Ribosomal RNA (rRNA) sequences from 33 globally distributed mosquito species for improved metagenomics and species identification
Figures
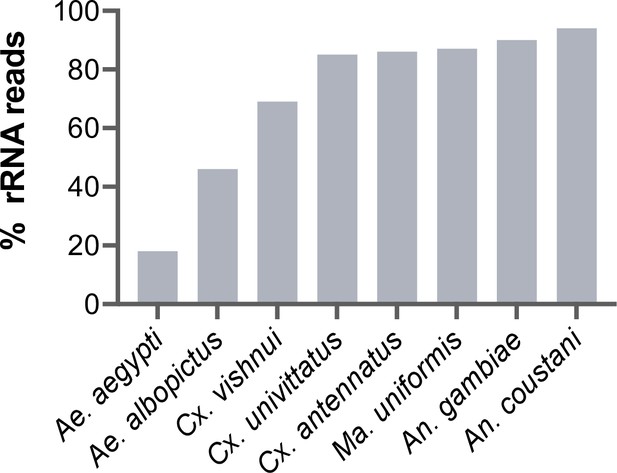
Percentage of rRNA reads in mosquito total RNA sequencing (RNA-seq) data after depletion using probes antisense to Aedes aegypti sequences.
Pools of five individual mosquitoes from genera Aedes (Ae), Culex (Cx), Mansonia (Ma), and Anopheles (An) were ribodepleted by probe hybridisation followed by RNase H digestion according to the protocol by Morlan et al., 2012. Y-axis depicts percentages of remaining rRNA reads calculated as the number of rRNA reads over total reads per sample pool. Depletion efficiency decreases with taxonomic distance from Ae. aegypti underlining the need for reference sequences for species of interest.
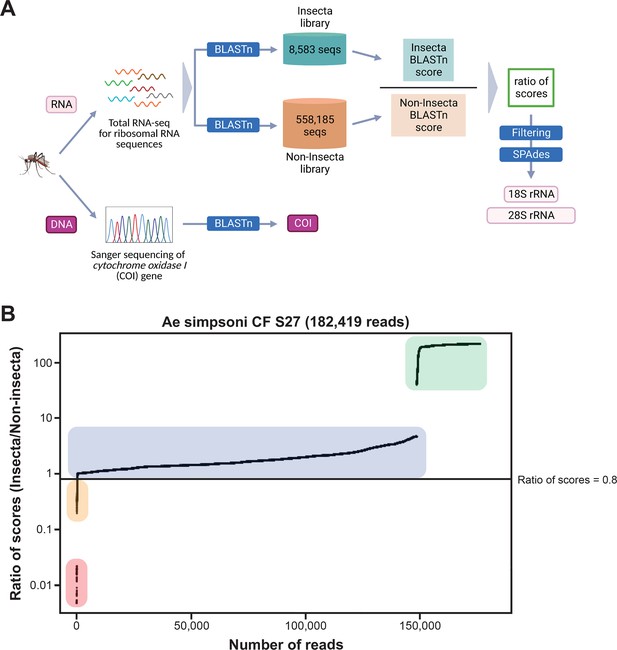
Novel mosquito rRNA sequences were obtained using a unique reads filtering method.
(A) Schematic of sequencing and bioinformatics analyses performed in this study to obtain full-length 18S and 28S rRNA sequences as well as cytochrome c oxidase I (COI) DNA sequences. Nucleic acids were isolated from mosquito specimens for next-generation (for rRNA) or Sanger (for COI) sequencing. Two in-house libraries were created from the SILVA rRNA gene database: Insecta and Non-Insecta, which comprises 8,585 sequences and 558,185 sequences, respectively. Following BLASTn analyses against these two libraries, each RNA-sequencing (RNA-seq) read is assigned a ratio of BLASTn scores to describe their relative nucleotide similarity to insect rRNA sequences. Based on these ratios of scores, RNA-seq reads can then be filtered to remove non-mosquito reads prior to assembly with SPAdes to give full-length 18S and 28S rRNA sequences. Image created with https://biorender.com/. (B) Based on their ratio of scores, reads can be segregated into four categories, as shown on this ratio of scores versus number of reads plot for the representative specimen ‘CF S27’: (i) reads with hits only in the Insecta library (shaded in green), (ii) reads with a higher score against the Insecta library (shaded in blue), (iii) reads with a higher score against the Non-Insecta library (shaded in yellow), and (iv) reads with no hits in the Insecta library (shaded in red). We applied a conservative threshold at 0.8, indicated by the black horizontal line, where only reads above this threshold are used in the assembly with SPAdes. For this given specimen, 175,671 reads (96.3% of total reads) passed the ≥0.8 cut-off, 325 reads (0.18% of total reads) had ratios of scores <0.8, while 6,423 reads (3.52%) did not have hits against the Insecta library.
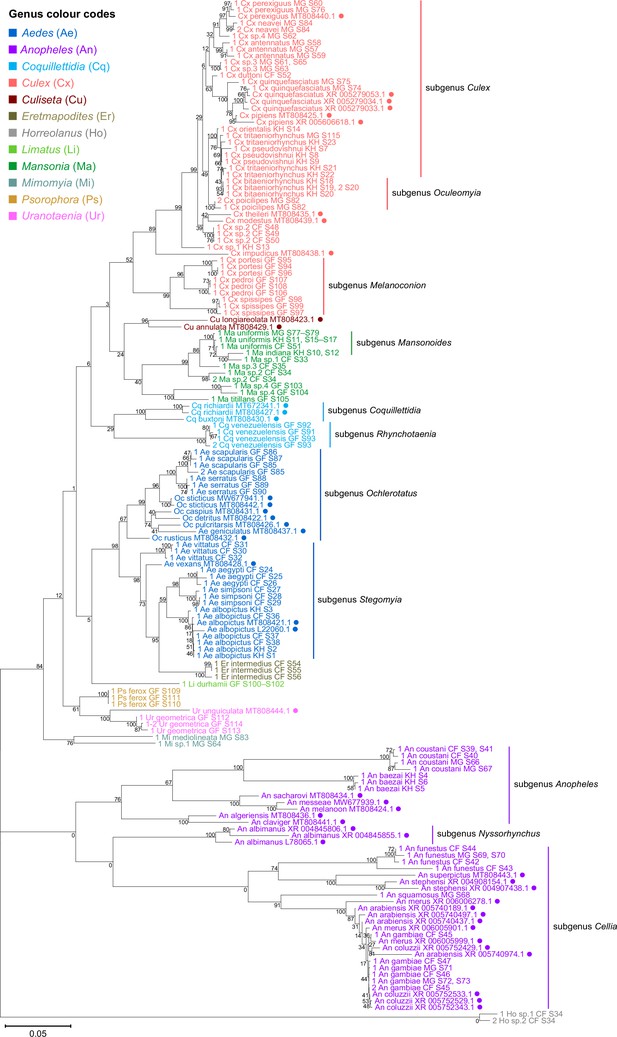
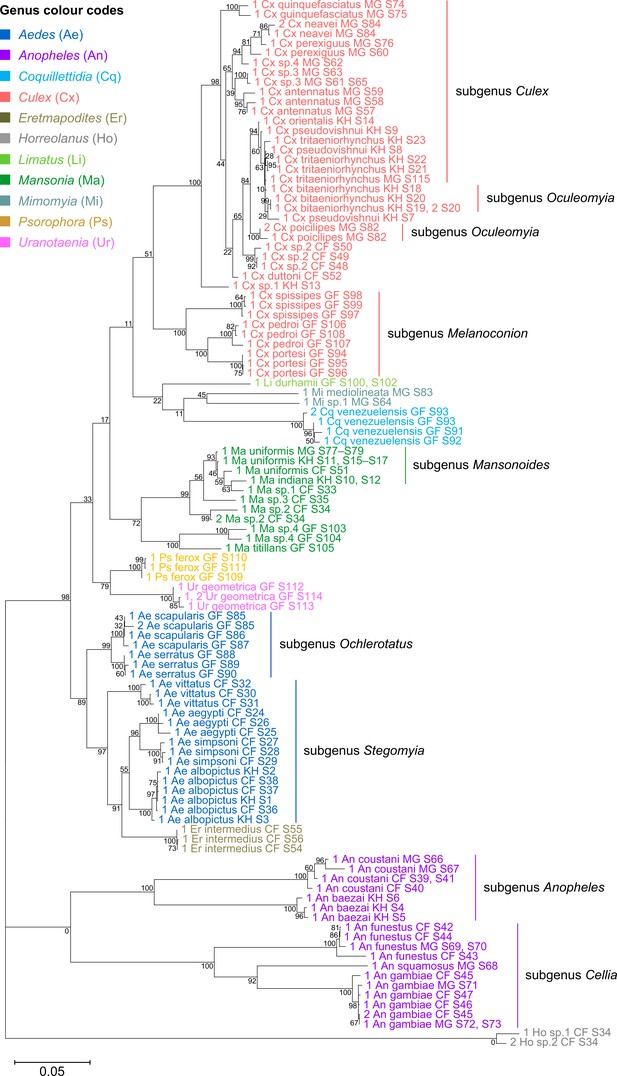
28S sequences generated from this study clustered with conspecifics or congenerics from existing GenBank records.
