pYtags enable spatiotemporal measurements of receptor tyrosine kinase signaling in living cells
Figures
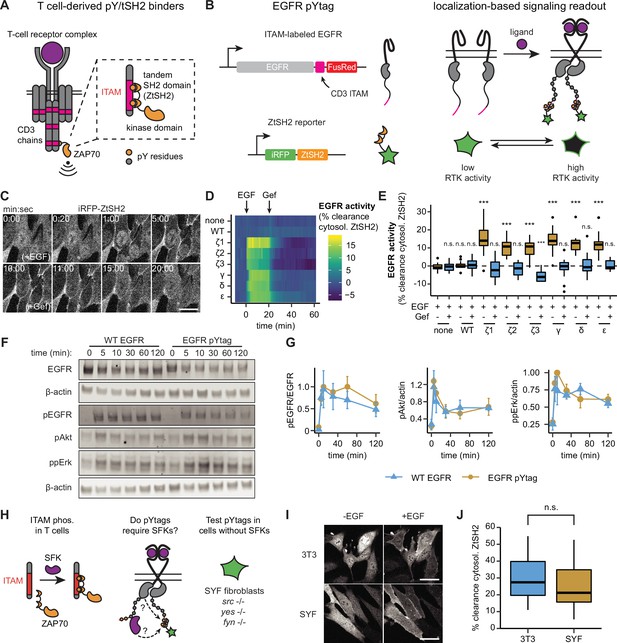
pYtags: a biosensing strategy to monitor receptor tyrosine kinase (RTK) activity in living cells.
(A) The T-cell receptor complex contains six immunoreceptor tyrosine-based activation motifs (ITAMs) from CD3 chains that, when phosphorylated, bind to the tSH2 domain of ZAP70 (ZtSH2). (B) Three repeats of CD3 ITAMs were appended to the C-terminus of epidermal growth factor receptor (EGFR) and clearance of ZtSH2 from the cytosol was assessed. (C) Timelapse images of NIH3T3 cells expressing EGFR pYtag (CD3ε variant), treated first with EGF (100 ng/mL) and then with gefitinib (10 µM). Scale bar, 20 µm. (D) Mean clearance of cytosolic ZtSH2 in cells co-expressing iRFP-ZtSH2 and EGFR C-terminally labeled with one of six CD3 ITAMs. EGF (100 ng/mL) and gefitinib (10 µM) were sequentially added at times denoted by arrows. n = 2 independent experiments. (E) Clearance of cytosolic ZtSH2 10 min post-EGF treatment and 40 min post-gefitinib treatment from (D). Lines denote mean values, boxes denote 25–75th percentiles, and whiskers denote minima and maxima. n ≥ 14 cells from two independent experiments. n.s., not significant, ***p<0.001 by Kolmogorov–Smirnov test with cells expressing no additional EGFR 10 min post-EGF. (F) Immunoblots of NIH3T3 cells expressing either WT EGFR or EGFR pYtag treated with EGF (100 ng/mL). (G) Mean ± SEM levels of EGFR, Akt, and Erk phosphorylation from (F). n = 3 independent experiments. (H) The EGFR pYtag was tested in SYF cells to determine whether SFKs are required for ITAM phosphorylation. (I) Representative images of NIH3T3 and SYF cells expressing EGFR pYtag, treated with EGF (100 ng/mL). Scale bars, 40 µm. (J) Mean clearance of cytosolic ZtSH2 in SYF and NIH3T3 cells 10 min after treatment with EGF. For each condition, n > 20 cells from three independent experiments. See also Figure 1—video 1.
-
Figure 1—source data 1
Uncropped gels for Figure 1F.
- https://cdn.elifesciences.org/articles/82863/elife-82863-fig1-data1-v1.zip
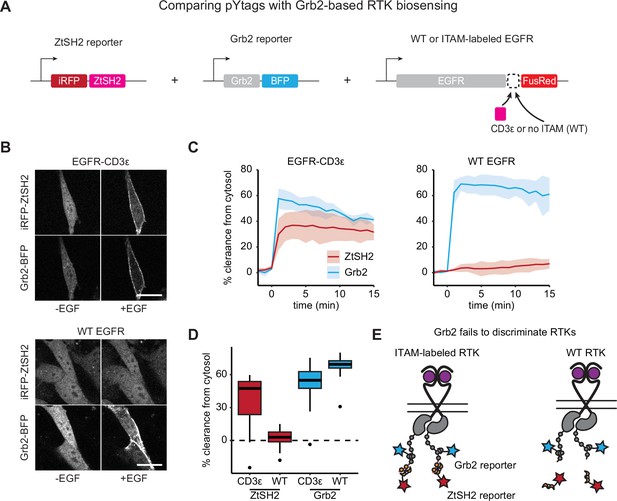
Grb2 fails to discriminate between immunoreceptor tyrosine-based activation motif (ITAM)-labeled and unlabeled RTKs.
(A) ZtSH2- and Grb2-based reporters of receptor tyrosine kinase (RTK) signaling were co-expressed in cells expressing either ITAM-labeled or WT epidermal growth factor receptor (EGFR). (B) NIH3T3 cells before and 5 min after treatment with EGF (100 ng/mL). Scale bars, 20 µm. (C) Mean ± SD clearance of ZtSH2 and Grb2 reporters from the cytosol following treatment with EGF (100 ng/mL). n = 3 independent experiments. (D) Clearance of reporters from the cytosol 5 min after treatment with EGF in (C). Lines denote mean values, boxes denote 25–75th percentiles, and whiskers denote minima and maxima. For each condition, n > 16 cells from three independent experiments. (E) ZtSH2 reports the signaling of ITAM-labeled RTKs, while Grb2 fails to discriminate between RTKs displaying or lacking ITAMs.
