Science Forum: A low-cost, open-source evolutionary bioreactor and its educational use
Figures
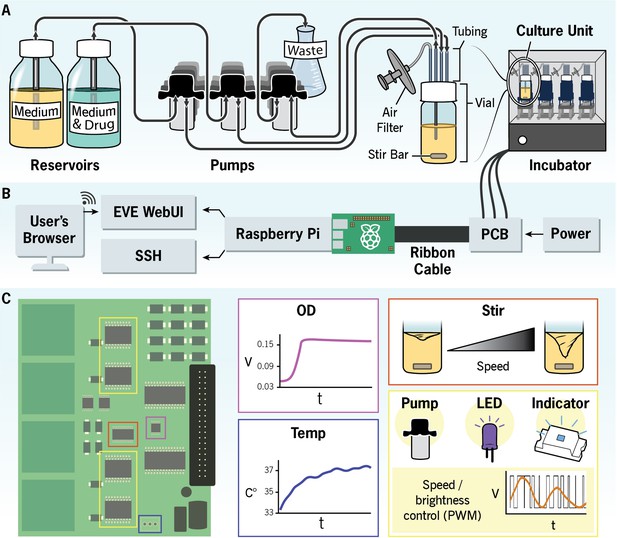
The EVolutionary bioreactor (EVE).
Schematic illustration of the EVE hardware and software architecture. (A) Reservoirs containing permissive (i.e., drug-free) and selective growth media are each connected to a culture unit (CU) through silicone tubing. Peristaltic pumps control the rate at which the media are added to the evolving bacterial population, and an additional pump removes waste. Multiple CUs can fit into an incubator, allowing users to simultaneously evolve several independent replicate populations. (B) A Raspberry Pi interfaces with a custom printed circuit board (PCB) to monitor culture growth and coordinate the addition and removal of growth media and waste. The user controls the EVE hardware with a web application. (C) The PCB chips measure voltage and temperature. These voltage measurements are proportional to optical density and thus are a way to estimate population size. The chips also control the fluidic pumps, diodes, onboard indicators, and stirring speed. The PCB diagram illustrates where these chips are located on the board.
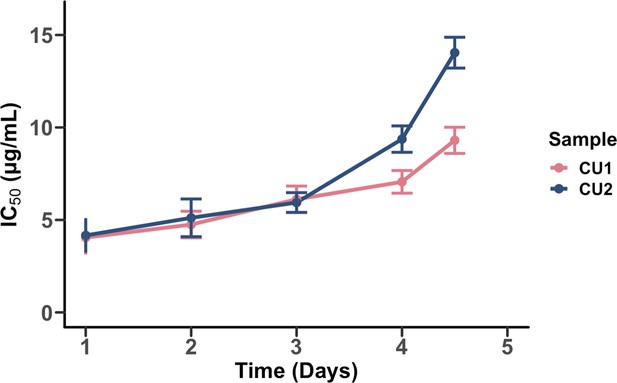
The evolution of chloramphenicol resistance over time.
Half-maximal inhibitory concentration (IC50) values for two biologically independent replicates (CU1 and CU2) are plotted against time. Points represent the estimated IC50 determined by fitting a Hill function to OD600 measurements across this dilution series after 18 hours of population growth. Error bars represent the variance in IC50. The data used to generate this figure are available in our Github repository: https://github.com/vishhvaan/eve-pi (Gopalakrishnan, 2022).