A rooted phylogenetic tree based on full-length 28S sequences (3,900 bp) from this study and from GenBank was inferred using the maximum-likelihood method and constructed to scale in MEGA X (Kumar et al., 2018) using an unknown Horreolanus species found among our samples as an outgroup. Values at each node indicate bootstrap support (%) from 500 replications. Sequences from GenBank are annotated with filled circles and their accession numbers are shown. For sequences from this study, each specimen label contains information on taxonomy, origin (in two-letter country codes), and specimen ID number. Some specimens produced up to two consensus 28S sequences; this is indicated by the numbers 1 or 2 at the beginning of the specimen label. Specimen genera are indicated by colour: Culex in coral, Anopheles in purple, Aedes in dark blue, Mansonia in dark green, Culiseta in maroon, Limatus in light green, Coquillettidia in light blue, Psorophora in yellow, Mimomyia in teal, Uranotaenia in pink, and Eretmapodites in brown. Scale bar at 0.05 is shown.
-
Figure 3—source data 1
Multiple sequence alignment of 169 28S rRNA sequences from this study and from GenBank (FASTA).
- https://cdn.elifesciences.org/articles/82762/elife-82762-fig3-data1-v2.zip
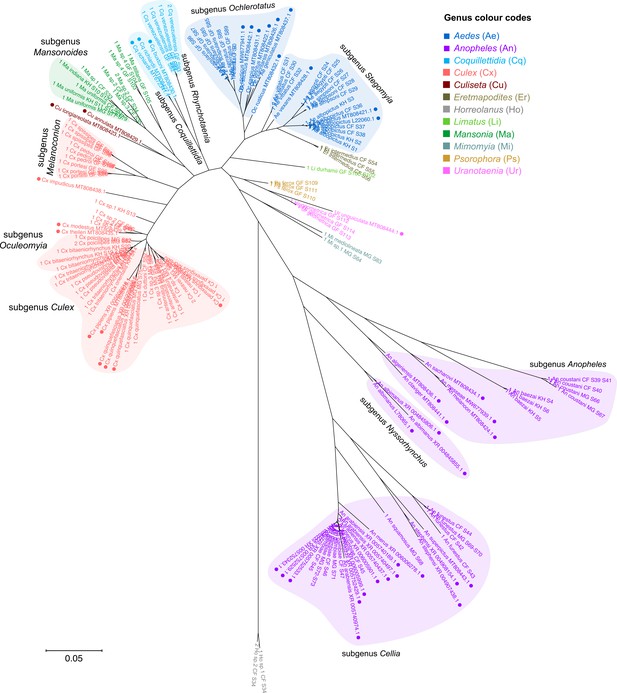
Interspecific and intersubgeneric distances within the genus Anopheles indicate a greater degree of divergence than those within any other genera of family Culicidae.
The phylogenetic tree presented in Figure 3 based on 28S sequences from this study and from GenBank (annotated with filled circles) is depicted here in radial format to illustrate how the branch lengths separating Anopheline taxa are longer relative to other members of family Culicidae. An unknown Horreolanus species found among our samples serves as an outgroup. For sequences from this study, each specimen label contains information on taxonomy, origin (in two-letter country codes), and specimen ID number. Some specimens produced up to two consensus 28S sequences; this is indicated by the numbers 1 or 2 at the beginning of the specimen label. Specimen genera are indicated by colour: Culex in coral, Anopheles in purple, Aedes in dark blue, Mansonia in dark green, Limatus in light green, Coquillettidia in light blue, Psorophora in yellow, Mimomyia in teal, Uranotaenia in pink, and Eretmapodites in brown. Scale bar at 0.05 is shown.

Sequence conservation among 169 28S rRNA sequences obtained from this study and from GenBank combined.
Multiple sequence alignment was performed on 28S rRNA sequences, 3,900 bp in length. Each bar represents a 25 bp sliding window of the 28S rRNA sequence alignment where the y-axis values are the lowest percentage nucleotide identity found.
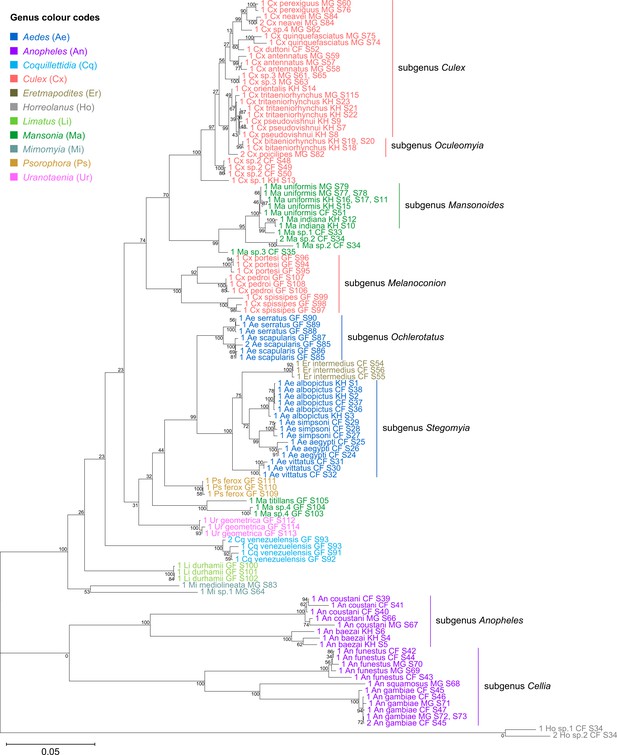
Concatenating 28S and 18S rRNA sequences produces phylogenetic relationships that are concordant with classical Culicidae systematics with higher bootstrap support than 28S sequences alone.
This phylogenetic tree based on concatenated 28S+18S rRNA sequences (3,900+1,900 bp) generated from this study was inferred using the maximum-likelihood method and constructed to scale using MEGA X (Kumar et al., 2018) using an unknown Horreolanus species found among our samples as an outgroup. Values at each node indicate bootstrap support (%) from 500 replications. Each specimen label contains information on taxonomy, origin (as indicated in two-letter country codes), and specimen ID number. Some specimens produced up to two consensus 28S+18S rRNA sequences; this is indicated by the numbers 1 or 2 at the beginning of the specimen label. Specimen genera are indicated by colour: Culex in coral, Anopheles in purple, Aedes in dark blue, Mansonia in dark green, Limatus in light green, Coquillettidia in light blue, Psorophora in yellow, Mimomyia in teal, Uranotaenia in pink, and Eretmapodites in brown. Scale bar at 0.05 is shown.
-
Figure 4—source data 1
Multiple sequence alignment of 122 28S rRNA sequences, including two sequences from Horreolanus sp. (FASTA).
- https://cdn.elifesciences.org/articles/82762/elife-82762-fig4-data1-v2.zip
-
Figure 4—source data 2
Multiple sequence alignment of 114 18S rRNA sequences, including two sequences from Horreolanus sp. (FASTA).
- https://cdn.elifesciences.org/articles/82762/elife-82762-fig4-data2-v2.zip
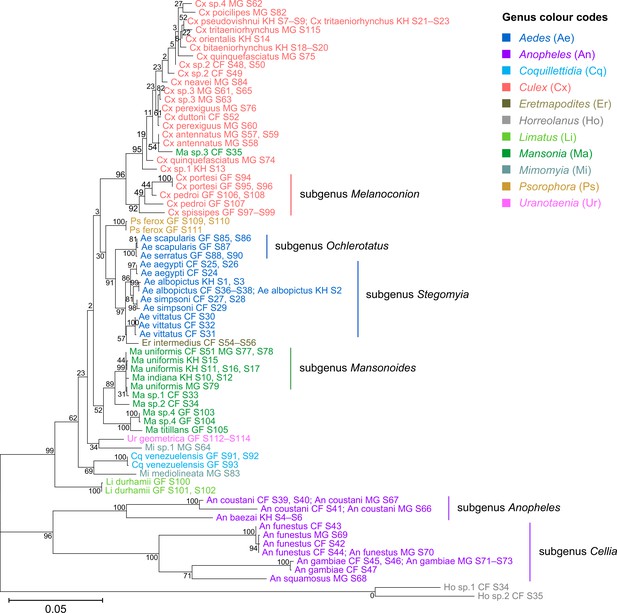
Phylogenetic tree based on 28S rRNA sequences generated from this study (3,900 bp).