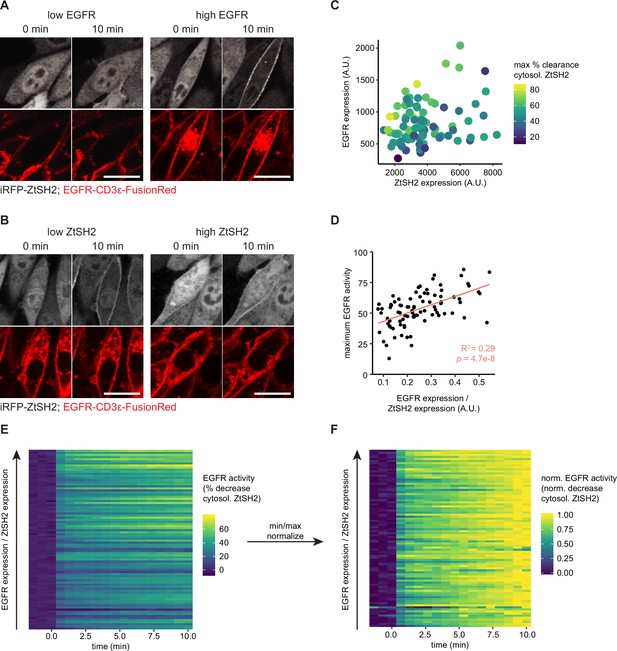
Effects of the expression levels of pYtag components in NIH3T3 cells.
Images of EGFR-pYtag expressing NIH3T3 cells expressing various levels of (A) EGFR-CD3ε-FusionRed or (B) iRFP-ZtSH2, treated with EGF (20 ng/mL). Scale bars, 20 µm. (C) Maximum % clearance of ZtSH2 from the cytosol in EGFR-pYtag expressing NIH3T3 cells treated with EGF (20 ng/mL). (D) Maximum % clearance of ZtSH2 from the cytosol as a function of the ratio of EGFR-CD3ε-FusionRed to iRFP-ZtSH2 (EGFR:ZtSH2 ratio). (E, F) Heatmaps of single-cell trajectories for all cells quantified in (C) and sorted in order of increasing EGFR:ZtSH2 ratio. (E) shows the % clearance of ZtSH2 from the cytosol; (F) shows min–max normalized responses to compare activation kinetics. Panels (C–E): points denote individual cells. n = 86 cells from three independent experiments.

Effects of the expression levels of pYtag components in HEK293T cells.
Images of EGFR-pYtag expressing HEK293T cells expressing variable levels of (A) EGFR-CD3ε-mNeonGreen or (B) mScarlet-ZtSH2, treated with EGF (100 ng/mL). Scale bars, 20 µm. (C) Maximum % clearance of ZtSH2 from the cytosol in EGFR-pYtag expressing HEK293T cells treated with EGF (100 ng/mL). (D) Maximum % clearance of ZtSH2 from the cytosol as a function of the ratio of EGFR-CD3ε-mNeonGreen to mScarlet-ZtSH2 (EGFR:ZtSH2 ratio). (E, F) Heatmaps of single-cell trajectories for all cells quantified in (C) and sorted in order of increasing EGFR:ZtSH2 ratio. (E) shows the % clearance of ZtSH2 from the cytosol; (F) shows min–max normalized responses to compare activation kinetics. Panels (C, D): points denote individual cells. n = 84 cells from three independent experiments.
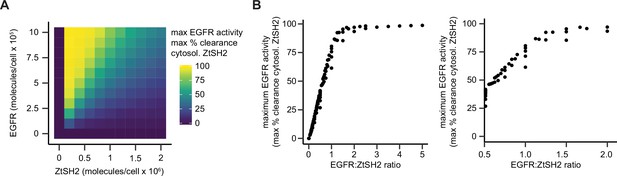
Mathematical modeling of the effects of pYtag component expression levels on biosensor readout.
(A) Heatmap of maximum % clearance of ZtSH2 in response to 20 ng/mL EGF. For each cell, rows and columns denote levels of EGFR-CD3ε and ZtSH2, respectively. (B) Each cell from (A) was plotted as a function of the ratio of EGFR to ZtSH2 (EGFR:ZtSH2 ratio). Right plot presents zoomed-in results for EGFR:ZtSH2 ratios between 0.5 and 2.
Timelapse of iRFP-ZtSH2 in NIH3T3 cells co-expressing iRFP-ZtSH2 and EGFR-CD3ε-FusionRed.
Cells were first treated with EGF (100 ng/mL) then treated with gefitinib (10 µM) at the times denoted in the video. Related to Figure 1C.
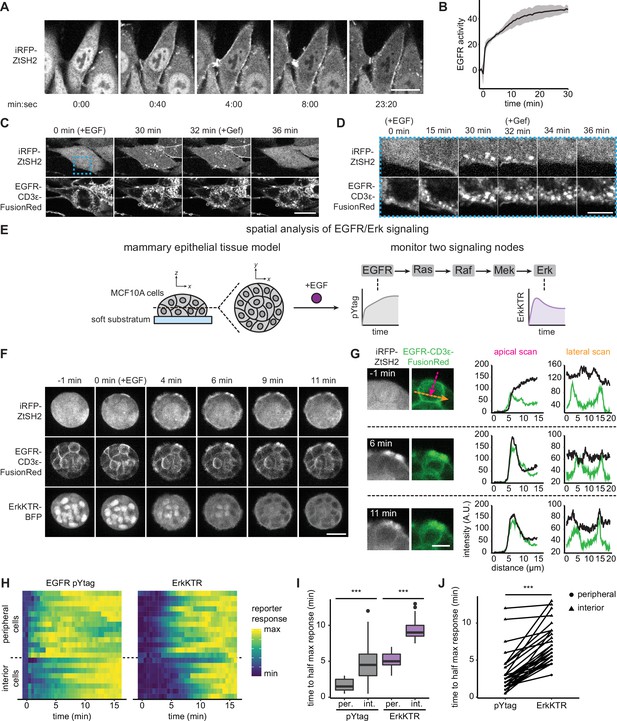
Monitoring epidermal growth factor receptor (EGFR) signaling at subcellular and multicellular length scales.
