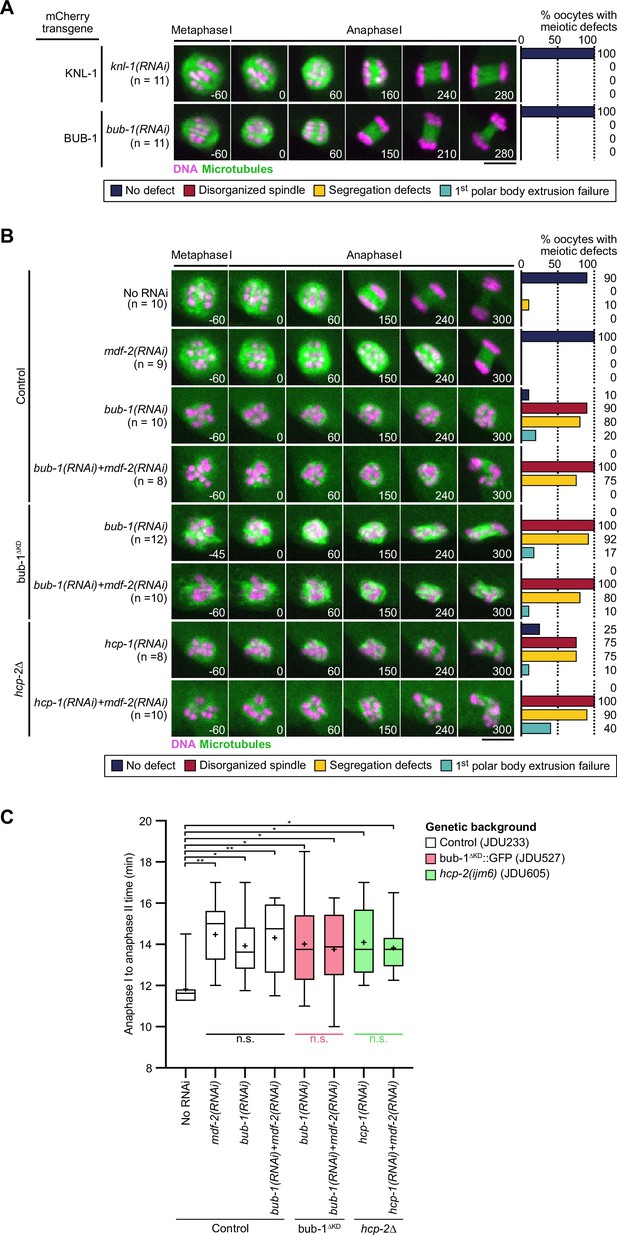
SAC activation is not responsible for defects observed upon BHC module perturbation.
(A–B) Stills from live imaging in indicated conditions. Microtubules (GFP::TBA-2α-tubulin or GFP::TBB-2β-tubulin) in green, chromosomes (mCherry::HIS-11H2B) in magenta. Time in seconds relative to anaphase I onset. Right panels indicate quantifications of meiotic defects as referred to in color key. Scale bars, 5 µm. (C) Box plot of time between anaphase I and II onsets in indicated conditions upon depletion of the spindle assembly checkpoint protein mdf-2 (left). Whiskers represent 5–95 percentile, (+) signs represent the mean. Kruskal-Wallis multiple comparisons, alpha = 0.05 (black bars), Mann-Whitney test, alpha = 0.05 (color bars), *p<0,05, **p<0.01, n.s. not significant.
-
Figure 3—figure supplement 1—source data 1
Panel A source data.
Quantifications of the meiotic defects in indicated conditions.
- https://cdn.elifesciences.org/articles/82579/elife-82579-fig3-figsupp1-data1-v2.xlsx
-
Figure 3—figure supplement 1—source data 2
Panel B-C source data.
Time measurement between anaphase I and anaphase II in indicated conditions (details of statistical tests are provided), and quantifications of meiotic defects observed.