This tree was inferred using maximum-likelihood method and constructed to scale in MEGA X (Kumar et al., 2018) using an unknown Horreolanus species found among our samples as an outgroup. Values at each node indicate bootstrap support (%) from 500 replications. For sequences from this study, each specimen label contains information on taxonomy, origin (in two-letter country codes), and specimen ID number. Some specimens produced up to two consensus 28S rRNA sequences; this is indicated by the numbers 1 or 2 at the beginning of the specimen label. Specimen genera are indicated by colour: Culex in coral, Anopheles in purple, Aedes in dark blue, Mansonia in dark green, Limatus in light green, Coquillettidia in light blue, Psorophora in yellow, Mimomyia in teal, Uranotaenia in pink, and Eretmapodites in brown. Scale bar at 0.05 is shown.
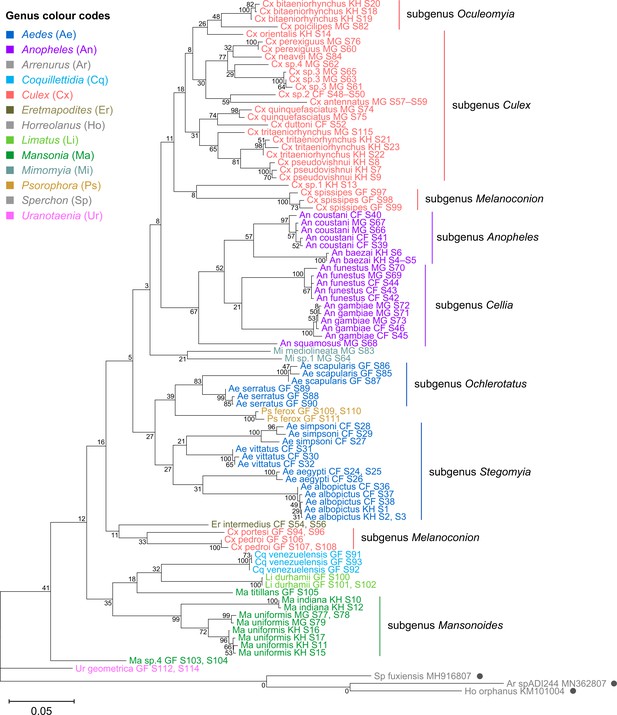
Phylogenetic tree based on 18S rRNA sequences (1,900 bp).
This tree was inferred using maximum-likelihood method and constructed to scale in MEGA X (Kumar et al., 2018) using an unknown Horreolanus species found among our samples as an outgroup. Values at each node indicate bootstrap support (%) from 500 replications. For sequences from this study, each specimen label contains information on taxonomy, origin (in two-letter country codes), and specimen ID number. One 18S rRNA sequence was obtain for each specimen. Specimen genera are indicated by colour: Culex in coral, Anopheles in purple, Aedes in dark blue, Mansonia in dark green, Limatus in light green, Coquillettidia in light blue, Psorophora in yellow, Mimomyia in teal, Uranotaenia in pink, and Eretmapodites in brown. Scale bar at 0.05 is shown.
Cytochrome c oxidase I (COI) sequences cluster by species but show phylogenetic relationships that contrast those derived from rRNA trees.
A phylogenetic tree based on COI sequences (621–699 bp) was inferred using the maximum-likelihood method and constructed to scale using MEGA X (Kumar et al., 2018) with three water mite species to serve as outgroups. Outgroup sequences obtained from GenBank are annotated with filled circles and their accession numbers are shown. Values at each node indicate bootstrap support (%) from 500 replications. Each specimen label contains information on taxonomy, origin (as indicated in two-letter country codes), and specimen ID. Specimen genera are indicted by colour: Culex in coral, Anopheles in purple, Aedes in dark blue, Mansonia in dark green, Limatus in light green, Coquillettidia in light blue, Psorophora in yellow, Mimomyia in teal, Uranotaenia in pink, and Eretmapodites in brown. Scale bar at 0.05 is shown.
-
Figure 5—source data 1
Multiple sequence alignment of 106 cytochrome c oxidase I (COI) sequences (FASTA).
- https://cdn.elifesciences.org/articles/82762/elife-82762-fig5-data1-v2.zip
Tables
Mosquito species represented in this study and their vector status.
| Mosquito taxonomy‡ | Origin* | Collection site (ecosystem type) | Vector for† | Reference |
|---|---|---|---|---|
| Aedes (Fredardsius) vittatus | CF | Rural (village) | ZIKV, CHIKV, YFV | Diallo et al., 2020 |
| Aedes (Ochlerotatus) scapularis | GF | Rural (village) | YFV | Vasconcelos et al., 2001 |
| Aedes (Ochlerotatus) serratus | GF | Rural (village) | YFV, OROV | Cardoso et al., 2010; Romero-Alvarez and Escobar, 2018 |
| Aedes (Stegomyia) aegypti | CF | Urban | DENV, ZIKV, CHIKV, YFV | Kraemer et al., 2019 |
| Aedes (Stegomyia) albopictus | CF, KH | Rural (village, nature reserve) | DENV, ZIKV, CHIKV, YFV, JEV | Auerswald et al., 2021; Kraemer et al., 2019 |
| Aedes (Stegomyia) simpsoni | CF | Rural (village) | YFV | Mukwaya et al., 2000 |
| Anopheles (Anopheles) baezai | KH | Rural (nature reserve) | Unreported | – |
| Anopheles (Anopheles) coustani | MG, CF | Rural (village) | RVFV, malaria | Mwangangi et al., 2013; Nepomichene et al., 2018; Ratovonjato et al., 2011 |
| Anopheles (Cellia) funestus | MG, CF | Rural (village) | ONNV, malaria | Lutomiah et al., 2013; Tabue et al., 2017 |
| Anopheles (Cellia) gambiae | MG, CF | Rural (village) | ONNV, malaria | Brault et al., 2004 |
| Anopheles (Cellia) squamosus | MG | Rural (village) | RVFV, malaria | Ratovonjato et al., 2011; Stevenson et al., 2016 |
| Coquillettidia (Rhynchotaenia) venezuelensis | GF | Rural (village) | OROV | Travassos da Rosa et al., 2017 |
| Culex (Culex) antennatus | MG | Rural (village) | RVFV | Nepomichene et al., 2018; Ratovonjato et al., 2011 |
| Culex (Culex) duttoni | CF | Rural (village) | Unreported | – |
| Culex (Culex) neavei | MG | Rural (village) | USUV | Nikolay et al., 2011 |
| Culex (Culex) orientalis | KH | Rural (nature reserve) | JEV | Kim et al., 2015 |
| Culex (Culex) perexiguus | MG | Rural (village) | WNV, USUV | Vezenegho et al., 2022 |
| Culex (Culex) pseudovishnui | KH | Rural (nature reserve) | JEV | Auerswald et al., 2021 |
| Culex (Culex) quinquefasciatus | MG, CF, KH | Rural (village, nature reserve) | ZIKV, JEV, WNV, DENV, SLEV, RVFV, Wuchereria bancrofti | Bhattacharya and Basu, 2016; Maquart et al., 2021; Ndiaye et al., 2016; Serra et al., 2016 |
| Culex (Culex) tritaeniorhynchus | MG, KH | Rural (village, nature reserve) | JEV, WNV, RVFV | Auerswald et al., 2021; Hayes et al., 1980; Jupp et al., 2002 |
| Culex (Melanoconion) spissipes | GF | Rural (village) | VEEV | Weaver et al., 2004 |
| Culex (Melanoconion) portesi | GF | Rural (village) | VEEV, TONV | Talaga et al., 2021; Weaver et al., 2004 |
| Culex (Melanoconion) pedroi | GF | Rural (village) | EEEV, VEEV, MADV | Talaga et al., 2021; Turell et al., 2008 |
| Culex (Oculeomyia) bitaeniorhynchus | MG, KH | Rural (village, nature reserve) | JEV | Auerswald et al., 2021 |
| Culex (Oculeomyia) poicilipes | MG | Rural (village) | RVFV | Ndiaye et al., 2016 |
| Eretmapodites intermedius | CF | Rural (village) | Unreported | – |
| Limatus durhamii | GF | Rural (village) | ZIKV | Barrio-Nuevo et al., 2020 |
| Mansonia (Mansonia) titillans | GF | Rural (village) | VEEV, SLEV | Hoyos-López et al., 2015; Turell, 1999 |
| Mansonia (Mansonioides) indiana | KH | Rural (nature reserve) | JEV | Arunachalam et al., 2004 |
| Mansonia (Mansonioides) uniformis | MG, CF, KH | Rural (village, nature reserve) | RVFV, Wuchereria bancrofti | Lutomiah et al., 2013; Ughasi et al., 2012 |
| Mimomyia (Etorleptiomyia) mediolineata | MG | Rural (village) | Unreported | – |
| Psorophora (Janthinosoma) ferox | GF | Rural (village) | ROCV | Mitchell et al., 1986 |
| Uranotaenia (Uranotaenia) geometrica | GF | Rural (village) | Unreported | – |
-
*
Dengue virus, DENV; Zika virus, ZIKV; chikungunya virus, CHIKV; Yellow Fever virus, YFV; Oropouche virus, OROV; Japanese encephalitis virus, JEV; Rift Valley Fever virus, RVFV; O’Nyong Nyong virus, ONNV; Usutu virus, USUV; West Nile virus, WNV; St Louis encephalitis virus, SLEV; Venezuelan equine encephalitis virus, VEEV; Tonate virus, TONV; Eastern equine encephalitis virus, EEEV; Madariaga virus, MADV; Rocio virus, ROCV.