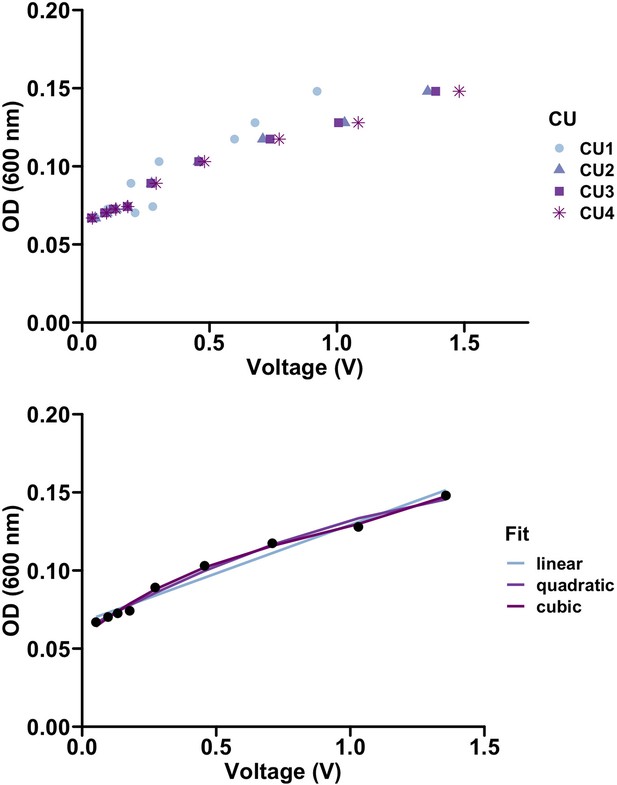
Calibration of culture optical density with reported voltage.
(Top) Culture units behave similarly when measuring voltage. (Bottom) Various regression methods were used to quantify the relationship between the optical density (OD) and voltage. R2 values of 0.97, 0.99, and 0.99 were calculated for the linear, quadratic, and cubic fits respectively. We used a linear model to translate voltage to optical density in our experiments.
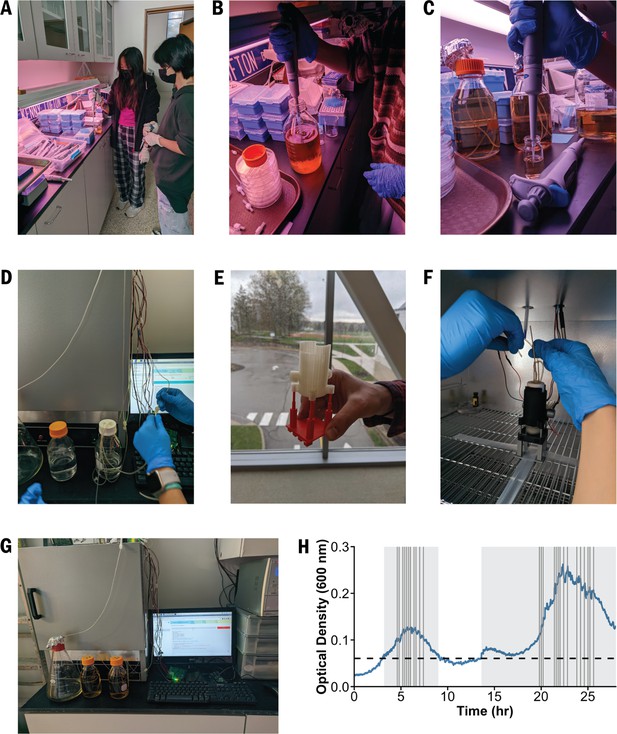
High school students at Hawken School built the EVE and performed an evolution experiment.
(A) Prior to experimentation, the students practiced pipetting with media. (B) They prepared the selective growth medium and (C) inoculated E. coli into a culture unit containing the permissive medium. (D) The system was sterilized with bleach and ethanol solutions. (E) Before the experiment, the students printed a culture unit stand in the maker-space at Hawken School. (F) The media and waste reservoirs were attached to the culture unit by silicon tubing, and then (G) the students began the experiment. (H) Bacterial growth and inhibition in the EVE over 28 hours. The grey boxes and vertical lines indicate when the EVE control algorithm added the permissive and selective media into the culture units, respectively.
Additional files
-
Supplementary file 1
Curriculum unit plan.
- https://cdn.elifesciences.org/articles/83067/elife-83067-supp1-v2.pdf
-
MDAR checklist
- https://cdn.elifesciences.org/articles/83067/elife-83067-mdarchecklist1-v2.pdf