(A) Images of EGFR pYtag-expressing NIH3T3 cells treated with EGF (20 ng/mL). Scale bar, 20 µm. (B) Mean ± SD clearance of cytosolic ZtSH2 from (A). n = 3 independent experiments. (C) EGFR pYtag-expressing NIH3T3 cells treated with EGF (20 ng/mL) were monitored for internalized ZtSH2-positive vesicles, and then treated with gefitinib (10 µM). Scale bar, 20 µm. (D) Timelapse images from the region denoted by the blue dashed border from (C). Scale bar, 10 µm. (E) MCF10A human mammary epithelial cells cultured on soft substrata form round, multilayered clusters. EGFR pYtag and ErkKTR were used to spatiotemporally monitor both EGFR and Erk responses after stimulation with EGF. (F) Images of MCF10A cells cultured on soft substrata and treated with EGF (100 ng/mL). Scale bar, 25 µm. (G) Apical and lateral enrichment of ZtSH2 was quantified by line scans denoted by magenta and orange vectors, respectively. Scale bar, 10 µm. (H) Heatmaps of EGFR pYtag and ErkKTR responses from (F). Rows denote individual cells. For each cell, signaling responses of each biosensor were normalized to their respective minima and maxima. (I) Time to half maximal response for EGFR pYtag and ErkKTR for cells from (H). n = 19 (periphery) and n = 11 (interior) cells from three biological replicates. (J) Time to half maximal response for EGFR pYtag and ErkKTR. Responses of individual cells are denoted by points and connected by lines. n > 30 cells from three biological replicates. ***p<0.001 by Kolmogorov–Smirnov test. See also Figure 2—video 1.
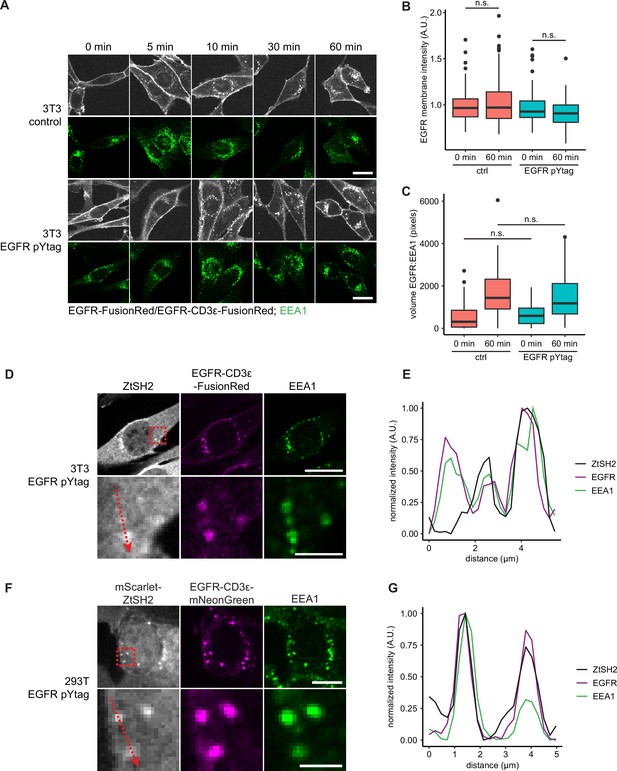
Analysis of epidermal growth factor receptor (EGFR) and ZtSH2 internalization.
(A) Images of control (EGFR-FusionRed) and EGFR pYtag (EGFR-CD3ε-FusionRed; iRFP-ZtSH2) NIH3T3 cells immunostained for EEA1 after treatment with EGF (100 ng/mL) for varying durations. Scale bars, 20 µm. (B) Intensity of EGFR-CD3ε-FusionRed at the cell membrane from (A). For each condition, n > 70 cells from three independent experiments. (C) Volume of pixels doubly positive for EGFR-CD3ε-FusionRed and EEA1 in individual cells. For each condition, n > 36 cells from three independent experiments. (D) Image of EGFR pYtag-expressing NIH3T3 cell treated with EGF (100 ng/mL) for 30 min and immunostained for ZtSH2 and EEA1. Scale bars, 20 µm (top) and 5 µm (bottom). (E) Fluorescence measurements along the red vector in (D). For each protein, intensities were normalized to the minimum and maximum values measured. (F) Image of EGFR pYtag-expressing HEK293T cell treated with EGF (100 ng/mL) for 5 min and immunostained for EEA1. Scale bars, 10 µm (top) and 3 µm (bottom). (G) Fluorescence measurements along the red vector in (F). For each protein, intensities were normalized to the minimum and maximum values measured.
Maximum intensity projection timelapse images of MCF10A cells co-expressing iRFP-ZtSH2 (left panel), EGFR-CD3ε-FusionRed (middle panel), and ErkKTR-BFP (right panel), cultured on soft substrata and treated with EGF (100 ng/mL).
Related to Figure 2F.
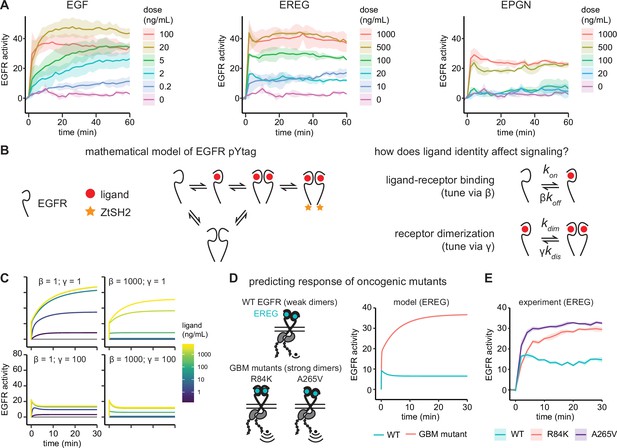
Epidermal growth factor receptor (EGFR) pYtag reveals dose- and ligand-dependent signaling dynamics.