-
†
Origin countries are listed as their ISO alpha-2 codes: Central African Republic, CF; Cambodia, KH; Madagascar, MG; French Guiana, GF.
-
‡
Subgenus indicated in brackets.
Summary of differences between rRNA and cytochrome c oxidase I (COI) phylogenies.
| Taxa | 28S+18S rRNA phylogeny (Figure 4) | COI phylogeny (Figure 5) |
|---|---|---|
| The Anopheles genus | Forms a clade that is basal to the all other members of family Culicidae; interspecies branch lengths are notably long | Forms a sister clade to the Culex genus, and is depicted to have diverged more recently; interspecies branch lengths are comparable to that of other genera |
| The Ur. geometrica species | Forms a clade within the Culicinae subfamily lineage | Forms a clade that is basal to the all other members of family Culicidae |
| The Aedini tribe | Forms a monophyletic clade comprising the genera Aedes, Eretmapodites, and Psorophora, with the latter being an early divergent lineage | Does not form a monophyletic clade; the Psorophora clade is placed among Aedes taxa and the Eretmapodites clade is sister to a Culex subgenus Melanoconion clade |
| The Culex genus | Splits into two monophyletic clades with the three French Guyanese species forming a closely related minor clade | Splits into two clades with two out of three French Guyanese species (Cx. pedroi and Cx. portesi) forming a distantly related minor clade, while the third (Cx. spissipes) is a part of the greater clade |
| The Mansonia genus | Is a polyphyletic group comprising two clades with the two French Guyanese taxa forming a distantly related minor clade; the major clade is placed among Culex taxa | Forms a subgenus Mansonoides clade as per the 28S+18S rRNA tree but the French Guyanese taxa do not cluster together; is depicted to have diverged earlier relative to other taxa in the assemblage |
| The Ma sp.4 species | Forms a sister clade to Ma. titillans as part of a minor French Guyanese Mansonia clade | Does not form a sister clade to Ma. titillans; instead is shown to have diverged earlier than all other members of family Culicidae after Ur. geometrica |
Comparison of 28S or concatenated 28S+18S rRNA and cytochrome c oxidase I (COI) sequences as molecular markers.
| 28S+18S rRNA | |
|---|---|
| Advantages | Disadvantages |
|
|
| COI | |
| Advantages | Disadvantages |
|
|
Taxonomic and sampling information on mosquito specimens and associated accession numbers of their cytochrome c oxidase I (COI), 18S rRNA, and 28S rRNA sequences (XLSX).
| Sequence ID | Taxonomy [Genus (subgenus) species] | Origin | Collection site | Collection period | Blood engorged (Y/N) | Sample ID | COI accession number | 18S rRNA accession number | 28S rRNA accession number |
|---|---|---|---|---|---|---|---|---|---|
| Ae_albopictus_KH_S1 | Aedes (Stegomyia) albopictus | Cambodia | Rattanakiri | Dec 2019 | N | 1 | OM630613 | OM350214 | OM542460 |
| Ae_albopictus_KH_S2 | Aedes (Stegomyia) albopictus | Cambodia | Rattanakiri | Dec 2019 | N | 2 | OM630614 | OM350220 | OM542373 |
| Ae_albopictus_KH_S3 | Aedes (Stegomyia) albopictus | Cambodia | Rattanakiri | Dec 2019 | N | 3 | OM630615 | OM350316 | OM542374 |
| An_baezai_KH_S4 | Anopheles (Anopheles) baezai | Cambodia | Koh Kong | Mar 2019 | N | 4 | OM630631 | OM350327 | OM542357 |
| An_baezai_KH_S5 | Anopheles (Anopheles) baezai | Cambodia | Koh Kong | Mar 2019 | N | 5 | OM630632 | OM350233 | OM542440 |
| An_baezai_KH_S6 | Anopheles (Anopheles) baezai | Cambodia | Koh Kong | Mar 2019 | N | 6 | OM630633 | OM350234 | OM542358 |
| Cx_pseudovishnui_KH_S7 | Culex (Culex) pseudovishnui | Cambodia | Rattanakiri | Dec 2019 | N | 7 | OM630689 | OM350285 | OM542413 |
| Cx_pseudovishnui_KH_S8 | Culex (Culex) pseudovishnui | Cambodia | Rattanakiri | Dec 2019 | N | 8 | OM630690 | OM350286 | OM542414 |
| Cx_pseudovishnui_KH_S9 | Culex (Culex) pseudovishnui | Cambodia | Rattanakiri | Dec 2019 | N | 9 | OM630691 | OM350287 | OM542415 |
| Ma_indiana_KH_S10 | Mansonia (Mansonioides) indiana | Cambodia | Battambong | Nov 2019 | N | 10 | OM630698 | OM350295 | OM542422 |
| Ma_uniformis_KH_S11 | Mansonia (Mansonioides) uniformis | Cambodia | Battambong | Nov 2019 | N | 11 | OM630699 | OM350296 | OM542423 |
| Ma_indiana_KH_S12 | Mansonia (Mansonioides) indiana | Cambodia | Battambong | Nov 2019 | N | 12 | OM630700 | OM350297 | OM542424 |
| Cx_sp.1_KH_S13 | Culex sp.1 | Cambodia | Prek Toal | Feb 2019 | N | 13 | OM630672 | OM350267 | OM542395 |
| Cx_orientalis_KH_S14 | Culex (Culex) orientalis | Cambodia | Prek Toal | Feb 2019 | N | 14 | OM630673 | OM350268 | OM542396 |
| Ma_uniformis_KH_S15 | Mansonia (Mansonioides) uniformis | Cambodia | Battambong | Nov 2019 | N | 15 | OM630705 | OM350303 | OM542430 |
| Ma_uniformis_KH_S16 | Mansonia (Mansonioides) uniformis | Cambodia | Battambong | Nov 2019 | N | 16 | OM630706 | OM350305 | OM542432 |
| Ma_uniformis_KH_S17 | Mansonia (Mansonioides) uniformis | Cambodia | Battambong | Nov 2019 | N | 17 | OM630707 | OM350304 | OM542431 |
| Cx_bitaeniorhynchus_KH_S18 | Culex (Oculeomyia) bitaeniorhynchus | Cambodia | Battambong | Nov 2019 | N | 18 | OM630656 | OM350255 | OM542381 |
| Cx_bitaeniorhynchus_KH_S19 | Culex (Oculeomyia) bitaeniorhynchus | Cambodia | Battambong | Nov 2019 | N | 19 | OM630657 | OM350256 | OM542382 |
| Cx_bitaeniorhynchus_KH_S20 | Culex (Oculeomyia) bitaeniorhynchus | Cambodia | Battambong | Nov 2019 | N | 20 | OM630658 | OM350257 | OM542383, OM542384 |
| Cx_tritaeniorhynchus_KH_S21 | Culex (Culex) tritaeniorhynchus | Cambodia | Battambong | Nov 2019 | N | 21 | OM630680 | OM350277 | OM542404 |
| Cx_tritaeniorhynchus_KH_S22 | Culex (Culex) tritaeniorhynchus | Cambodia | Battambong | Nov 2019 | N | 22 | OM630681 | OM350278 | OM542405 |
| Cx_tritaeniorhynchus_KH_S23 | Culex (Culex) tritaeniorhynchus | Cambodia | Battambong | Nov 2019 | N | 23 | OM630682 | OM350279 | OM542406 |
| Ae_aegypti_CF_S24 | Aedes (Stegomyia) aegypti | Central African Republic | Pissa | Jun 2019 | N | 24 | OM630610 | OM350314 | OM542339 |
| Ae_aegypti_CF_S25 | Aedes (Stegomyia) aegypti | Central African Republic | Pissa | Jun 2019 | N | 25 | OM630611 | OM350215 | OM542340 |