- https://cdn.elifesciences.org/articles/82579/elife-82579-fig3-figsupp1-data2-v2.xlsx
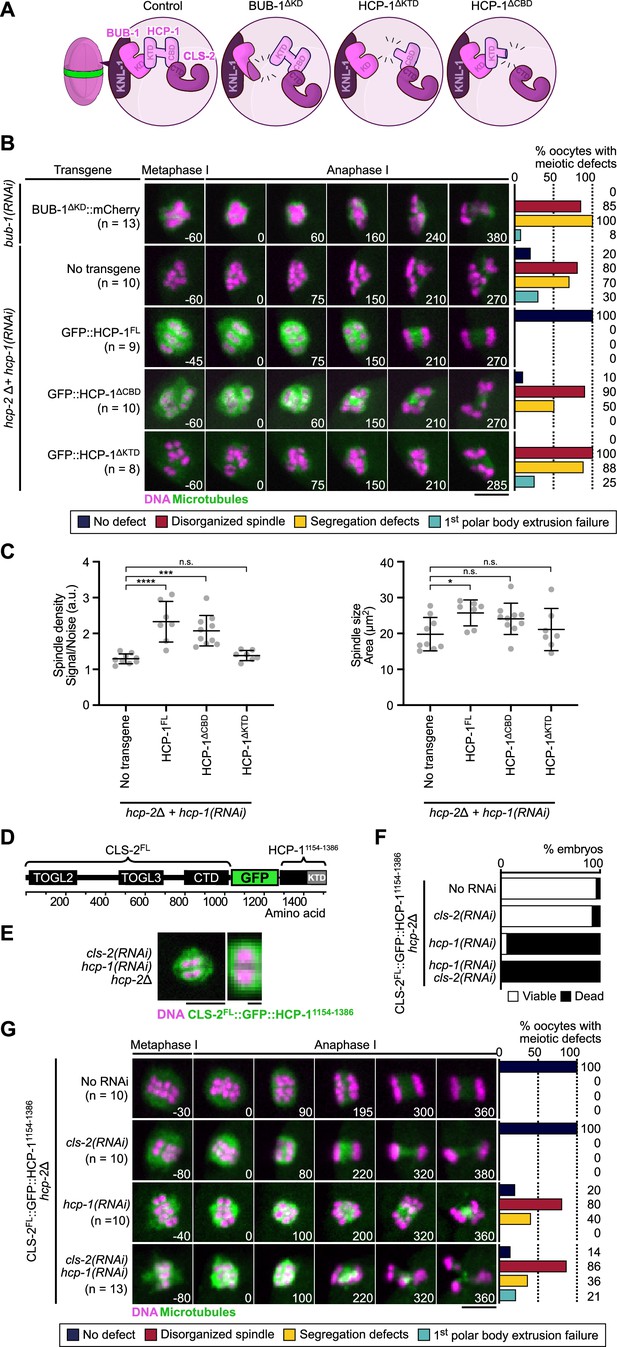
BHC module integrity is essential for spindle assembly and accurate chromosome segregation.
(A) Schematic of BHC-module integrity mutants. (B) Stills from live imaging of meiosis I in indicated conditions. Microtubules (GFP::TBA-2α-tubulin or GFP::TBB-2β-tubulin) in green, chromosomes (mCherry::HIS-11H2B) in magenta. Time in seconds relative to anaphase I onset. Graphs indicate quantifications as referred to in color key. (C) Plots of spindle density (corrected GFP intensity, left) and spindle area (right) 45 s before anaphase I onset. Kruskal-Wallis multiple comparisons, alpha = 0.05, *p<0.05, ***p<0.001, ****p<0.0001, n.s. not significant. Error bars, Mean with standard deviation. (D) Schematic of the CLS-2FL::GFP::HCP-11154-1386 protein fusion. The HCP-111154-1386 fragment contains the KTD. (E) Localization of CLS-2FL::GFP::HCP-11154-1386 (green) in metaphase I, and magnification of a single meiosis I chromosome. DNA (mCherry::HIS-11H2B) in magenta (n=13). (F) Embryonic lethality and (G) meiotic defects rescue assays of indicated depletions by CLS-2FL::GFP::HCP-11154-1386. Scale bars, full spindle 5 µm, single chromosome details 1 µm.