(A) Mean ± SD responses of EGFR pYtag-expressing NIH3T3 cells to varying doses of EGF, epiregulin (EREG), and epigen (EPGN). The same 0 ng/mL control was used for each ligand. Data were collected from 475 cells across four independent experiments with each dose tested at least twice. (B) Dose–response profiles from (A) were analyzed using a mathematical model of EGFR pYtag. (C) Simulations of EGFR pYtag responses to ligand of varying doses for different values of β and γ. (D) GBM-associated mutants of EGFR that form strong EREG-bound dimers were predicted to exhibit stronger pYtag responses to 20 ng/mL EREG compared to WT EGFR. (E) Mean ± SD pYtag response of WT and GBM-associated mutant EGFRs in NIH3T3 cells after EREG treatment (20 ng/mL). n = 3 independent experiments.
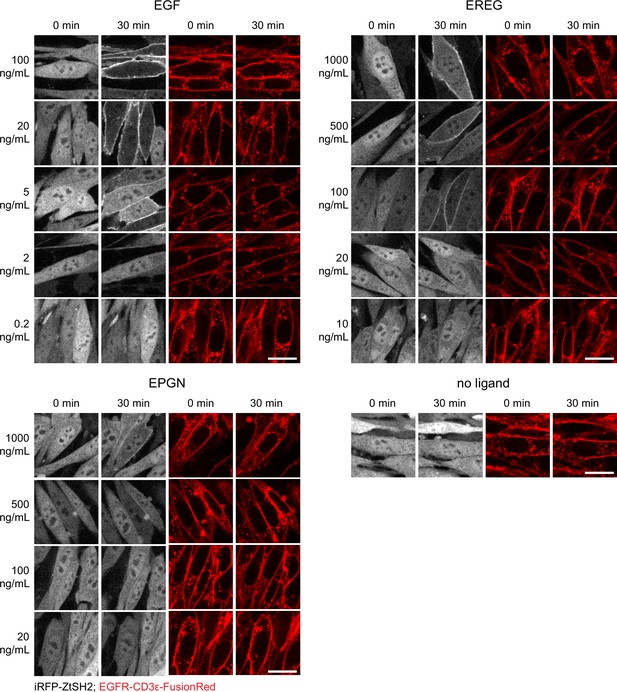
ZtSH2 and epidermal growth factor receptor (EGFR) localization in response to different ligands.
Representative images of EGFR pYtag-expressing NIH3T3 cells treated with different concentrations of EGF, EREG, or EPGN. Scale bars, 20 µm.

Ligand-free dimers in mathematical model recapitulate biphasic signaling response of epidermal growth factor receptor (EGFR).
(A) EGFR pYtag response to EGF (20 ng/mL) shown in Figure 2B. (B) The effect of ligand-free dimers on EGFR pYtag responses was simulated by tuning the dissociation rate of ligand-free dimers k6. (C) Percentage of receptors existing as ligand-free dimers before ligand stimulation as a function of k6. Color of data points corresponds to color of curves in (D). (D) Simulated EGFR pYtag responses to EGF for varying values of k6.
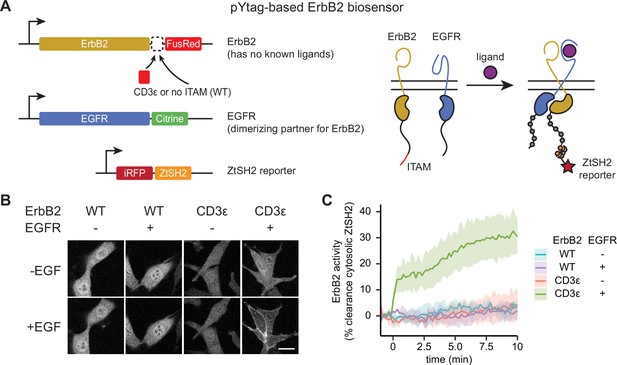
Monitoring distinct receptor tyrosine kinases (RTKs) in heterodimeric complexes.
(A) In order to signal, the ligandless ErbB2 must heterodimerize with a ligand-binding member of the ErbB family. The pYtag strategy enables measurements of ErbB2’s activity despite the co-activation of epidermal growth factor receptor (EGFR). (B) Representative images of NIH3T3 cells treated with EGF (100 ng/mL). Scale bar, 20 µm. (C) Mean ± SD clearance of cytosolic ZtSH2 after treatment with EGF (100 ng/mL). n = 3 independent experiments.
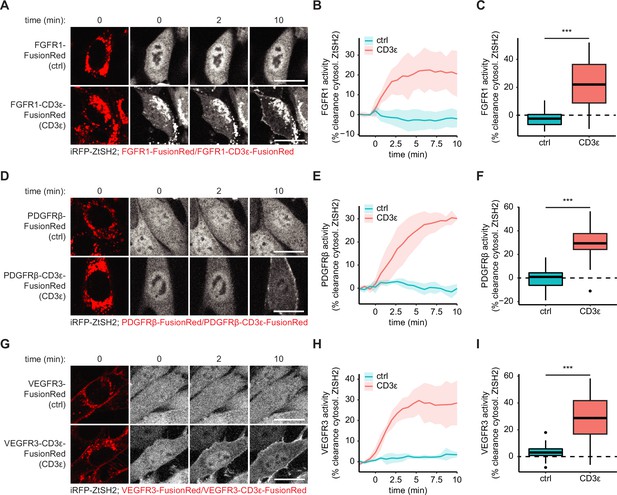
pYtag biosensors of additional receptor tyrosine kinases (RTKs).