| Ae_aegypti_CF_S26 | Aedes (Stegomyia) aegypti | Central African Republic | Pissa | Jun 2019 | N | 26 | OM630612 | OM350216 | OM542341 |
| Ae_simpsoni_CF_S27 | Aedes (Stegomyia) simpsoni | Central African Republic | Pissa | Jun 2019 | N | 27 | OM630619 | OM350221 | OM542345 |
| Ae_simpsoni_CF_S28 | Aedes (Stegomyia) simpsoni | Central African Republic | Pissa | Jun 2019 | N | 28 | OM630620 | OM350222 | OM542346 |
| Ae_simpsoni_CF_S29 | Aedes (Stegomyia) simpsoni | Central African Republic | Pissa | Jun 2019 | N | 29 | OM630621 | OM350223 | OM542347 |
| Ae_vittatus_CF_S30 | Aedes (Fredwardsius) vittatus | Central African Republic | Gbozo | Aug 2019 | Y | 30 | OM630628 | OM350230 | OM542439 |
| Ae_vittatus_CF_S31 | Aedes (Fredwardsius) vittatus | Central African Republic | Gbozo | Aug 2019 | N | 31 | OM630629 | OM350231 | OM542355 |
| Ae_vittatus_CF_S32 | Aedes (Fredwardsius) vittatus | Central African Republic | Gbozo | Aug 2019 | N | 32 | OM630630 | OM350232 | OM542356 |
| Ma_sp.1_CF_S33 | Mansonia sp.1 | Central African Republic | Bayanga | Nov 2019 | Y | 33 | N/A | OM350294 | OM542449 |
| Ma_sp.2_CF_S34 | Mansonia sp.2 | Central African Republic | Bayanga | Nov 2019 | Y | 34 | N/A | OM350322 | OM542450, OM542456 |
| Ho_sp.1_CF_S34 | Horreolanus sp.1 | Central African Republic | Bayanga | Nov 2019 | – | 34 | N/A | OM350325 | OM542457 |
| Ho_sp.2_CF_S34 | Horreolanus sp.2 | Central African Republic | Bayanga | Nov 2019 | – | 34 | N/A | OM350326 | OM542458 |
| Ma_sp.3_CF_S35 | Mansonia sp.3 | Central African Republic | Bayanga | Nov 2019 | Y | 35 | N/A | OM350323 | OM542451 |
| Ae_albopictus_CF_S36 | Aedes (Stegomyia) albopictus | Central African Republic | Pissa | Jun 2019 | N | 36 | OM630616 | OM350217 | OM542342 |
| Ae_albopictus_CF_S37 | Aedes (Stegomyia) albopictus | Central African Republic | Pissa | Jun 2019 | N | 37 | OM630617 | OM350218 | OM542343 |
| Ae_albopictus_CF_S38 | Aedes (Stegomyia) albopictus | Central African Republic | Pissa | Jun 2019 | N | 38 | OM630618 | OM350219 | OM542344 |
| An_coustani_CF_S39 | Anopheles (Anopheles) coustani | Central African Republic | Pissa | Jan 2020 | N | 39 | OM630634 | OM350235 | OM542359 |
| An_coustani_CF_S40 | Anopheles (Anopheles) coustani | Central African Republic | Pissa | Jan 2020 | N | 40 | OM630635 | OM350236 | OM542360 |
| An_coustani_CF_S41 | Anopheles (Anopheles) coustani | Central African Republic | Pissa | Jan 2020 | N | 41 | OM630636 | OM350237 | OM542361 |
| An_funestus_CF_S42 | Anopheles (Cellia) funestus | Central African Republic | Pissa | Jun 2019 | Y | 42 | OM630640 | OM350241 | OM542365 |
| An_funestus_CF_S43 | Anopheles (Cellia) funestus | Central African Republic | Pissa | Jun 2019 | Y | 43 | OM630641 | OM350242 | OM542366 |
| An_funestus_CF_S44 | Anopheles (Cellia) funestus | Central African Republic | Pissa | Jun 2019 | Y | 44 | OM630642 | OM350243 | OM542367 |
| An_gambiae_CF_S45 | Anopheles (Cellia) gambiae | Central African Republic | Pissa | Jun 2019 | Y | 45 | OM630645 | OM350245 | OM542369, OM542370 |
| An_gambiae_CF_S46 | Anopheles (Cellia) gambiae | Central African Republic | Pissa | Jun 2019 | Y | 46 | OM630646 | OM350246 | OM542371 |
| An_gambiae_CF_S47 | Anopheles (Cellia) gambiae | Central African Republic | Pissa | Jun 2019 | Y | 47 | N/A | OM350247 | OM542372 |
| Cx_sp.2_CF_S48 | Culex sp.2 | Central African Republic | Bayanga | Nov 2019 | Y | 48 | OM630669 | OM350269 | OM542446 |
| Cx_sp.2_CF_S49 | Culex sp.2 | Central African Republic | Bayanga | Nov 2019 | Y | 49 | OM630670 | OM350315 | OM542397 |
| Cx_sp.2_CF_S50 | Culex sp.2 | Central African Republic | Bayanga | Nov 2019 | Y | 50 | OM630671 | OM350270 | OM542398 |
| Ma_uniformis_CF_S51 | Mansonia (Mansonioides) uniformis | Central African Republic | Bouar | May 2019 | Y | 51 | N/A | OM350301 | OM542428 |
| Cx_duttoni_CF_S52 | Culex (Culex) duttoni | Central African Republic | Mbaiki | Jan 2019 | Y | 52 | OM630704 | OM350302 | OM542429 |
| Er_intermedius_CF_S54 | Eretmapodites intermedius | Central African Republic | Pissa | Jun 2019 | N | 54 | OM630692 | OM350288 | OM542416 |
| Er_intermedius_CF_S55 | Eretmapodites intermedius | Central African Republic | Pissa | Jun 2019 | N | 55 | OM630693 | OM350289 | OM542417 |
| Er_intermedius_CF_S56 | Eretmapodites intermedius | Central African Republic | Pissa | Jun 2019 | N | 56 | OM630694 | OM350290 | OM542418 |
| Cx_antennatus_MG_S57 | Culex (Culex) antennatus | Madagascar | Ambato Boeny | Feb 2019 | N | 57 | OM630653 | OM350253 | OM542379 |
| Cx_antennatus_MG_S58 | Culex (Culex) antennatus | Madagascar | Ambato Boeny | Feb 2019 | N | 58 | OM630654 | OM350319 | OM542444 |
| Cx_antennatus_MG_S59 | Culex (Culex) antennatus | Madagascar | Ambato Boeny | Feb 2019 | N | 59 | OM630655 | OM350254 | OM542380 |
| Cx_perexiguus_MG_S60 | Culex (Culex) perexiguus | Madagascar | Amparafaravola | Feb 2019 | N | 60 | OM630660 | OM350258 | OM542386 |
| Cx_sp.3_MG_S61 | Culex sp.3 | Madagascar | Ambato Boeny | Aug 2019 | N | 61 | OM630661 | OM350259 | OM542387 |
| Cx_sp.4_MG_S62 | Culex sp.4 | Madagascar | Ambato Boeny | Aug 2019 | N | 62 | OM630662 | OM350260 | OM542388 |
| Cx_sp.3_MG_S63 | Culex sp.3 | Madagascar | Ambato Boeny | Feb 2019 | N | 63 | OM630686 | OM350282 | OM542410 |
| Mi_sp.1_MG_S64 | Mimomyia sp.1 | Madagascar | Ambato Boeny | Feb 2019 | N | 64 | OM630687 | OM350283 | OM542411 |
| Cx_sp.3_MG_S65 | Culex sp.3 | Madagascar | Ambato Boeny | Feb 2019 | N | 65 | OM630688 | OM350284 | OM542412 |
| An_coustani_MG_S66 | Anopheles (Anopheles) coustani | Madagascar | Ambato Boeny | Feb 2019 | N | 66 | OM630637 | OM350238 | OM542362 |
| An_coustani_MG_S67 | Anopheles (Anopheles) coustani | Madagascar | Ambato Boeny | Feb 2019 | N | 67 | OM630638 | OM350239 | OM542363 |
| An_squamosus_MG_S68 | Anopheles (Cellia) squamosus | Madagascar | Ambato Boeny | Feb 2019 | N | 68 | OM630639 | OM350240 | OM542364 |
| An_funestus_MG_S69 | Anopheles (Cellia) funestus | Madagascar | Ambato Boeny | Feb 2019 | N | 69 | OM630643 | OM350244 | OM542368 |
| An_funestus_MG_S70 | Anopheles (Cellia) funestus | Madagascar | Ambato Boeny | Feb 2020 | N | 70 | OM630644 | OM350317 | OM542441 |