-
Figure 4—source data 1
Panel B source data.
Quantifications of the meiotic defects observed in indicated conditions.
- https://cdn.elifesciences.org/articles/82579/elife-82579-fig4-data1-v2.xlsx
-
Figure 4—source data 2
Panel C source data.
Quantifications of spindle density and spindle size during meiosis I in indicated hcp-2(ijm6) mutants upon depletion of endogenous hcp-1. Raw quantifications and corrected intensities of GFP::TBA-2α-tubulin signal (GFP Integrated Intensity/Background Noise). Details of statistical analyses are provided.
- https://cdn.elifesciences.org/articles/82579/elife-82579-fig4-data2-v2.xlsx
-
Figure 4—source data 3
Panel F source data.
Embryonic viability assay of hcp-2(ijm6) mutant, cls-2FL::gfp::hcp-111541154-1386 transgenic worms upon depletion of hcp-1 and/or cls-2.
- https://cdn.elifesciences.org/articles/82579/elife-82579-fig4-data3-v2.xlsx
-
Figure 4—source data 4
Panel G source data.
Quantifications of the meiotic defects in hcp-2(ijm6) mutant, cls-2FL::gfp::hcp-111541154-1386 transgenic worms upon depletion of hcp-1 and/or cls-2.
- https://cdn.elifesciences.org/articles/82579/elife-82579-fig4-data4-v2.xlsx
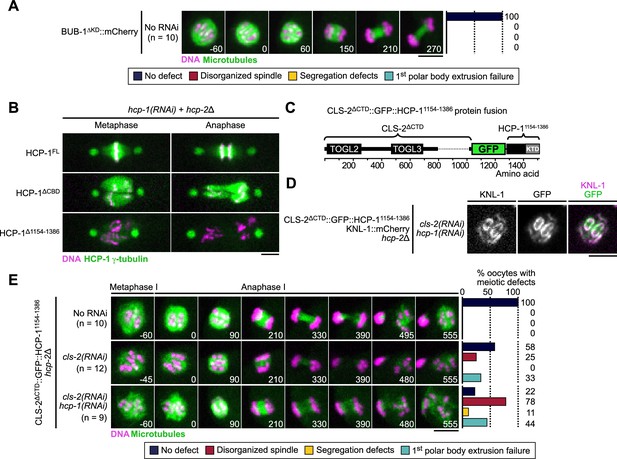
Artificial kinetochore and ring localization of CLS-2 is not sufficient to rescue loss of BHC module integrity.
(A) Stills from live imaging of BUB-1∆KD::GFP transgenic oocytes. Microtubules (GFP::TBB-2β-tubulin) in green, chromosomes (mCherry::HIS-11H2B) in magenta. Right panels indicate quantifications of meiotic defects shown in the color key. Time in seconds relative to anaphase I onset. (B) Localization of indicated GFP::HCP-1 fusion proteins (green) during the first zygotic mitosis (n≥9). Centrosomes (GFP::TBG-1ɣ-tubulin) and indicated GFP::HCP-1 deletion mutants in green, chromosomes (mCherry::HIS-11H2B) in magenta. (C) Schematic of the CLS-2∆CTD::GFP::HCP-11154-1386 protein fusion. (D) Localization of CLS-2∆CTD::GFP::HCP-11154-1386 in metaphase I, relative to KNL-1 marked kinetochores (KNL-1::mCherry, magenta), in hcp-2∆ mutants depleted of endogenous hcp-1 and cls-2 (n=11). (E) Stills from live imaging of meiosis I (left) and color-coded meiotic defects scored (right) in CLS-2∆CTD::GFP::HCP-11154-1386, hcp-2∆ worms, in indicated conditions. Time in seconds relative to anaphase I onset. Scale bars, 5 µm.
-
Figure 4—figure supplement 1—source data 1
Panel A source data.
Quantifications of the meiotic defects in bub-1∆KD::mCherry transgenic worms.