(A) Images of NIH3T3 cells co-expressing iRFP-ZtSH2 with either FGFR1-FusionRed or FGFR1-CD3ε-FusionRed, treated with FGF4 (100 ng/mL). Scale bars, 20 µm. (B) Mean ± SD clearance of cytosolic ZtSH2 in cells expressing FGFR1-FusionRed (ctrl) or FGFR1-CD3ε-FusionRed (CD3ε) from (A). n = 3 independent experiments. (C) Clearance of ZtSH2 from the cytosol 10 min after treatment with FGF4 in (B). (D) Images of NIH3T3 cells co-expressing iRFP-ZtSH2 with either PDGFRβ-FusionRed or PDGFRβ-CD3ε-FusionRed, treated with PDGF-BB (100 ng/mL). Scale bars, 20 µm. (E) Mean ± SD clearance of cytosolic ZtSH2 in cells expressing PDGFRβ-FusionRed (ctrl) or PDGFRβ-CD3ε-FusionRed (CD3ε) from (D). n = 3 independent experiments. (F) Clearance of ZtSH2 from the cytosol 10 min after treatment with PDGF-BB in (E). (G) Images of NIH3T3 cells co-expressing iRFP-ZtSH2 with either VEGFR3-FusionRed or VEGFR3-CD3ε-FusionRed, treated with VEGF-C (500 ng/mL). Scale bars, 20 µm. (H) Mean ± SD clearance of cytosolic ZtSH2 in cells expressing VEGFR3-FusionRed (ctrl) or VEGFR3-CD3ε-FusionRed (CD3ε) from (G). n = 3 independent experiments. (I) Clearance of ZtSH2 from the cytosol 10 min after treatment with VEGF-C in (H). Lines denote mean values, boxes denote 25–75th percentiles, and whiskers denote minima and maxima. For each condition, n > 38 cells from three independent experiments. ***p<0.001 by Kolmogorov–Smirnov test.
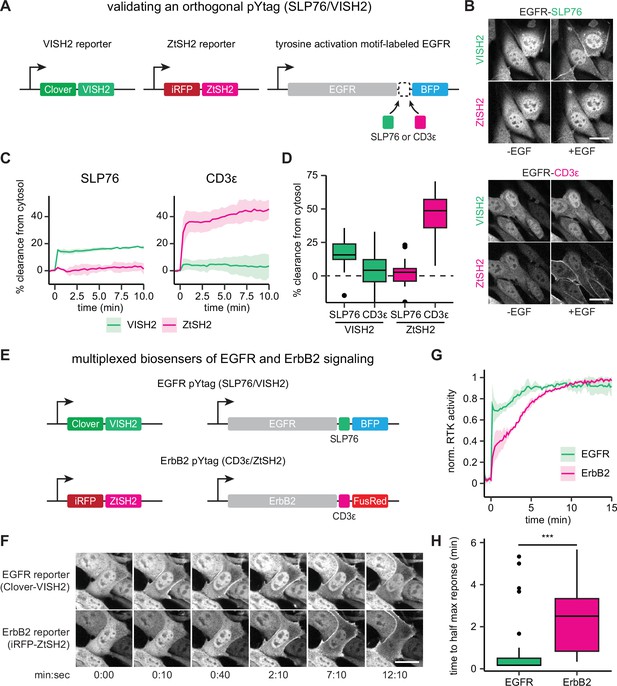
Orthogonal pYtags enable multiplexed receptor tyrosine kinase (RTK) biosensing.
(A) To assess the performance of the VISH2/SLP76 system as a pYtag-based biosensor, VISH2 and ZtSH2 reporters were co-expressed in NIH3T3 cells along with either SLP76- or CD3ε-labeled epidermal growth factor receptor (EGFR). (B) NIH3T3 cells co-expressing VISH2 and ZtSH2 reporters before and 3 min after treatment with EGF (100 ng/mL). Scale bars, 20 µm. (C) Mean ± SD clearances of VISH2 and ZtSH2 from the cytosol, expressed with either SLP76- or CD3ε-labeled EGFR and stimulated with EGF (100 ng/mL). n = 3 independent experiments. (D) Response of VISH2 and ZtSH2 reporters 10 min after EGF treatment in (C). n > 30 cells from three independent experiments. (E) Orthogonal pYtags can be multiplexed to monitor the activity of multiple RTKs in the same cell. (F) Images of cells expressing orthogonal reporters for EGFR and ErbB2, treated with EGF (100 ng/mL). Scale bar, 20 µm. (G) Mean ± SD trajectories for EGFR and ErbB2 activity using multiplexed pYtags. For each reporter, the mean response was normalized to its minimum and maximum measured values. n = 3 independent experiments. (H) Time to half maximal response for individual cells from (G). Lines denote mean values, boxes denote 25–75th percentiles, and whiskers denote minima and maxima. n > 30 cells from three independent experiments. ***p<0.001 by Kolmogorov–Smirnov test. See also Figure 5—video 1 and Figure 5—video 2.
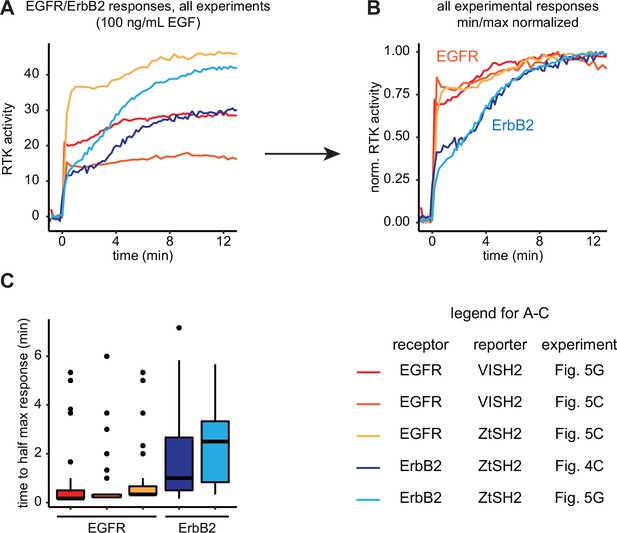
Comparison of epidermal growth factor receptor (EGFR) and ErbB2 responses across experiments.