| An_gambiae_MG_S71 | Anopheles (Cellia) gambiae | Madagascar | Ambato Boeny | Feb 2019 | N | 71 | OM630647 | OM350249 | OM542442 |
| An_gambiae_MG_S72 | Anopheles (Cellia) gambiae | Madagascar | Ambato Boeny | Feb 2019 | N | 72 | OM630648 | OM350248 | OM542443 |
| An_gambiae_MG_S73 | Anopheles (Cellia) gambiae | Madagascar | Ambato Boeny | Feb 2019 | N | 73 | OM630649 | OM350318 | OM542459 |
| Cx_quinquefasciatus_MG_S74 | Culex (Culex) quinquefasciatus | Madagascar | Amparafaravola | Feb 2019 | N | 74 | OM630674 | OM350271 | OM542399 |
| Cx_quinquefasciatus_MG_S75 | Culex (Culex) quinquefasciatus | Madagascar | Amparafaravola | Feb 2019 | N | 75 | OM630675 | OM350272 | OM542447 |
| Cx_perexiguus_MG_S76 | Culex (Culex) perexiguus | Madagascar | Mampikony | Aug 2019 | N | 76 | OM630676 | OM350273 | OM542400 |
| Ma_uniformis_MG_S77 | Mansonia (Mansonioides) uniformis | Madagascar | Ambato Boeny | Feb 2019 | N | 77 | OM630708 | OM350306 | OM542433 |
| Ma_uniformis_MG_S78 | Mansonia (Mansonioides) uniformis | Madagascar | Ambato Boeny | Feb 2019 | N | 78 | OM630709 | OM350307 | OM542434 |
| Ma_uniformis_MG_S79 | Mansonia (Mansonioides) uniformis | Madagascar | Ambato Boeny | Feb 2019 | N | 79 | OM630710 | OM350308 | OM542435 |
| Cx_poicilipes_MG_S82 | Culex poicilipes | Madagascar | Mampikony | Feb 2019 | N | 82 | OM630659 | OM350320 | OM542385, OM542445 |
| Mi_mediolineata_MG_S83 | Mimomyia mediolineata | Madagascar | Ambato Boeny | Feb 2019 | N | 83 | OM630683 | OM350280 | OM542407 |
| Cx_neavei_MG_S84 | Culex (Culex) neavei | Madagascar | Ambato Boeny | Feb 2019 | N | 84 | OM630684 | OM350281 | OM542408, OM542409 |
| Ae_scapularis_GF_S85 | Aedes (Ochlerotatus) scapularis | French Guiana | Hameau Prefontaine | Jul 2019 | N | 85 | OM630624 | OM350224 | OM542348, OM542349 |
| Ae_scapularis_GF_S86 | Aedes (Ochlerotatus) scapularis | French Guiana | Hameau Prefontaine | Jul 2019 | N | 86 | OM630622 | OM350225 | OM542350 |
| Ae_scapularis_GF_S87 | Aedes (Ochlerotatus) scapularis | French Guiana | Hameau Prefontaine | Jul 2019 | N | 87 | OM630623 | OM350226 | OM542351 |
| Ae_serratus_GF_S88 | Aedes (Ochlerotatus) serratus | French Guiana | Hameau Prefontaine | Nov 2020 | N | 88 | OM630625 | OM350227 | OM542352 |
| Ae_serratus_GF_S89 | Aedes (Ochlerotatus) serratus | French Guiana | Hameau Prefontaine | Nov 2020 | N | 89 | OM630626 | OM350228 | OM542353 |
| Ae_serratus_GF_S90 | Aedes (Ochlerotatus) serratus | French Guiana | Hameau Prefontaine | Nov 2020 | N | 90 | OM630627 | OM350229 | OM542354 |
| Cq_venezuelensis_GF_S91 | Coquillettidia venezuelensis | French Guiana | Hameau Prefontaine | Jul 2019 | N | 91 | OM630650 | OM350250 | OM542375 |
| Cq_venezuelensis_GF_S92 | Coquillettidia venezuelensis | French Guiana | Hameau Prefontaine | Jul 2019 | N | 92 | OM630651 | OM350251 | OM542376 |
| Cq_venezuelensis_GF_S93 | Coquillettidia venezuelensis | French Guiana | Hameau Prefontaine | Jul 2019 | N | 93 | OM630652 | OM350252 | OM542377, OM542378 |
| Cx_portesi_GF_S94 | Culex sp. BTLHVDV-2014 | French Guiana | Hameau Prefontaine | Jul 2019 | N | 94 | OM630666 | OM350264 | OM542392 |
| Cx_portesi_GF_S95 | Culex sp. BTLHVDV-2014 | French Guiana | Hameau Prefontaine | Jul 2019 | N | 95 | OM630667 | OM350265 | OM542393 |
| Cx_portesi_GF_S96 | Culex sp. BTLHVDV-2014 | French Guiana | Hameau Prefontaine | Jul 2019 | N | 96 | OM630668 | OM350266 | OM542394 |
| Cx_spissipes_GF_S97 | Culex (Melanoconion) sp. DJS-2020 | French Guiana | Hameau Prefontaine | Jul 2019 | N | 97 | OM630677 | OM350274 | OM542401 |
| Cx_spissipes_GF_S98 | Culex (Melanoconion) sp. DJS-2020 | French Guiana | Hameau Prefontaine | Jul 2019 | N | 98 | OM630678 | OM350275 | OM542402 |
| Cx_spissipes_GF_S99 | Culex (Melanoconion) sp. DJS-2020 | French Guiana | Hameau Prefontaine | Jul 2019 | N | 99 | OM630679 | OM350276 | OM542403 |
| Li_durhamii_GF_S100 | Limatus durhamii | French Guiana | Hameau Prefontaine | Jul 2019 | N | 100 | OM630695 | OM350291 | OM542419 |
| Li_durhamii_GF_S101 | Limatus durhamii | French Guiana | Hameau Prefontaine | Jul 2019 | N | 101 | OM630696 | OM350292 | OM542420 |
| Li_durhamii_GF_S102 | Limatus durhamii | French Guiana | Hameau Prefontaine | Jul 2019 | N | 102 | OM630697 | OM350293 | OM542421 |
| Ma_sp.4_GF_S103 | Mansonia sp.4 | French Guiana | Hameau Prefontaine | Jan 2020 | N | 103 | OM630701 | OM350298 | OM542425 |
| Ma_sp.4_GF_S104 | Mansonia sp.4 | French Guiana | Hameau Prefontaine | Jan 2020 | N | 104 | OM630702 | OM350299 | OM542426 |
| Ma_titillans_GF_S105 | Mansonia (Mansonia) titillans | French Guiana | Hameau Prefontaine | Jan 2020 | N | 105 | OM630703 | OM350300 | OM542427 |
| Cx_pedroi_GF_S106 | Culex (Melanoconion) pedroi | French Guiana | Hameau Prefontaine | Nov 2020 | N | 106 | OM630663 | OM350261 | OM542389 |
| Cx_pedroi_GF_S107 | Culex (Melanoconion) pedroi | French Guiana | Hameau Prefontaine | Nov 2020 | N | 107 | OM630664 | OM350262 | OM542390 |
| Cx_pedroi_GF_S108 | Culex (Melanoconion) pedroi | French Guiana | Hameau Prefontaine | Nov 2020 | N | 108 | OM630665 | OM350263 | OM542391 |
| Ps_ferox_GF_S109 | Psorophora ferox | French Guiana | Iracoubo | 2009 | N | 109 | OM630711 | OM350309 | OM542436 |
| Ps_ferox_GF_S110 | Psorophora ferox | French Guiana | Iracoubo | 2009 | N | 110 | OM630712 | OM350310 | OM542437 |
| Ps_ferox_GF_S111 | Psorophora ferox | French Guiana | Iracoubo | 2009 | N | 111 | OM630713 | OM350324 | OM542452 |
| Ur_geometrica_GF_S112 | Uranotaenia (Uranotaenia) geometrica | French Guiana | 2010 | N | 112 | OM630714 | OM350311 | OM542453 | |
| Ur_geometrica_GF_S113 | Uranotaenia (Uranotaenia) geometrica | French Guiana | 2010 | N | 113 | N/A | OM350312 | OM542454 | |
| Ur_geometrica_GF_S114 | Uranotaenia (Uranotaenia) geometrica | French Guiana | 2010 | N | 114 | OM630715 | OM350313 | OM542438, OM542455 | |
| Cx_tritaeniorhynchus_MG_S115 | Culex (Culex) tritaeniorhynchus | Madagascar | Ambato Boeny | Feb 2019 | N | 115 | OM630685 | OM350321 | OM542448 |
Cytochrome c oxidase I (COI) sequence BLAST analyses summary (XLSX).