- https://cdn.elifesciences.org/articles/82579/elife-82579-fig4-figsupp1-data1-v2.xlsx
-
Figure 4—figure supplement 1—source data 2
Panel E source data.
Quantifications of the meiotic defects observed in hcp-2(ijm6) mutant, cls-2∆CTD::gfp::hcp-111541154-1386 transgenic worms upon depletion of cls-2 or both hcp-1 and cls-2.
- https://cdn.elifesciences.org/articles/82579/elife-82579-fig4-figsupp1-data2-v2.xlsx
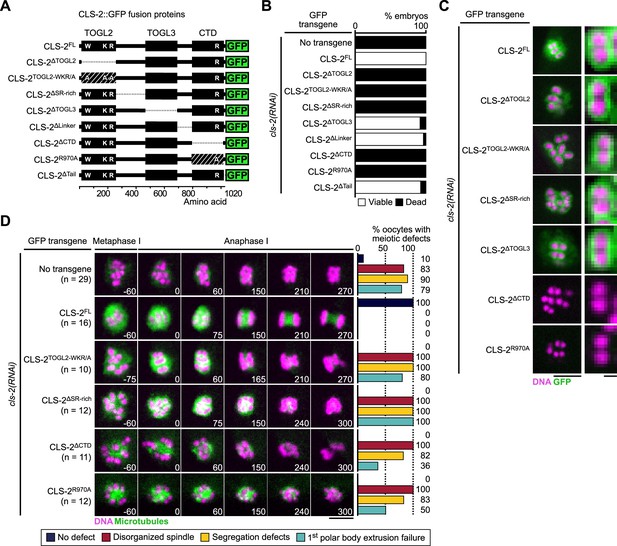
CLS-2 function does not require TOGL3.
(A) Schematic of CLS-2::GFP truncated fusions. (B) Embryonic viability assay upon depletion of endogenous cls-2 in the presence of indicated transgene. (C) Localization of CLS-2::GFP truncations (green) during metaphase I in indicated conditions. DNA (mCherry::HIS-11H2B) in magenta (n≥9). (D) Stills from live imaging of meiosis I. Microtubules (GFP::TBA-2α -tubulin) in green, chromosomes (mCherry::HIS-11H2B) in magenta. Time in seconds relative to anaphase I onset. Graphs indicate quantifications of meiotic defects. Scale bars, full spindles 5 µm, single chromosome details 1 µm.
-
Figure 5—source data 1
Panel B source data.
Embryonic viability assay of cls-2::gfp transgenic variants upon depletion of endogenous cls-2.
- https://cdn.elifesciences.org/articles/82579/elife-82579-fig5-data1-v2.xlsx
-
Figure 5—source data 2
Panel D source data.
Quantifications of the meiotic defects in cls-2::gfp transgenic variants upon depletion of endogenous cls-2.
- https://cdn.elifesciences.org/articles/82579/elife-82579-fig5-data2-v2.xlsx
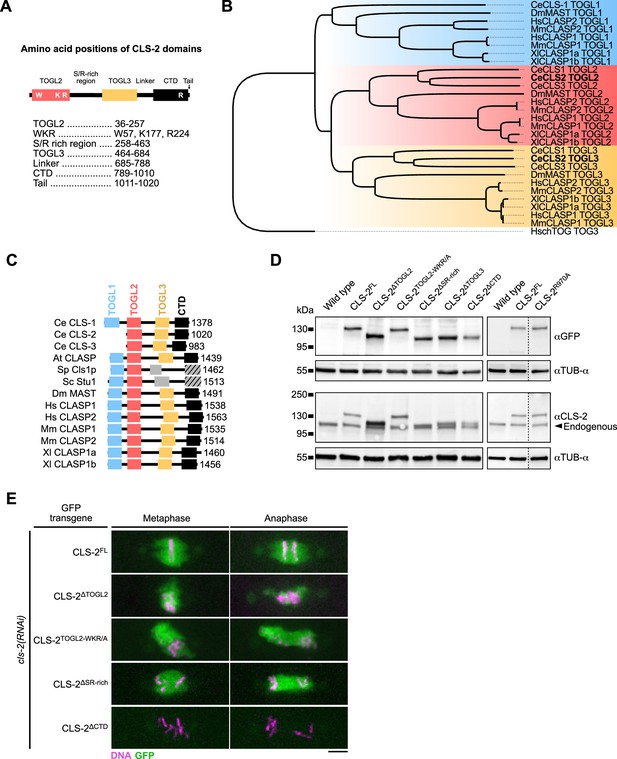
Protein domains essential for CLS-2 function.