(A) Mean responses of EGFR and ErbB2 to EGF (100 ng/mL) were collected from experiments with or without multiplexed biosensors (see legend at bottom right of figure) and (B) normalized to their minimum and maximum responses for comparison. (C) Time to half maximal response from (B). Lines denote mean values, boxes denote 25–75th percentiles, and whiskers denote minima and maxima. For each condition, n ≥ 30 cells from three independent experiments.
Timelapse of NIH3T3 cells co-expressing VISH2 and ZtSH2 reporters, and either SLP76- or CD3ε-labeled epidermal growth factor receptor (EGFR), treated with EGF (100 ng/mL).
Related to Figure 5B.
Timelapse of NIH3T3 cells co-expressing pYtag biosensors for epidermal growth factor receptor (EGFR) and ErbB2, treated with EGF (100 ng/mL).
Related to Figure 5F.
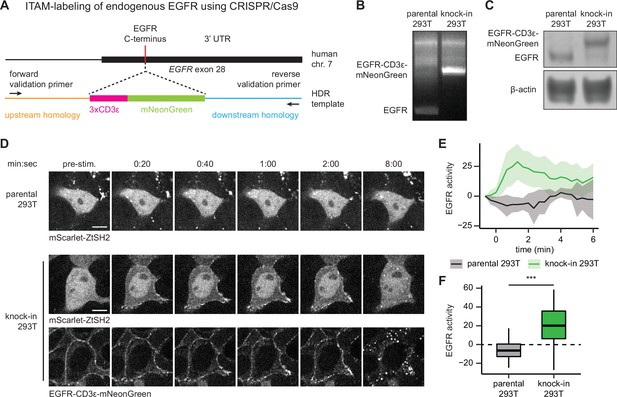
pYtags can be used to monitor the activity of endogenous receptor tyrosine kinases (RTKs).
(A) Schematic of EGFR locus containing the C-terminus of epidermal growth factor receptor (EGFR), where CRISPR/Cas9 was used to label the receptor with CD3ε-mNeonGreen via homology-directed repair. (B) PCR of genomic DNA from parental or knock-in HEK293T cells. Validation primers targeting homology regions upstream and downstream of the CD3ε-mNeonGreen insert are labeled by black arrows in (A). (C) Immunoblots of EGFR in parental or knock-in HEK293T cells. (D) Images of parental or knock-in HEK293T cells treated with EGF (100 ng/mL). mScarlet-ZtSH2 images show averages of two successive frames to decrease background noise; full raw movie is included as Figure 6—video 1. Scale bar, 10 µm. (E) Mean ± SD clearance of ZtSH2 from the cytosol following treatment with EGF (100 ng/mL). parental HEK293T, n = 3 independent experiments; knock-in HEK293T, n = 4 independent experiments. (F) Clearance of ZtSH2 from the cytosol 1 min after treatment with EGF in (E). Lines denote mean values, boxes denote 25–75th percentiles, and whiskers denote minima and maxima. parental HEK293T, n = 23 cells from three independent experiments; knock-in HEK293T, n = 46 cells from four independent experiments. ***p<0.001 by Kolmogorov–Smirnov test. See also Figure 6—video 1.
-
Figure 6—source data 1
Uncropped gel for Figure 6B.
- https://cdn.elifesciences.org/articles/82863/elife-82863-fig6-data1-v1.zip
-
Figure 6—source data 2
Uncropped gel for Figure 6C.
- https://cdn.elifesciences.org/articles/82863/elife-82863-fig6-data2-v1.zip
Timelapse of HEK293T cells expressing mScarlet-ZtSH2 and an endogenously labeled EGFR-CD3ε-mNeonGreen, treated with EGF (100 ng/mL).
Left panel shows mScarlet fluorescence; right panel shows mNeonGreen fluorescence at the indicate time points. Scale bar, 20 μm. Related to Figure 6D.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (human) | MCF10A-5E | Janes et al., 2010 | RRID:CVCL_0598 | |
| Cell line (human) | HEK293T LX | ClonTech Laboratories | Cat # 632180 | |
| Cell line (mouse) | NIH3T3 | ATCC | Cat # CRL-1658 | |
| Cell line (mouse) | SYF mouse embryonic fibroblasts (MEFs) | ATCC | Cat # CRL-2459 | |
| Cell line (Escherichia coli) | Stellar chemically competent cells | ClonTech Laboratories | Cat # 636763 | |
| Recombinant DNA reagent | pCMV-dR8.91 lentivirus packaging plasmid | Gift from Prof. Didier Trono, EPFL | Addgene # 12263 | |
| Recombinant DNA reagent | pMD2.G lenti helper plasmid | Gift from Prof. Didier Trono, EPFL | Addgene # 12259 | |
| Recombinant DNA reagent | pHR EGFR-FusionRed | Yang et al., 2021 | Addgene # 179263 | |
| Recombinant DNA reagent | pHR EGFR-CD3ζ1-FusionRed | This paper | N/A | EGFR-ITAM construct |
| Recombinant DNA reagent | pHR EGFR-CD3ζ2-FusionRed | This paper | N/A | EGFR-ITAM construct |
| Recombinant DNA reagent | pHR EGFR-CD3ζ3-FusionRed | This paper | N/A | EGFR-ITAM construct |
| Recombinant DNA reagent | pHR EGFR-CD3γ-FusionRed | This paper | N/A | EGFR-ITAM construct |
| Recombinant DNA reagent | pHR EGFR-CD3δ-FusionRed | This paper | N/A | EGFR-ITAM construct |
| Recombinant DNA reagent | pHR EGFR-CD3ε-FusionRed | This paper | Addgene # 188626 | EGFR-ITAM construct |
| Recombinant DNA reagent | pHR EGFR- CD3ε-mNeonGreen | This paper | N/A | EGFR-ITAM construct |
| Recombinant DNA reagent | pHR iRFP-ZtSH2 | This paper | Addgene # 188627 | ZtSH2 biosensor |
| Recombinant DNA reagent | pHR mScarlet-ZtSH2 | This paper | N/A | ZtSH2 biosensor |