| Sequence ID | Sequence length | Morphological identification | BLASTn top hit species | BLASTn top hit accession | Query coverage | % identity | Comments |
|---|---|---|---|---|---|---|---|
| Ae_albopictus_KH_S1 | 699 | Aedes albopictus | Aedes albopictus | MK714006.1 | 99% | 99.71% | |
| Ae_albopictus_KH_S2 | 695 | Aedes albopictus | Aedes albopictus | MK714006.1 | 100% | 99.71% | |
| Ae_albopictus_KH_S3 | 695 | Aedes albopictus | Aedes albopictus | MK714006.1 | 100% | 99.71% | |
| An_baezai_KH_S4 | 658 | Anopheles baezai | Anopheles darlingi | MF381626.1 | 100% | 92.71% | Anopheles baezai not found in GenBank |
| An_baezai_KH_S5 | 670 | Anopheles baezai | Anopheles darlingi | MF381626.1 | 99% | 92.81% | Anopheles baezai not found in GenBank |
| An_baezai_KH_S6 | 659 | Anopheles baezai | Anopheles darlingi | MF381626.1 | 100% | 92.72% | Anopheles baezai not found in GenBank |
| Cx_pseudovishnui_KH_S7 | 660 | Culex vishnui | Culex pseudovishnui | MW321882.1 | 98% | 98.92% | 95% similarity to Culex vishnui, 94% similarity to Culex tritaeniorhynchus |
| Cx_pseudovishnui_KH_S8 | 659 | Culex vishnui | Culex pseudovishnui | MW321882.1 | 98% | 99.38% | 95% similarity to Culex vishnui, 94% similarity to Culex tritaeniorhynchus |
| Cx_pseudovishnui_KH_S9 | 659 | Culex vishnui | Culex pseudovishnui | MW321882.1 | 98% | 98.92% | 95% similarity to Culex vishnui, 94% similarity to Culex tritaeniorhynchus |
| Ma_indiana_KH_S10 | 660 | Mansonia indiana | Mansonia indiana | MK637632.1 | 98.00% | 99.54% | |
| Ma_uniformis_KH_S11 | 686 | Mansonia indiana | Mansonia uniformis | MK757484.1 | 99% | 99.71% | 89.99% similarity to Mansonia indiana MK637632.1 |
| Ma_indiana_KH_S12 | 693 | Mansonia indiana | Mansonia indiana | MK637632.1 | 97% | 99.41% | |
| Cx_sp.1_KH_S13 | 687 | Culex quinquefasciatus | Culex (Lophoceraomyia) sp.5 HY-2020 | MW321904.1 | 98% | 94.39% | 90% similarity to Culex quinquefasciatus GU188856.2 |
| Cx_orientalis_KH_S14 | 662 | Culex quinquefasciatus | Culex orientalis | MW228488.1 | 97% | 98.29% | |
| Ma_uniformis_KH_S15 | 658 | Mansonia uniformis | Mansonia uniformis | MK757484.1 | 100.00% | 99.54% | |
| Ma_uniformis_KH_S16 | 654 | Mansonia uniformis | Mansonia uniformis | MK757484.1 | 100.00% | 99.39% | |
| Ma_uniformis_KH_S17 | 657 | Mansonia uniformis | Mansonia uniformis | MK757484.1 | 99.00% | 99.54% | |
| Cx_bitaeniorhynchus_KH_S18 | 658 | Culex bitaeniorhynchus | Culex bitaeniorhynchus | HQ398898.1 | 97.00% | 99.69% | |
| Cx_bitaeniorhynchus_KH_S19 | 650 | Culex bitaeniorhynchus | Culex bitaeniorhynchus | HQ398898.1 | 98.00% | 99.84% | |
| Cx_bitaeniorhynchus_KH_S20 | 652 | Culex bitaeniorhynchus | Culex bitaeniorhynchus | HQ398898.1 | 98.00% | 99.38% | |
| Cx_tritaeniorhynchus_KH_S21 | 695 | Culex tritaeniorhynchus | Culex vishnui or Culex tritaeniorhynchus | MH374857.1 | 100% | 99.57% | 99.69% similarity to Culex tritaeniorhynchus MF179213.1 |
| Cx_tritaeniorhynchus_KH_S22 | 690 | Culex tritaeniorhynchus | Culex vishnui or Culex tritaeniorhynchus | MT876103.1 | 100% | 99.57% | |
| Cx_tritaeniorhynchus_KH_S23 | 663 | Culex tritaeniorhynchus | Culex tritaeniorhynchus | MT876103.1 | 99% | 98.79% | |
| Ae_aegypti_CF_S24 | 689 | Aedes aegypti | Aedes aegypti | MN299016.1 | 100% | 99.56% | |
| Ae_aegypti_CF_S25 | 660 | Aedes aegypti | Aedes aegypti | MN299024.1 | 100.00% | 99.70% | |
| Ae_aegypti_CF_S26 | 660 | Aedes aegypti | Aedes aegypti | MN299024.1 | 100.00% | 99.70% | |
| Ae_simpsoni_CF_S27 | 644 | Aedes opok | Aedes simpsoni | LC473669.1 | 97.00% | 97.77% | Aedes opok not found in GenBank, sequence has 90% and 89% similarity to Aedes luteocephalus and Aedes africanus, sister species of Aedes opok. |
| Ae_simpsoni_CF_S28 | 649 | Aedes opok | Aedes simpsoni | MN552302.1 | 99.00% | 100.00% | Aedes. opok not found in GenBank, sequence has 90% and 89% similarity to Aedes luteocephalus and Aedes africanus, sister species of Aedes opok. |
| Ae_simpsoni_CF_S29 | 627 | Aedes opok | Aedes simpsoni | MN552302.1 | 98.00% | 98.87% | Aedes opok not found in GenBank, sequence has 90% and 89% similarity to Aedes luteocephalus and Aedes africanus, sister species of Aedes opok. |
| Ae_vittatus_CF_S30 | 623 | Aedes vittatus | Aedes vittatus | MN552298.1 | 100.00% | 99.84% | |
| Ae_vittatus_CF_S31 | 622 | Aedes vittatus | Aedes vittatus | MN552298.1 | 100.00% | 99.68% | |
| Ae_vittatus_CF_S32 | 621 | Aedes vittatus | Aedes vittatus | MN552298.1 | 100.00% | 99.68% | |
| Ma_sp.1_CF_S33 | – | Mansonia africana | – | – | – | – | No COI obtained |
| Ma_sp.2_CF_S34 | – | Mansonia africana | – | – | – | – | No COI obtained |
| Ma_sp.3_CF_S35 | – | Mansonia africana | – | – | – | – | No COI obtained |
| Ae_albopictus_CF_S36 | 627 | Aedes albopictus | Aedes albopictus | MK995332.1 | 100.00% | 99.84% | |
| Ae_albopictus_CF_S37 | 621 | Aedes albopictus | Aedes albopictus | MK995332.1 | 100.00% | 100.00% | |
| Ae_albopictus_CF_S38 | 621 | Aedes albopictus | Aedes albopictus | MK995332.1 | 100.00% | 100.00% | |
| An_coustani_CF_S39 | 621 | Anopheles coustani | Anopheles coustani | MK585968.1 | 100.00% | 99.84% | |
| An_coustani_CF_S40 | 621 | Anopheles coustani | Anopheles coustani | MK585959.1 | 100.00% | 99.03% | |
| An_coustani_CF_S41 | 699 | Anopheles coustani | Anopheles coustani | MK585968.1 | 94.00% | 99.70% | |
| An_funestus_CF_S42 | 696 | Anopheles funestus | Anopheles funestus | MK300231.1 | 100.00% | 99.71% | |
| An_funestus_CF_S43 | 660 | Anopheles funestus | Anopheles funestus | MT375215.1 | 100.00% | 99.85% | |
| An_funestus_CF_S44 | 658 | Anopheles funestus | Anopheles funestus | MT375215.1 | 100.00% | 99.70% | |
| An_gambiae_CF_S45 | 660 | Anopheles gambiae | Anopheles gambiae | MG930895.1 | 86.00% | 99.79% | |
| An_gambiae_CF_S46 | 659 | Anopheles gambiae | Anopheles gambiae | MT375223.1 | 89.00% | 100.00% | |
| An_gambiae_CF_S47 | – | Anopheles gambiae | – | – | – | – | No COI obtained |
| Cx_sp.2_CF_S48 | 653 | Culex quinquefasciatus | Culex corniger | KM593015.1 | 100.00% | 94.95% | 94% similarity to all other Culex species |
| Cx_sp.2_CF_S49 | 660 | Culex quinquefasciatus | Culex nigripalpus | KM593058.1 | 99.00% | 94.65% | 94% similarity to all other Culex species |
| Cx_sp.2_CF_S50 | 658 | Culex quinquefasciatus | Culex bidens | MH931446.1 | 100.00% | 94.68% | 94% similarity to all other Culex species |
| Ma_uniformis_CF_S51 | – | Mansonia uniformis | – | – | – | – | No COI obtained |
| Cx_duttoni_CF_S52 | 621 | Mansonia uniformis | Culex duttoni | LC473629.1 | 100.00% | 99.68% | |
| Er_intermedius_CF_S54 | 620 | Eretmapodites sp. | Eretmapodites intermedius | MN552305.1 | 100.00% | 99.52% | |
| Er_intermedius_CF_S55 | 621 | Eretmapodites sp. | Eretmapodites intermedius | MN552305.1 | 100.00% | 99.68% | |