(A) Positions of CLS-2 domains and conserved amino-acids. (B) Maximum Likelihood phylogenetic tree of eukaryotic CLASP TOG-like domains, rooted on human chTOG TOG3. (C) Comparative schematics of eukaryotic CLASP protein structures (Ce: Caenorhabditis elegans, At: Arabidopsis thaliana, Sp: Schizosaccharomyces pombe, Sc: Saccharomyces cerevisiae, Dm: Drosophila melanogaster, Hs: Homo sapiens, Mm: Mus musculus, Xl: Xenopus laevis). Sizes in amino-acids are indicated on the right. Grey boxes and hatched grey boxes represent respectively the helical and dimerization domains of Sc and Sp CLASPs. (D) Western blot of full-protein extracts (50 worms per lane) of indicated genotypes. Arrowhead indicates endogenous CLS-2. (E) Mitotic localization of indicated CLS-2::GFP fusion proteins upon depletion of endogenous cls-2 (green). DNA (mCherry::HIS-11) in magenta (n≥10). Scale bar, 5 µm.
-
Figure 5—figure supplement 1—source data 1
Panel D source data.
Raw images and uncropped annotated image of western blots of CLS-2::GFP fusion protein variants in full-protein worm extracts.
- https://cdn.elifesciences.org/articles/82579/elife-82579-fig5-figsupp1-data1-v2.zip

Protein sequence alignment of indicated eukaryotic TOGL domains.
Conserved amino acids mutated in the CLS-2TOGL2-WKR/A mutant are shown in red.
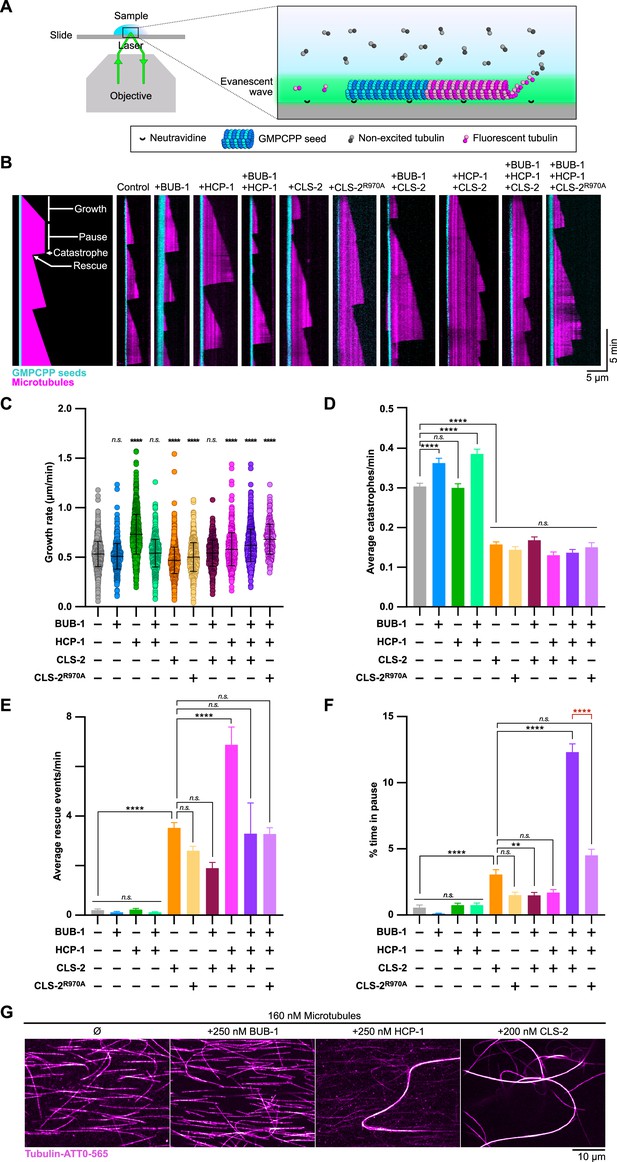
BHC module components synergistically stabilize microtubules in vitro.