| Recombinant DNA reagent | pHR EGFR(R84K)-CD3ε-FusionRed | This paper | N/A | GBM-mutant EGFR construct (Figure 3) |
| Recombinant DNA reagent | pHR EGFR(A265V)-CD3ε-FusionRed | This paper | N/A | GBM-mutant EGFR construct (Figure 3) |
| Recombinant DNA reagent | pHR ErbB2-FusionRed | This paper | N/A | ITAM-less ErbB2 construct (Figure 4) |
| Recombinant DNA reagent | pHR ErbB2- CD3ε-FusionRed | This paper | Addgene # 188628 | ITAM-tagged ErbB2 (Figure 4) |
| Recombinant DNA reagent | pHR EGFR-Citrine | This paper | N/A | Fluorescent EGFR construct (Figure 4) |
| Recombinant DNA reagent | pHR Clover-VISH2 | This paper | Addgene # 188629 | tSH2 biosensor (Figure 5) |
| Recombinant DNA reagent | pHR EGFR-CD3ε-TagBFP | This paper | N/A | ITAM-tagged EGFR (Figure 5) |
| Recombinant DNA reagent | pHR EGFR-SLP76-TagBFP | This paper | Addgene # 188630 | ITAM-tagged EGFR (Figure 5) |
| Recombinant DNA reagent | pHR Grb2-TagBFP | This paper | Addgene # 188631 | Grb2-based biosensor (Figure 1) |
| Recombinant DNA reagent | pHR FGFR1-CD3ε-FusionRed | This paper | Addgene # 188632 | ITAM-tagged FGFR1 (Figure 4) |
| Recombinant DNA reagent | pHR FGFR1-FusionRed | This paper | N/A | ITAM-less FGFR1 (Figure 4) |
| Recombinant DNA reagent | pHR PDGFRβ-CD3ε-FusionRed | This paper | N/A | ITAM-tagged PDGFR (Figure 4) |
| Recombinant DNA reagent | pHR PDGFRβ-FusionRed | This paper | N/A | ITAM-less PDGFR (Figure 4) |
| Recombinant DNA reagent | pHR VEGFR3-CD3ε-FusionRed | This paper | N/A | ITAM-tagged VEGFR (Figure 4) |
| Recombinant DNA reagent | pHR VEGFR3-FusionRed | This paper | N/A | ITAM-less VEGFR (Figure 4) |
| Recombinant DNA reagent | pHR ErkKTR-TagBFP | 9 | N/A | |
| Recombinant DNA reagent | pX330 EGFR-sgRNA | This paper, using a plasmid from Feng Zhang, MIT | Addgene # 188633 | EGFR-targeting gRNA (Figure 6) |
| Recombinant DNA reagent | pUC19 EGFRup-CD3ε-mNeonGreen-EGFRdown | This paper | Addgene # 188634 | CRISPR plasmid for EGFR modification (Figure 6) |
| Sequence-based reagent | PEF122 forward primer | This paper | 5′- TTCTTTTGCAGCAACAGCAAGAGGGCCCTCCC-3′ | Used to verify CRISPR tagging; see ‘Methods’ |
| Sequence-based reagent | PEF123 reverse primer | This paper | 5’- TCCGTTTCTTCTTTGCCCAGGAAGGGACAGAGTGGCTTATCC-3’ | Used to verify CRISPR tagging; see ‘Methods’ |
| Antibody | Anti-EGFR antibody (rabbit monoclonal) | Cell Signaling Technology | Cat # 4267 | Used at 1:1000 for western blotting |
| Antibody | Anti-pEGFR antibody (rabbit monoclonal) | Cell Signaling Technology | Cat # 3777 | Used at 1:1000 for western blotting |
| Antibody | Anti-β-actin antibody (mouse monoclonal) | Cell Signaling Technology | Cat # 3700 | Used at 1:1000 for western blotting |
| Antibody | Anti-pAkt antibody (rabbit polyclonal) | Cell Signaling Technology | Cat # 9271 | Used at 1:1000 for western blotting |
| Antibody | Anti-ppErk antibody (rabbit polyclonal) | Cell Signaling Technology | Cat # 9101 | Used at 1:1000 for western blotting |
| Antibody | Anti-EEA1 antibody (mouse monoclonal) | Cell Signaling Technology | Cat # 48453 | Used at 1:100 for immunostaining |
| Antibody | Anti-ZAP70 antibody (rabbit monoclonal) | Cell Signaling Technology | Cat # 3165 | Used at 1:1000 for western blotting |
| Antibody | Goat anti-Mouse IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 (goat polyclonal) | Invitrogen | Cat # A-11001 | Used at 1:500 for immunostaining |
| Antibody | Goat anti-Rabbit IgG (Heavy chain), Superclonal Recombinant Secondary Antibody, Alexa Fluor 647 (goat polyclonal) | Invitrogen | Cat # A27040 | Used at 1:500 for immunostaining |
| Antibody | Goat anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 (goat polyclonal) | Invitrogen | Cat # A-21236 | Used at 1:500 for immunostaining |
| Antibody | IRDye 680RD Goat anti-Mouse IgG antibody (goat polyclonal) | LI-COR | Cat # 926-68070 | Used at 1:10,000 for western blotting |
| Antibody | IRDye 800CW Goat anti-Rabbit IgG antibody (goat polyclonal) | LI-COR | Cat # 926-32211 | Used at 1:10,000 for western blotting |
| Peptide, recombinant protein | Bovine serum albumin | Sigma-Aldrich | Cat # 12659 | |
| Peptide, recombinant protein | Fibronectin | Corning | Cat # CB-40008A | Cell adhesion coating |
| Peptide, recombinant protein | ClonAmp HiFi PCR polymerase | ClonTech Laboratories | Cat # 639298 | Polymerase |
| Peptide, recombinant protein | Insulin | Sigma-Aldrich | Cat # I6634 | |
| Peptide, recombinant protein | Cholera toxin | Sigma-Aldrich | Cat # C8052 | |