| Er_intermedius_CF_S56 | 621 | Eretmapodites sp. | Eretmapodites intermedius | MN552305.1 | 100.00% | 99.68% | |
| Cx_antennatus_MG_S57 | 621 | Culex antennatus | Culex antennatus | LC473659.1 | 100.00% | 100.00% | |
| Cx_antennatus_MG_S58 | 621 | Culex antennatus | Culex antennatus | LC473659.1 | 100.00% | 100.00% | |
| Cx_antennatus_MG_S59 | 621 | Culex. antennatus | Culex antennatus | LC473659.1 | 100.00% | 100.00% | |
| Cx_perexiguus_MG_S60 | 621 | Culex decens | Culex perexiguus | LC473634.1 | 100.00% | 99.84% | |
| Cx_sp.3_MG_S61 | 685 | Culex decens | Unknown Culex species | KU380436.1 | 96.00% | 96.05% | |
| Cx_sp.4_MG_S62 | 687 | Culex decens | Unknown Culex species | MT993494.1 | 99.00% | 95.63% | |
| Cx_sp.3_MG_S63 | 687 | Culex univittatus | Unknown Culex species | KU380436.1 | 95.00% | 96.50% | |
| Mi_sp.1_MG_S64 | 694 | Culex univittatus | Mimomyia mimomyiaformis | LC473719.1 | 94.00% | 92.55% | Unknown Mimomyia species |
| Cx_sp.3_MG_S65 | 691 | Culex univittatus | Unknown Culex species | KU380436.1 | 95.00% | 96.66% | |
| An_coustani_MG_S66 | 669 | Anopheles coustani | Anopheles coustani | NC_050693.1 | 99.00% | 99.40% | |
| An_coustani_MG_S67 | 659 | Anopheles coustani | Anopheles coustani | NC_050693.1 | 99.00% | 99.08% | |
| An_squamosus_MG_S68 | 653 | Anopheles coustani | Anopheles squamosus | MK776741.1 | 100.00% | 100.00% | |
| An_funestus_MG_S69 | 654 | Anopheles funestus | Anopheles funestus | MT375215.1 | 100.00% | 99.85% | |
| An_funestus_MG_S70 | 654 | Anopheles funestus | Anopheles funestus | MG742199.1 | 100.00% | 99.69% | |
| An_gambiae_MG_S71 | 654 | Anopheles gambiae | Anopheles gambiae | MT375222.1 | 100.00% | 99.85% | |
| An_gambiae_MG_S72 | 654 | Anopheles gambiae | Anopheles gambiae | MT375222.1 | 100.00% | 99.85% | |
| An_gambiae_MG_S73 | 622 | Anopheles gambiae | Anopheles gambiae | MT375222.1 | 100.00% | 100.00% | |
| Cx_quinquefasciatus_MG_S74 | 654 | Culex quinquefasciatus | Culex pipiens | MT199095.1 | 100.00% | 100.00% | 99.85% similarity to Culex quinquefasciatus |
| Cx_quinquefasciatus_MG_S75 | 647 | Culex quinquefasciatus | Culex quinquefasciatus | MH423504.1 | 100.00% | 98.15% | Also 98% similarity to Culex pipiens |
| Cx_perexiguus_MG_S76 | 621 | Culex quinquefasciatus | Culex perexiguus | LC473634.1 | 100.00% | 99.52% | Same SNPs to Culex pipiens MH374861.1 |
| Ma_uniformis_MG_S77 | 621 | Mansonia uniformis | Mansonia uniformis | KU187165.1 | 100.00% | 100.00% | |
| Ma_uniformis_MG_S78 | 621 | Mansonia uniformis | Mansonia uniformis | KU187165.1 | 100.00% | 100.00% | |
| Ma_uniformis_MG_S79 | 626 | Mansonia uniformis | Mansonia uniformis | KU187157.1 | 100.00% | 99.68% | |
| Cx_poicilipes_MG_S82 | 689 | Culex bitaeniorhynchus | Culex poicilipes | LC473618.1 | 95.00% | 99.70% | |
| Mi_mediolineata_MG_S83 | 694 | Culex tritaeniorhynchus | Mimomyia mediolineata | LC473723.1 | 94.00% | 99.39% | |
| Cx_neavei_MG_S84 | 671 | Culex tritaeniorhynchus | Culex neavei | LC473635.1 | 98.00% | 99.85% | |
| Ae_scapularis_GF_S85 | 659 | Aedes scapularis | Aedes scapularis | MN997484.1 | 97.00% | 98.76% | |
| Ae_scapularis_GF_S86 | 658 | Aedes scapularis | Aedes scapularis | MF172265.1 | 97.00% | 99.38% | |
| Ae_scapularis_GF_S87 | 654 | Aedes scapularis | Aedes scapularis | MF172265.1 | 98.00% | 99.22% | |
| Ae_serratus_GF_S88 | 660 | Aedes serratus | Aedes serratus | MF172269.1 | 97.00% | 98.91% | |
| Ae_serratus_GF_S89 | 660 | Aedes serratus | Aedes serratus | MF172268.1 | 97.00% | 99.22% | |
| Ae_serratus_GF_S90 | 654 | Aedes serratus | Aedes serratus | MF172268.1 | 98.00% | 99.07% | |
| Cq_venezuelensis_GF_S91 | 658 | Coquillettidia venezuelensis | Coquillettidia venezuelensis | MN997703.1 | 97.00% | 97.98% | |
| Cq_venezuelensis_GF_S92 | 621 | Coquillettidia venezuelensis | Coquillettidia. venezuelensis | MN997703.1 | 100.00% | 98.07% | |
| Cq_venezuelensis_GF_S93 | 621 | Coquillettidia venezuelensis | Coquillettidia venezuelensis | MN997703.1 | 100.00% | 97.75% | |
| Cx_portesi_GF_S94 | 653 | Culex portesi | Culex portesi | in-house reference library | 98.5–100% | Reference sequence provided by Amandine Guidez, IP Guyane | |
| Cx_portesi_GF_S95 | 693 | Culex portesi | Culex portesi | in-house reference library | 98.5–100% | Reference sequence provided by Amandine Guidez, IP Guyane | |
| Cx_portesi_GF_S96 | 687 | Culex portesi | Culex portesi | in-house reference library | 98.5–100% | Reference sequence provided by Amandine Guidez, IP Guyane | |
| Cx_spissipes_GF_S97 | 672 | Culex spissipes | Culex spissipes | in-house reference library | 98.5–100% | Reference sequence provided by Amandine Guidez, IP Guyane | |
| Cx_spissipes_GF_S98 | 663 | Culex spissipes | Culex spissipes | in-house reference library | 98.5–100% | Reference sequence provided by Amandine Guidez, IP Guyane | |
| Cx_spissipes_GF_S99 | 660 | Culex spissipes | Culex spissipes | in-house reference library | 98.5–100% | Reference sequence provided by Amandine Guidez, IP Guyane | |
| Li_durhamii_GF_S100 | 653 | Lmatus durhamii | Limatus durhamii | MF172330.1 | 98.00% | 99.84% | |
| Li_durhamii_GF_S101 | 621 | Limatus durhamii | Limatus durhamii | MF172330.1 | 100.00% | 100.00% | |
| Li_durhamii_GF_S102 | 699 | Limatus durhamii | Limatus durhamii | MF172330.1 | 94.00% | 100.00% | |
| Ma_sp.4_GF_S103 | 621 | Mansonia titillans | Mansonia sp. | MT329066.1 | 100.00% | 99.84% | 87.12% similarity to Mansonia titillans MN968244.1 |
| Ma_sp.4_GF_S104 | 695 | Mansonia titillans | Mansonia sp. | MT329066.1 | 95.00% | 99.85% | 87.39% to Mansonia titillans MN968244.1 |
| Ma_titillans_GF_S105 | 669 | Mansonia titillans | Mansonia titillans | MN968244.1 | 98.00% | 99.70% | |
| Cx_pedroi_GF_S106 | 653 | Culex pedroi | Culex pedroi | KX779887.1 | 98.00% | 98.60% | |
| Cx_pedroi_GF_S107 | 661 | Culex pedroi | Culex pedroi | KX779887.1 | 97.00% | 98.76% | |
| Cx_pedroi_GF_S108 | 621 | Culex pedroi | Culex pedroi | KX779887.1 | 99.00% | 98.87% | |
| Ps_ferox_GF_S109 | 633 | Psorophora ferox | Psorophora ferox | MF172349.1 | 100.00% | 99.68% | |
| Ps_ferox_GF_S110 | 621 | Psorophora ferox | Psorophora ferox | MF172349.1 | 100.00% | 99.68% | |
| Ps_ferox_GF_S111 | 621 | Psorophora ferox | Psorophora ferox | MF172347.1 | 99.00% | 99.51% | |
| Ur_geometrica_GF_S112 | 621 | Uranotaenia geometrica | Uranotaenia geometrica | NC_044662.1 | 100.00% | 100.00% | |
| Ur_geometrica_GF_S113 | – | Uranotaenia geometrica | – | – | – | – | No COI obtained |
| Ur_geometrica_GF_S114 | 621 | Uranotaenia geometrica | Uranotaenia geometrica | NC_044662.1 | 100.00% | 100.00% | |
| Cx_tritaeniorhynchus_MG_S115 | 653 | Culex tritaeniorhynchus | Culex tritaeniorhynchus | MK861440.1 | 100.00% | 98.77% |