(A) Schematic of the TIRF microscopy-based microtubule assay. Labeled tubulin (ATTO-565, magenta) fluoresces only when close to the surface of the coverslip. Microtubules polymerize from biotinylated GMPCPP seeds (tubulin-ATTO-488, cyan) bound to a Neutravidine-coated glass coverslip. (B) Representative kymographs of microtubules (magenta) growing from GMPCPP seeds (cyan) in the presence or absence of BUB-1, HCP-1, CLS-2-GFP and/or CLS-2R970A-GFP (100 nM each). Schematics on the left highlights the different microtubule dynamics events observed. (C–F) Dot plot showing the quantification of growth rate (C), and histograms showing the average catastrophe (D), rescue (F), and pause (D) events per microtubule. Dunnett’s multiple comparison tests, alpha = 0.01, **p<0.01, ****p<0.0001, n.s. not significant. Error bars, Mean and standard deviation (C) or standard error of the mean (D–F). (G) Microtubule bundling assay. Organization of microtubules (magenta) observed in indicated conditions.
-
Figure 6—source data 1
Panel B-F source data.
Raw measurements and calculations for quantification of microtubule dynamics. Parameters were extracted from kymographs using the ImageJ software (https://imagej.nih.gov/ij/index.html). Each sheet corresponds to an experiment. The first sheet (read me) provides details about data extraction.
- https://cdn.elifesciences.org/articles/82579/elife-82579-fig6-data1-v2.xlsx
-
Figure 6—source data 2
B-F statistics source data.
Processed data for the analysis of microtubule dynamics. Details of statistical analyses are provided.
- https://cdn.elifesciences.org/articles/82579/elife-82579-fig6-data2-v2.xlsx
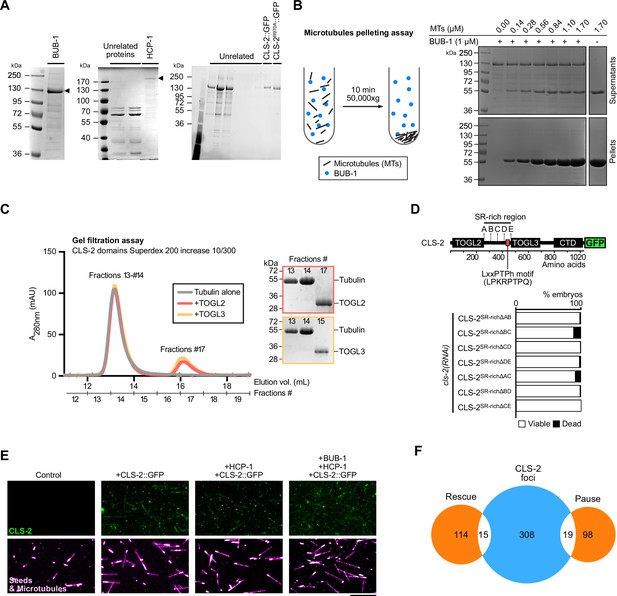
CLS-2 decorates the microtubule lattice in vitro.