| Peptide, recombinant protein | L-glutamine | Gibco | Cat # 25030-081 | |
| Peptide, recombinant protein | EGF | R&D Systems | Cat # 236-EG-200 | |
| Peptide, recombinant protein | Epiregulin | R&D Systems | Cat # 1195-EP-025 | |
| Peptide, recombinant protein | Epigen | R&D Systems | Cat # 6629-EP-025 | |
| Peptide, recombinant protein | FGF4 | R&D Systems | Cat # 235-F4-025 | |
| Peptide, recombinant protein | PDGF-BB | Millipore Sigma | Cat # P3201 | |
| Peptide, recombinant protein | VEGF-C | R&D Systems | Cat # 9199-VC-025 | |
| Chemical compound, drug | Gefitinib | Cell Signaling Technology | Cat # 4765 | |
| Chemical compound, drug | Hydrocortisone | Sigma-Aldrich | Cat # H0888 | |
| Chemical compound, drug | Penicillin/ streptomycin | Gibco | Cat # 15140–122 | |
| Chemical compound, drug | TrypLE Express | Gibco | Cat # 12605-028 | |
| Chemical compound, drug | FuGENE HD | Promega | Cat # E2311 | |
| Chemical compound, drug | Lipofectamine 3000 | Thermo Fisher Scientific | Cat # L3000015 | |
| Chemical compound, drug | Aminopropyl trimethoxysilane | Sigma-Aldrich | Cat # 281778 | |
| Chemical compound, drug | Glutaraldehyde | Sigma-Aldrich | Cat # 340855 | |
| Chemical compound, drug | 40% acrylamide solution | Bio-Rad | Cat # 1610140 | |
| Chemical compound, drug | 2% bis-acrylamide solution | Bio-Rad | Cat # 161-0142 | |
| Chemical compound, drug | N,N,N’,N’-Tetramethyl ethylenediamine (TEMED) | Sigma-Aldrich | Cat # T9281 | |
| Chemical compound, drug | Ammonium persulfate (APS) | Sigma-Aldrich | Cat # A3678 | |
| Commercial assay or kit | inFusion HD cloning kit | ClonTech Laboratories | Cat # 638911 | Cloning kit |
| Other | DMEM/F12 | Gibco | Cat # 11320033 | Culture media |
| Other | Horse serum | Gibco | Cat # 16050122 | Serum for culture media |
| Other | DMEM | Gibco | Cat # 11995-065 | Culture media |
| Other | Fetal bovine serum | R&D Systems | Cat # S11150 | Serum for culture media |
| Software, algorithm | FIJI | Schindelin et al., 2012 | http://fiji.sc; RRID:SCR_00228 | |
| Software, algorithm | Python code for computational model; analysis code for raw data | This paper | https://github.com/toettchlab/Farahani2022/ (copy archived at toettchlab, 2023) | |
| Software, algorithm | R Studio 1.1.456 | RStudio | rstudio.com; RRID:SCR_000432 |
Equations used in the mathematical model.
L-EGFR: ligand-bound EGFR; EGFR:EGFR: EGFR in dimeric form; EGFR: EGFR bound to ZtSH2.
| Species | Notation | Equation |
|---|---|---|
| Soluble ligand (L) | N1 | (A1) |
| Unbound ZtSH2 (*) | N2 | (A2) |
| EGFR | N3 | (A3) |
| L-EGFR | N4 | (A4) |
| EGFR:EGFR | N5 | (A5) |
| L-EGFR:EGFR | N6 | (A6) |
| L-EGFR:L-EGFR | N7 | (A7) |
| EGFR*:EGFR | N8 | (A8) |
| L-EGFR*:EGFR | N9 | (A9) |
| L-EGFR:EGFR* | N10 | (A10) |
| L-EGFR*:L-EGFR | N11 | (A11) |
| EGFR*:EGFR* | N12 | (A12) |
| L-EGFR*:EGFR* | N13 | (A13) |
| L-EGFR*:L-EGFR* | N14 | (A14) |
| EGFR* | N15 | (A15) |
| L-EGFR* | N16 | (A16) |
Parameters used in the mathematical model.
| Parameter | Notation | Value | Units | Notes |
|---|---|---|---|---|
| Receptor–ligand binding | k1 | 0.03 | nM–1 s–1 | Schoeberl et al., 2002 |
| Ligand dissociating from L-EGFR (β = 1 corresponds to EGF) | k2 | β*6.6e-3 | s–1 | Macdonald and Pike, 2008 |
| Ligand dissociating from L-EGFR:EGFR (β = 1 corresponds to EGF) | k3 | β*5.7e-3 | s–1 | Macdonald and Pike, 2008 |
| Ligand dissociating from L-EGFR:L-EGFR | k4 | 0.087 | s–1 | Macdonald and Pike, 2008 |
| Receptor dimerization and activation | k5 | 1e-5 | nM–1 s–1 | Estimated |
| EGFR:EGFR dissociation | k6 | 5e-3; variable values in Figure 3—figure supplement 2. | s–1 | Estimated |
| L-EGFR:EGFR dissociation (γ = 1 corresponds to EGF) | k7 | γ*1e-4 | s–1 | Estimated |
| ZtSH2 binding to receptor | k8 | 5 | nM–1 s–1 | Kd from Ottinger et al., 1998; kinetics set to be ~10 s based on our experimental measurements of ZtSH2 translocation |
| ZtSH2 dissociating from receptor | k9 | 16.67 | s–1 | Kd from Ottinger et al., 1998; kinetics set to be ~10 s based on our experimental measurements of ZtSH2 translocation |
| Scaling parameter for ligand–receptor binding | β | 1 for EGF; 50 for low-affinity ligands; variable values in Figure 3C | Unitless | Freed et al., 2017 |
| Scaling parameter for dimerization of ligand-bound receptors | γ | 1 for EGF; 100 for low-affinity ligands; variable values in Figure 3C | Unitless | Freed et al., 2017; Hu et al., 2022 |