(A) Coomassie-stained gels of purified proteins used for in vitro assays. Arrowheads indicate the protein of interest. (B) Microtubule pelleting assay in the presence of 1 µM purified BUB-1 protein. Schematic of the experiment principle (left) and Coomassie staining of supernatant and pellet fractions (right) with indicated concentrations of microtubules (MTs). (C) Gel filtration assay of tubulin in the presence of indicated CLS-2 TOGL domains. Coomassie staining of the fractions of interest are shown on the right. (D) Embryonic viability assay of worms carrying cls-2::gfp transgenes with indicated truncations in the S/R-rich region, and upon depletion of endogenous cls-2. Position of the divergent LxxPTPh motif is indicated on the protein fusion diagram. (E–F) TIRF-microscopy localization of 100 nM purified CLS-2::GFP protein (green) on microtubules (magenta). Stills from imaging in indicated conditions (E) and Venn diagram of CLS-2::GFP foci associated with microtubule rescue and pause events (F). Scale bar 10 µm.
-
Figure 6—figure supplement 1—source data 1
Panels A-B source data.
Raw images and uncropped annotated image of Coomassie-stained gels for purification of BUB-1, HCP-1, CLS-2::GFP and CLS-2R970A::GFP proteins, and of microtubule/BUB-1 pelleting assay.
- https://cdn.elifesciences.org/articles/82579/elife-82579-fig6-figsupp1-data1-v2.zip
-
Figure 6—figure supplement 1—source data 2
Panel C source data.
Raw images and uncropped annotated image of Coomassie-stained gels for protein fractions of gel filtration assay.
- https://cdn.elifesciences.org/articles/82579/elife-82579-fig6-figsupp1-data2-v2.zip
-
Figure 6—figure supplement 1—source data 3
Panel D source data.
Embryonic viability assay of worms carrying a cls-2::gfp transgene with mutations in the S/R-rich domain, upon depletion of cls-2 compared to non-depleted controls.
- https://cdn.elifesciences.org/articles/82579/elife-82579-fig6-figsupp1-data3-v2.xlsx
Videos
Live imaging of meiosis I in indicated conditions.
Microtubules (GFP::TBA-2α-tubulin) in green, DNA (mCherry::HIS-11H2B) in magenta. Time in seconds relative to anaphase I onset. Scale bar 5 µm.
Live imaging of meiosis I in knl-1∆85–505::mCherry transgenic worms in indicated conditions.
Microtubules (GFP::TBA-2α-tubulin) in green, DNA (mCherry::HIS-11H2B) in magenta. Time in seconds relative to anaphase I onset. Scale bar 5 µm.
Live imaging of meiosis I in indicated transgenic hcp-2-mutant worms upon depletion of endogenous hcp-1.
Microtubules (GFP::TBA-2α-tubulin) in green, DNA (mCherry::HIS-11H2B) in magenta. Time in seconds relative to anaphase I onset. Scale bar 5 µm.
Live imaging of meiosis I in hcp-2-mutant worms expressing CLS-2::GFP::HCP-111154-1386 fusion protein in indicated conditions.
Microtubules (GFP::TBA-2α-tubulin) in green, DNA (mCherry::HIS-11H2B) in magenta. Time in seconds relative to anaphase I onset. Scale bar 5 µm.
Live imaging of meiosis I in hcp-2-mutant worms expressing CLS-2∆CTD::GFP:: HCP-11154-1386 fusion protein in indicated conditions.
Microtubules (GFP::TBA-2α-tubulin) in green, DNA (mCherry::HIS-11H2B) in magenta. Time in seconds relative to anaphase I onset. Scale bar 5 µm.
Live imaging of meiosis I in indicated transgenic worms upon depletion of endogenous cls-2.
Microtubules (GFP::TBA-2α-tubulin) in green, DNA (mCherry::HIS-11H2B) in magenta. Time in seconds relative to anaphase I onset. Scale bar 5 µm.
TIRF microscopy-mediated live imaging of in vitro microtubule (magenta) polymerization dynamics from GMPCPP seeds (cyan) in the presence of indicated protein (100 nm each).
Scale bar 10 µm.
Additional files
-
MDAR checklist
- https://cdn.elifesciences.org/articles/82579/elife-82579-mdarchecklist1-v2.pdf
-
Supplementary file 1
List of C. elegans strains used in this study.
- https://cdn.elifesciences.org/articles/82579/elife-82579-supp1-v2.docx





